Abstract
Both IgCAMs and the actin cytoskeleton play critical roles in neuronal growth cone motility and guidance. However, it is unclear how IgCAM receptors transduce signals from the plasma membrane to induce actin remodeling. Previous studies have shown that local clustering and immobilization of apCAM, the Aplysia homolog of NCAM, induces Src kinase activity and F-actin polymerization in the peripheral domain of cultured Aplysia bag cell growth cones. Therefore, we wanted to test whether the Src kinase substrate and actin regulator cortactin could be a molecular link between Src activity and actin assembly during apCAM-mediated growth cone guidance. Here, we cloned Aplysia cortactin and showed that it is abundant in the nervous system. Immunostaining of growth cones revealed a strong colocalization of cortactin with F-actin in filopodial bundles and at the leading edge of lamellipodia. Perturbation of the cytoskeleton indicated that cortactin distribution largely depends on actin filaments. Furthermore, active Src colocalized with cortactin in regions of actin assembly, including leading edge and filopodia tips. Finally, we observed that cortactin, like F-actin, localizes to apCAM adhesion sites mediating growth cone guidance. Altogether, these data suggest that cortactin is a mediator of IgCAM-triggered actin assembly in the growth cone and involved in motility and guidance.
Keywords: cortactin, F-actin, filopodia, growth cone, IgCAM
Introduction
Neurite outgrowth and guidance are driven by the specialized structure at the tip of neurites, the growth cone. Growth cones reach their target by navigating through complex environments composed of diffusible factors, extracellular matrix components, and cell adhesion molecules (Huber et al., 2003). Cell adhesion molecules (CAMs) in the nervous system mainly comprise integrin, cadherin and immunoglobulin superfamily (IgCAM) receptors, which mediate interactions between adhesive substrates and the underlying dynamic F-actin cytoskeleton, thereby leading to growth cone outgrowth, turning or retraction. While substantial data have been accumulated for the functions of the integrin and cadherin families, relatively little is known about how IgCAMs, and in particular the neural cell adhesion molecule (NCAM), connect to the actin cytoskeleton and regulate growth cone behavior.
The study of NCAM signaling pathways has been hampered by the lack of simple in vitro assays which can accurately recapitulate NCAM-specific neurite outgrowth and signaling cascades of neurons in vivo (Ditlevsen et al., 2008). To circumvent this problem, we have developed a novel in vitro growth cone guidance assay that mimics cell adhesion sites presented to large Aplysia growth cones. In this so-called restrained bead interaction (RBI) assay, microbeads coated with the Aplysia cell adhesion molecule (apCAM), the Aplysia homolog of NCAM (Mayford et al., 1992), are placed onto the periphery of bag cell growth cones and physically restrained from moving with retrograde actin flow (Suter et al., 1998). Such manipulation results in morphological and cytoskeletal rearrangements typical of growth cone guidance responses to cellular adhesion substrates, including enrichment of F-actin around the beads, attenuation of actin flow, generation of traction force, and microtubule extension to the adhesion site (Suter et al., 1998). Src tyrosine kinase activity is required for strong apCAM-actin linkages (Suter and Forscher, 2001); however, the molecular components linking apCAM-activated Src to remodeling of the actin cytoskeleton are currently unknown. A potential effector in this pathway might be the Src substrate and actin-binding protein, cortactin.
Cortactin was initially identified as a Src substrate (Wu et al., 1991), and has recently gained attention because of its role in actin assembly regulation (Uruno et al., 2001; Weaver et al., 2001) and cancer (Buday and Downward, 2007). Cortactin contains an N-terminal acidic region (NTA) that binds and activates the Arp2/3 complex, followed by a series of tandem repeats that bind to actin filaments, a helical structure of unknown function, a proline-rich region containing Src tyrosine phosphorylation sites, and a C-terminal SH3 domain interacting with actin assembly regulators, endocytosis factors and scaffolding proteins (Cosen-Binker and Kapus, 2006; Daly, 2004; Weed and Parsons, 2001). Through its multiple interactions, cortactin functions mainly as a stimulator of actin nucleation and branching by recruiting other proteins to sites of actin polymerization. Consequently, it is involved in a plethora of cellular processes including cell motility, cell-cell adhesion, endocytosis, synaptogenesis, axon guidance, and neuronal morphogenesis (Daly, 2004), as well as in pathologies like microbial invasion (Selbach and Backert, 2005) and tumorigenesis (Buday and Downward, 2007; Weaver, 2008).
