Abstract
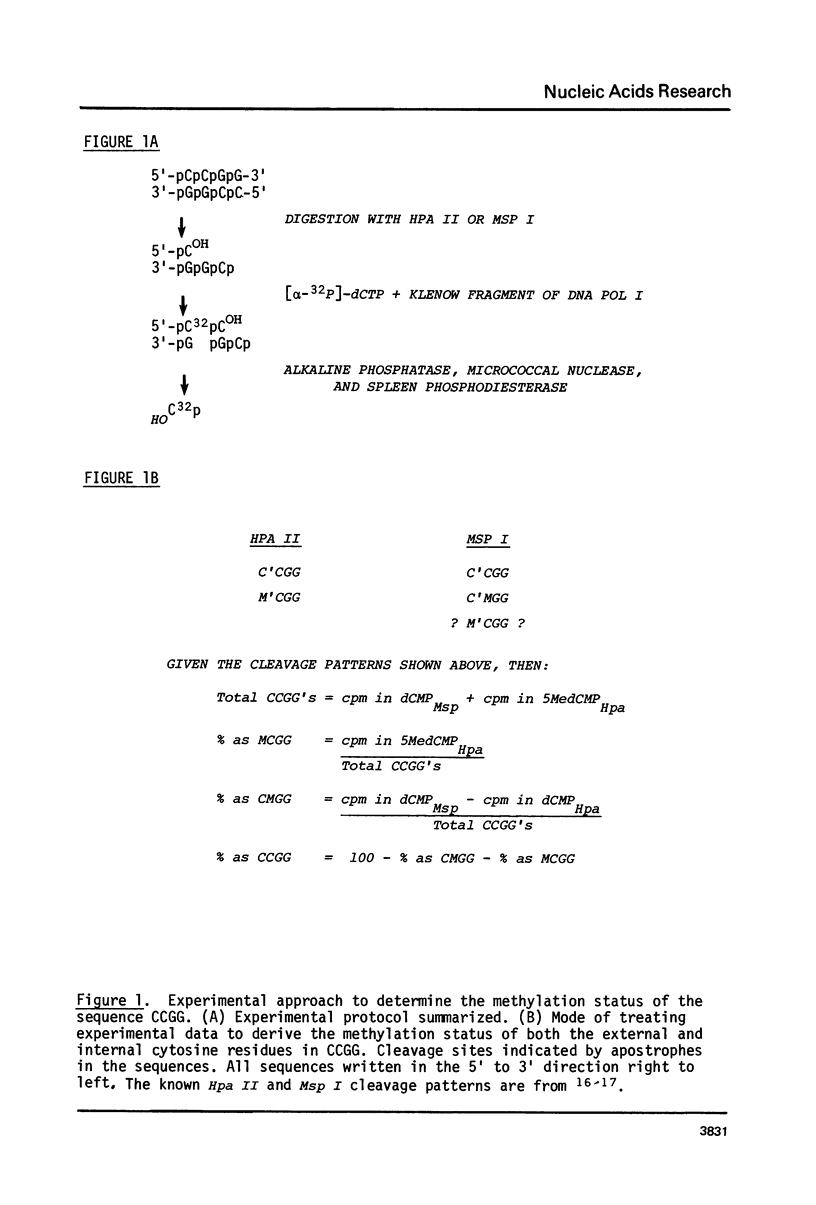
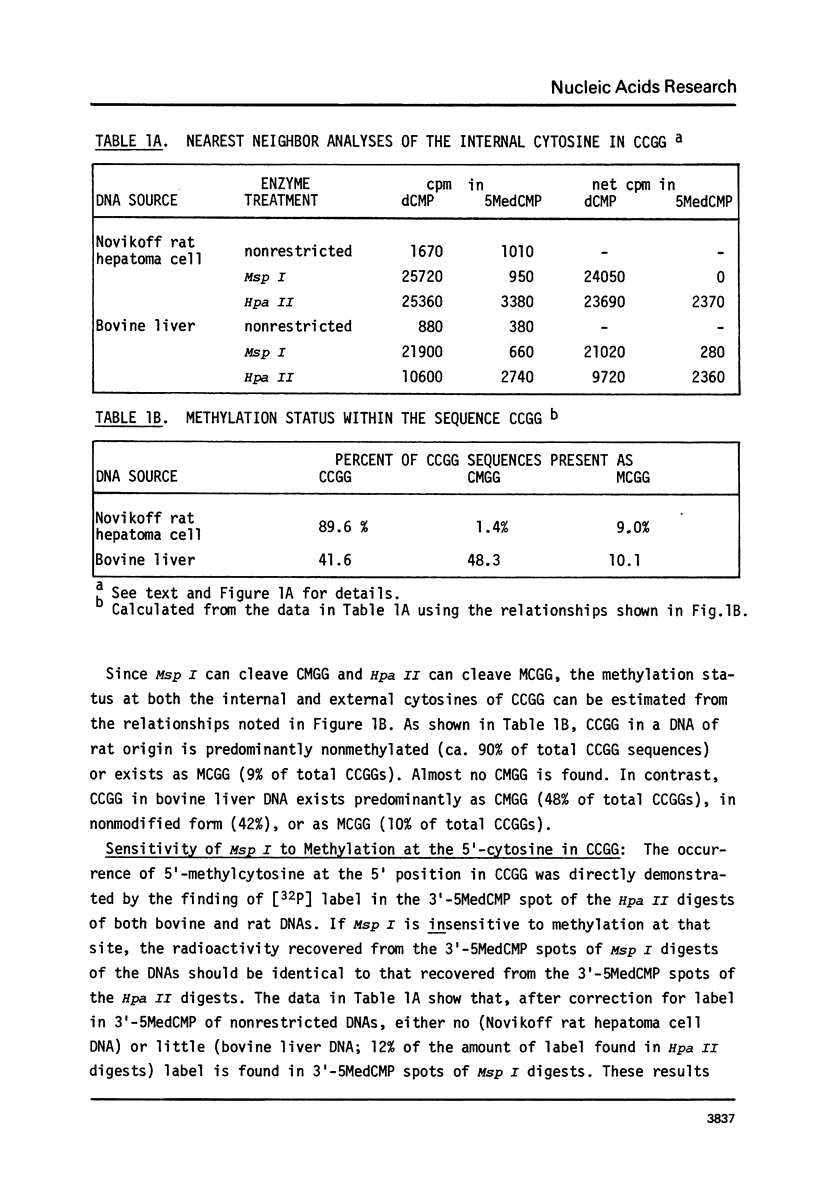
Novikoff rat hepatoma and bovine liver DNAs were digested with Msp I or Hpa II. Restriction fragments were end-labeled using [alpha-32P]-dCTP and the Klenow fragment of E. coli DNA polymerase I and then digested to 2'-deoxyribonucleoside-3'-monophosphates using micrococcal nuclease and spleen phosphodiesterase. Mononucleotides were separated by two-dimensional thin layer chromatography, localized by radioautography, and the [32P]-label quantitated by scintillation spectrometry. This method, based on known specificities of Msp I and Hpa II, shows that CCGG, CMGG, and MCGG (M refers to 5-methylcytosine) occur at frequencies of 89.6%, 1.4%, and 9.0%, respectively, in the rat DNA and at 41.6%, 48.3%, and 10.0%, respectively, in the bovine DNA. [32P] recovery in 3'-5-MedCMP from end-labeled Msp I digests was negligible compared to recovery from Hpa II digests. Hence, Msp I is sensitive to methylation at the 5' cytosine in the sequence CCGG.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird A. P., Southern E. M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978 Jan 5;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Bird A. P., Taggart M. H., Smith B. A. Methylated and unmethylated DNA compartments in the sea urchin genome. Cell. 1979 Aug;17(4):889–901. doi: 10.1016/0092-8674(79)90329-5. [DOI] [PubMed] [Google Scholar]
- Bird A. P. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J Mol Biol. 1978 Jan 5;118(1):49–60. doi: 10.1016/0022-2836(78)90243-7. [DOI] [PubMed] [Google Scholar]