Cortactin has been localized to neuronal somata, dendrites and dendritic spines (Decourt et al., 2005; Hering and Sheng, 2003; Racz and Weinberg, 2004) as well as neuronal growth cones (Du et al., 1998; Korobova and Svitkina, 2008). In addition, cortactin co-precipitated with NCAM in neurite-like structures formed by β-cells of Rip1Tag2 transgenic mice (Cavallaro et al., 2001). To test the hypothesis that cortactin is involved in IgCAM-mediated actin remodeling, we performed a detailed immunolocalization study of cortactin in both steady state and apCAM-stimulated Aplysia growth cones. We report here that cortactin colocalizes with F-actin in both filopodial bundles and lamellipodial networks as well as at apCAM adhesion sites.
Material and Methods
Reagents and antibodies
All reagents were from Sigma-Aldrich, unless otherwise specified. PCR Primers were purchased from Integrated DNA Technology. Monoclonal anti-cortactin antibody 4F11 was from Millipore. The following Aplysia Src peptide antibodies were prepared by Pacific Immunology: rabbit anti-Src1, rabbit anti-activated Src2 (pSrc2), and goat anti-Src2 (Wu et al., 2008). Alexa 488 phalloidin, Alexa 568 goat anti-rabbit, Alexa 568 donkey anti-goat, Alexa 647 goat anti-mouse, Alexa 680 goat anti-mouse antibodies, and fixable Texas Red-labeled dextran (3 kDa) were purchased from Molecular Probes.
Cloning of Aplysia cortactin
Total RNA was extracted from the entire nervous system of three Aplysia californica (Marinus) using TRIzol reagent following manufacturer’s instructions (Invitrogen). Reverse transcription was conducted using the SMART RACE cDNA amplification kit (Clontech). We took advantage of the fact that a few 20–25 bp segments of the cortactin nucleotide sequence are very similar between many species to prepare a first set of primers that could hybridize with the Aplysia sequence. A portion between the end of the NTA domain and beginning of the actin-binding repeats was first amplified using the forward primer: 5′-gagcaggaacagagatggggttccaagac -3′ and reverse primer: 5′-cccttggaataatctttctgtgactcgtgc -3′. Next, 5′ and 3′ segments of cortactin cDNA were amplified by RACE using the universal primers provided in the SMART RACE cDNA amplification kit, and the Aplysia cortactin-specific primers 5′-cagttgcgagagcaagtgtctatgg -3′ (reverse primer for the 5′ RACE) and 5′-gtttggttaggtcctccttgtgctc -3′ (forward primer for the 3′ RACE). Finally, the full length sequence was obtained using the forward primer 5′-gctggtgaccttctggtaagtgc -3′ and reverse primer 5′-gcttgtttagctatccgactttccag -3′ and inserted into pGEM-T vector (Promega). Several clones were sequenced to confirm the nucleotide sequence. To confirm the number of unique sequence features of Aplysia cortactin compared to other species (see Fig. 1), we have carried out additional RT-PCR experiments with different batches of Aplysia nervous system RNA and reagents, using the iScript Select cDNA synthesis kit (Bio-Rad). Identical Aplysia cortactin sequences were obtained with the two different RT-PCR approaches. Sequence analysis was performed with Vector NTI software (Invitrogen).
Figure 1. Cortactin is expressed in Aplysia nervous system.
A: Deduced amino acid sequence of Aplysia cortactin. It contains the Arp2/3 binding motif DEW (box), seven and a half tandem repeats (delimited by brackets) followed by a helix, a proline-rich region (underlined), and an SH3 domain. B: Domain-based and full-length cortactin amino-acid sequence comparison between Aplysia and other species (Nematostella: GI # 156408902; Sea Urchin: 47551203; Sponge: 37951188; Drosophila: 24648611; Zebrafish: 51870938; Xenopus laevis: 148222482; Chicken: 45382633; Mouse: 75677414; Human: 20357552). The percentages of sequence homology are indicated. C: Phylogenetic tree built from full length cortactin sequence from several species (same sequences as in B plus Xenopus tropicalis: 58332478; Fever mosquito: 157131030; Malaria mosquito: 158293853) using Vector NTI software. D: Cortactin Western blotting of mouse brain (Mo), Aplysia nervous system (Ap) and Drosophila S2 cell line (S2) extracts. Twenty micrograms of total protein was loaded for each tissue. The doublet in the Aplysia tissue could be clearly seen following shorter exposure. Molecular weights of standards (in kilo daltons) are indicated on the left.
Western Blotting
Aplysia were sacrificed by injection of 0.5 M MgCl2, and the entire nervous system was isolated. Two mice were anaesthetized with CO2 according to Purdue University animal care protocols, and their brains were removed. In both cases, tissues were homogenized in a lysis solution [50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 2 mM EDTA; 2 mM EGTA; 1% Triton X-100; 0.5 mM Pefabloc SC Plus (Roche); 1% protease inhibitor cocktail; 10mM NaF;1 mM Na3VO4; and 20 mM beta glycero-phosphate], and cleared at 10,000xg for 30 min at 4°C. A lysate made from Drosophila S2 cells was prepared using the same procedure and kindly provided by Dr. H. Chang (Purdue University). Lysates were mixed with Laemmli buffer, heated at 70°C for 10 min, separated by SDS-PAGE using 10% gels, and blotted onto polyvinylidene fluoride membranes. Membranes were blocked with PBS containing 5% low fat dry milk for 1 hour at RT, washed three times with PBS and three times with PBST (PBS plus 1% Tween 20), and incubated with the anti-cortactin antibody 4F11 diluted 1 μg/mL in antibody buffer (PBS plus 0.5% Tween 20 and 0.1% milk) for over night at 4°C. After washing, Alexa 680-labeled goat anti-mouse secondary antibody diluted 0.2 μg/mL in antibody buffer was applied for 1h at RT, and washed extensively. Signal detection was performed with Odyssey imaging system (LI-COR).
Aplysia bag cell cultures
Aplysia bag cell neurons were cultured in L15 medium (Invitrogen) supplemented with artificial sea water (400 mM NaCl; 9 mM CaCl2; 27 mM MgSO4; 28 mM MgCl2; 4 mM L-glutamine; 50 μg/mL gentamicin; 5 mM HEPES; pH 7.9) on glass coverslips coated with 20 μg/mL poly-L-lysine (70–150 kD), as previously described (Lee et al., 2008; Suter et al., 1998). For cytoskeletal perturbations, cells were treated with 1 μM cytochalasin B, 5 μM nocodazole, or 0.005% DMSO for 1h, respectively, and processed for immunocytochemistry.