- Browne M. J., Burdon R. H. The sequence specificity of vertebrate DNA methylation. Nucleic Acids Res. 1977 Apr;4(4):1025–1037. doi: 10.1093/nar/4.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne M. J., Cato A. C., Burdon R. H. The distribution of modified and non-modified C-G doublets in BHK-21 cell DNA. FEBS Lett. 1978 Jul 1;91(1):69–73. doi: 10.1016/0014-5793(78)80019-2. [DOI] [PubMed] [Google Scholar]
- Cedar H., Solage A., Glaser G., Razin A. Direct detection of methylated cytosine in DNA by use of the restriction enzyme MspI. Nucleic Acids Res. 1979;6(6):2125–2132. doi: 10.1093/nar/6.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOSKOCIL J., SORM F. Distribution of 5-methylcytosine in pyrimidine sequences of deoxyribonucleic acids. Biochim Biophys Acta. 1962 Jun 11;55:953–959. doi: 10.1016/0006-3002(62)90909-5. [DOI] [PubMed] [Google Scholar]
- Gautier F., Bünemann H., Grotjahn L. Analysis of calf-thymus satellite DNA: evidence for specific methylation of cytosine in C-G sequences. Eur J Biochem. 1977 Oct 17;80(1):175–183. doi: 10.1111/j.1432-1033.1977.tb11869.x. [DOI] [PubMed] [Google Scholar]
- JOSSE J., KAISER A. D., KORNBERG A. Enzymatic synthesis of deoxyribonucleic acid. VIII. Frequencies of nearest neighbor base sequences in deoxyribonucleic acid. J Biol Chem. 1961 Mar;236:864–875. [PubMed] [Google Scholar]
- Kaput J., Sneider T. W. Methylation of somatic vs germ cell DNAs analyzed by restriction endonuclease digestions. Nucleic Acids Res. 1979 Dec 20;7(8):2303–2322. doi: 10.1093/nar/7.8.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel J. L., Chambon P. DNA methylation: organ specific variations in the methylation pattern within and around ovalbumin and other chicken genes. Nucleic Acids Res. 1979 Dec 20;7(8):2081–2103. doi: 10.1093/nar/7.8.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M. B., Smith H. O. Specificity of Hpa II and Hae III DNA methylases. Nucleic Acids Res. 1977 Dec;4(12):4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Ginder G. D. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature. 1979 Aug 2;280(5721):419–420. doi: 10.1038/280419a0. [DOI] [PubMed] [Google Scholar]
- Rae P. M., Steele R. E. Absence of cytosine methylation at C-C-G-G and G-C-G-C sites in the rDNA coding regions and intervening sequences of Drosophila and the rDNA of other insects. Nucleic Acids Res. 1979 Jul 11;6(9):2987–2995. doi: 10.1093/nar/6.9.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SINSHEIMER R. L. The action of pancreatic desoxyribonuclease. I. Isolation of mono- and dinucleotides. J Biol Chem. 1954 May;208(1):445–459. [PubMed] [Google Scholar]
- Salomon R., Kaye A. M. Methylation of mouse DNA in vivo: di- and tripyrimidine sequences containing 5-methylcytosine. Biochim Biophys Acta. 1970 Apr 15;204(2):340–351. [PubMed] [Google Scholar]
- Singer J., Roberts-Ems J., Riggs A. D. Methylation of mouse liver DNA studied by means of the restriction enzymes msp I and hpa II. Science. 1979 Mar 9;203(4384):1019–1021. doi: 10.1126/science.424726. [DOI] [PubMed] [Google Scholar]
- Sneider T. W. Methylation of mammalian deoxyribonucleic acid. II. The distribution of 5-methylcytosine in pyrimidine deoxyribonucleotide clusters in Novikoff hepatoma cell deoxyribonucleic acid. J Biol Chem. 1971 Aug 10;246(15):4774–4783. [PubMed] [Google Scholar]
- Sneider T. Methylation of mammalian deoxyribonucleic acid. 3. Terminal versus internal location of 5-methylcytosine in oligodeoxyribonucleotides from Novikoff hepatoma cell deoxyribonucleic acid. J Biol Chem. 1972 May 10;247(9):2872–2875. [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Res. 1978 Sep;5(9):3231–3236. doi: 10.1093/nar/5.9.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]