Immunocytochemistry
Cultured cells were fixed with 3.7% formaldehyde in artificial sea water plus 400 mM sucrose for 30 min at RT, followed by permeabilization with 0.05% saponin or 1% Triton X-100 (in Fig. 4D) in fixative for 10 min and rinsing with a wash buffer (PBS plus 0.005% saponin or 0.1% Triton X-100, respectively). Alexa 488 phalloidin at 2 units/ml in wash buffer was applied for 30 min. Blocking was carried out with 5% BSA (cortactin staining) or 10% horse serum (cortactin/Src double staining) in wash buffer for 30 min. Primary antibodies diluted in blocking solution were incubated for 1 hour at RT (4F11: 2 μg/ml; Src antibodies: 2.5 μg/ml). Corresponding Alexa 568 (for Src) and 647 (for cortactin) conjugated secondary antibodies were sequentially incubated at 1 μg/ml in wash buffer for 30 min. After several washes, cells were observed by fluorescent microscopy in an anti-fading medium (20 mM n-propyl-gallate in PBS / 80% glycerol, pH 8.5). Control experiments were conducted by omitting primary antibodies and resulted in very weak background staining (data not shown). For some cortactin/Src colocalization experiments, a live cell extraction protocol was followed. Briefly, the culture medium was replaced by a Ca2+-free low ionic strength artificial sea water (100mM NaCl; 10 mM KCl; 5 mM MgCl2; 15 mM HEPES; 60 g/L glycine; 5 mM EGTA; pH 7.9) for 2 min. Cells were then permeabilized for 1 min with a cytoskeleton stabilization buffer (80 mM PIPES; 1 mM MgCl2; 5 mM EGTA; 4% polyethylene glycol, MW 35,000Da; 10 μM taxol; 1 μM phalloidin; pH 7.9) including 1% Triton X-100 and fixed with 3.7% formaldehyde in stabilization buffer for 30 min before labeling with Alexa 488 phalloidin and immunostaining. All images were acquired with a 60x Plan Apo 1.4 NA oil immersion objective on a Nikon TE2000 Eclipse microscope using a Cascade II CCD camera (Photometrics) controlled by MetaMorph software (Molecular Devices).
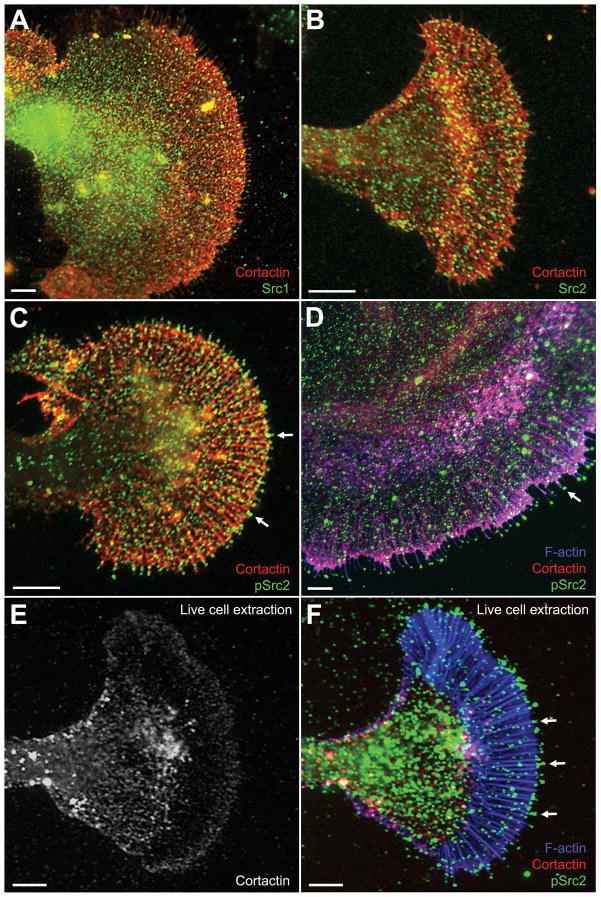
Figure 4. Cortactin and Src colocalization in growth cones.
Cultured Aplysia bag cells were fixed, extracted, and double labeled for cortactin together with Src1 (A), Src2 (B), activated Src2 (pSrc2; C), or triple labeled for cortactin together with F-actin and activated Src2 (D). Regions in magenta denote the colocalization of F-actin and cortactin. Note the enriched pSrc2 staining at the tip of filopodia when compared to total Src2 (arrows in C and D). E–F: Triple staining for cortactin, F-actin, and pSrc2 after live cell extraction. Cortactin was strongly removed from actin bundles in the P domain, but remained in the ruffling zone (E). Activated Src2, on the other hand, remained at the tip of filopodia (arrows in F), which were identified by F-actin staining. The pSrc2 staining outside the growth cone (mostly at the top of image F) is due to cell debris accumulated during the plasma membrane removal. Bars: 10 μm.
RBI assay
For RBI experiments, cells were first injected with 2 mg/mL lysine-fixable Texas Red dextran (3 kDa) using a Femto-Jet microinjection device and an NP-2 micromanipulator (both from Eppendorf). Two to five hours later, silica beads (5 μm diameter) coated with recombinant apCAM were deposited on the growth cone periphery and restrained using glass pipettes controlled by a hydraulic micromanipulator (Narishige) (Suter et al., 1998; Suter et al., 2004). Growth cone guidance responses were monitored with differential interference contrast (DIC) time lapse microscopy. Upon completion of central domain extension towards apCAM beads, growth cones were fixed and stained for both F-actin and cortactin, as indicated above.
Staining quantification, statistical analysis, and image processing
Fluorescence intensity quantification of at least three independent experiments was performed using MetaMorph. Background-subtracted average intensity values were determined for the different growth cone regions. For RBI experiments, background subtracted average intensity values of cortactin and F-actin, respectively, around the beads and in adjacent regions were divided by the corresponding Texas Red dextran value to correct each signal for the volume in the selected area. Then, the volume-corrected values around the beads were divided by the corrected values in adjacent regions to determine the fold change of cortactin and F-actin density around the beads compared to the peripheral domain (Fig. 5I). Statistical differences were determined using two-tailed t-tests for samples with unequal sizes and variances. Image processing including level adjustment, conversion into 8 bit files, cropping, size adjustment, conversion to 600 dpi, brightness and contrast adjustment, and application of unsharp mask filters, plus figure preparation were done in Photoshop software (Adobe).
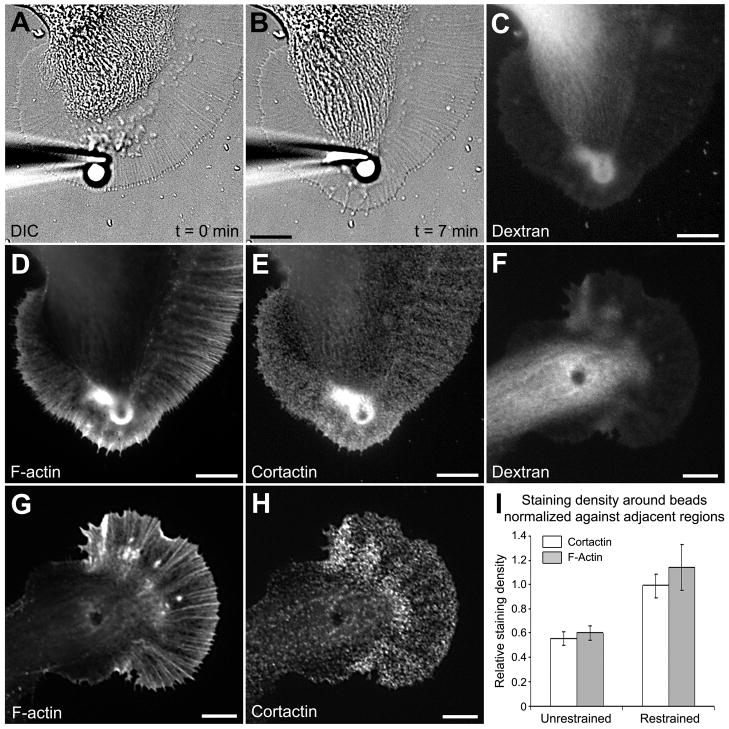
Figure 5. Cortactin accumulates in the vicinity of apCAM beads.
A: DIC image of a growth cone right after apCAM-bead placement. Growth cone guidance was induced by placing beads in the P domain and preventing them from undergoing retrograde movements by restraining them with glass pipettes. B: After a latency period of 5 min, the C domain advanced towards the beads, while leading edge protrusion occurred in front of the beads. C: A Growth cone shown in B was fixed at the end of C domain extension and Texas Red-dextran image was acquired to assess the volume distribution of subcellular regions. F-actin (D) and cortactin (E) labeling of the growth cone shown in B revealed an increase of both signals. Dextran (F), F-actin (G), and cortactin (H) images of unrestrained beads that were coupled to retrograde actin flow and generally localized in the T zone or the C domain during fixation. I: F-actin and cortactin stainings were quantified around the beads and in adjacent areas, and then normalized against the dextran signal (volume). Then, volume-corrected signals around the beads were divided by the respective volume-corrected signal in adjacent regions for both unrestrained (n = 11 growth cones) and restrained (n = 9) beads. Histograms represent means ± SEM. The value 1 on the y axis refers to the actin or cortactin staining density in the growth cone P domain in the absence of beads. Bars = 10 μm.
Results
Identification of cortactin in Aplysia
To study the role of cortactin in growth cones, we first cloned Aplysia cortactin by a combination of RT-PCR and RACE. We obtained a cDNA of 1850 bp which contained the 1734 bp long cortactin coding region plus a short portion of the 5′ and 3′ untranslated regions. The 577 amino acid-long deduced protein sequence shows an overall domain organization very similar to cortactin from other species (Fig. 1A). However, some differences were noticed. First, the N-terminal acidic (NTA) domain contained the Arp2/3 binding sequence DEW instead of the DDW found in all other cortactin sequences. Interestingly, the DEW sequence is identical to the motif present in other Arp2/3 complex regulators such as WASP or N-WASP (Weaver et al., 2003). Second, Aplysia cortactin contains seven and a half tandem repeats, which is one repeat more than mammalian cortactins. These 37 amino-acid repeats contain 16 highly conserved residues, resulting in an overall homology of 94% between the repeats. Third, the proline-rich region comprises six and a half repetitions of the motif PEPVREPS, which, once more, is unique to Aplysia as no other species arbors such a repeat in the proline-rich region. Fourth, only three tyrosine residues are found at the end of the proline-rich region (versus eight in mouse cortactin, for example), one of them (Y499) sharing an evolutionary conserved position for Src phosphorylation.
Besides the full length amino-acid sequence, we also compared individual domains between cortactin from Aplysia and other species (Fig. 1B). The NTA, tandem repeat and SH3 domains are the most conserved, the helix and proline-rich regions exhibit less homology with cortactin sequences from both invertebrates and vertebrates. Surprisingly, the SH3 domain of Aplysia cortactin has a higher identity with vertebrates (>85%) when compared with other invertebrates (<80%; Fig. 1B). Full-length cortactin sequences were used to prepare a phylogenetic tree (Fig. 1C), which revealed that Aplysia cortactin, together with Nematostella cortactin, form a distinctive invertebrate subgroup between other invertebrate and vertebrate species.
Next, we tested whether cortactin is present in the Aplysia nervous system. We performed Western blotting using the monoclonal antibody 4F11 (Wu et al., 1991), which binds to the fifth tandem repeat of mammalian cortactin (van Rossum et al., 2003), and compared Aplysia nervous system to mouse brain and Drosophila S2 cell lysates (Fig. 1D). The antibody detected a doublet in Aplysia nervous system tissue similarly to mouse brain but with much higher intensity. The apparent molecular weight (MW) of Aplysia cortactin is 95–100 kDa (theoretical MW: 65 kDa), which is higher than that of mouse cortactin (80–85 kDa), but very similar to Drosophila cortactin (100 kDa). We speculate that the difference between apparent and theoretical molecular weights is due to conformational effects and/or phosphorylation status of the helix/proline-rich region. Indeed, SDS-PAGE in the presence of 8 M urea resulted in only one band at 95 kDa (data not shown), providing evidence for conformational effects, which is in agreement with previous findings obtained on mouse (Huang et al., 1997b) and human cortactin (Campbell et al., 1999). The band around 40 kDa likely results from calpain protease-mediated degradation (Huang et al., 1997b). Expression of recombinant Aplysia cortactin in bacteria also resulted in two 4F11-positive bands at 95 and 100 kDa while no signal was obtained for bacteria transformed with an empty plasmid (data not shown), providing further evidence that the 4F11 antibody recognizes Aplysia cortactin. Strikingly, cortactin is highly enriched in Aplysia nervous system tissue when compared to mouse brain or Drosophila S2 cells.
Cortactin localization in growth cones
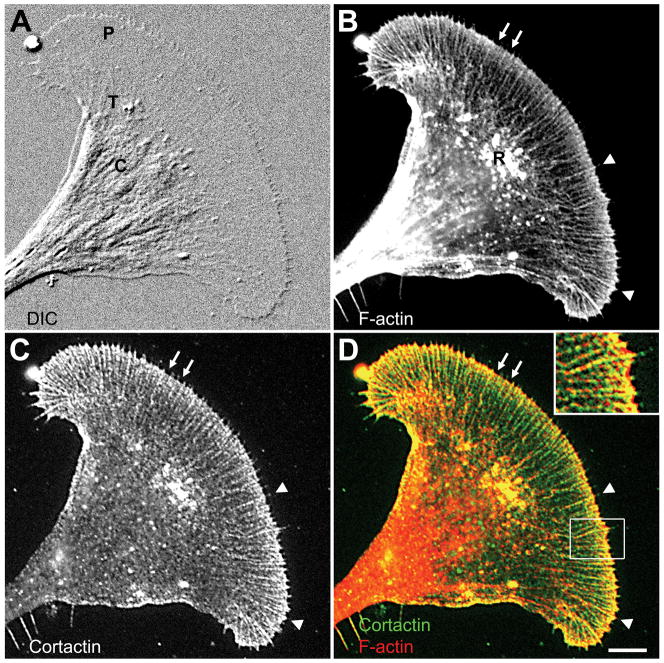
Although cortactin has been identified in growth cones (Du et al., 1998), its precise subcellular localization has not yet been investigated. Therefore, we studied its distribution in Aplysia bag cell growth cones in relationship to F-actin (Fig. 2). Consistent with previous studies (Forscher and Smith, 1988; Schaefer et al., 2002), phalloidin labeled the F-actin bundles forming filopodia in the growth cone peripheral domain (P), and produced a more diffuse staining of the lamellipodial actin at the leading edge of growth cones (Fig. 2B). In the transition (T) zone, dynamic F-actin is very prominent in the ruffling structures termed intrapodia (Rochlin et al., 1999), while the central (C) domain contains more stable actin bundles (Zhang et al., 2003). Cortactin, detected with the 4F11 antibody, presented a pattern very similar to F-actin in the growth cone P and T regions, but more punctate than the phalloidin labeling (Fig. 2C). The staining intensity in lamellipodial regions varied somewhat among growth cones, probably depending on growth cone activity (see Fig. 5E for example of dense lamellipodial staining). Overlays of both stainings revealed that cortactin is highly colocalized with F-actin in the growth cone periphery (Fig. 2D). A strong colocalization was detected in filopodial bundles, lamellipodial veil regions at the leading edge between filopodia (arrows in Fig. 2C–D) and in T zone ruffles. Cortactin was also enriched in filopodia tips (arrowheads and inset in Fig. 2C–D). The least actin colocalization was detected in the C domain where cortactin exhibited a more random distribution. In summary, our data indicate that cortactin localizes to regions of dynamic actin assembly in the growth cone, including filopodia tips, leading edge, and T zone ruffles.
Figure 2. Cortactin colocalizes with F-actin in Aplysia growth cones.
Cortactin and F-actin show a strong colocalization in the growth cone peripheral domain (P) and transition zone (T) when compared to the central domain (C). A: DIC image of a fixed growth cone. B: F-actin staining of the same growth cone using Alexa 488 phalloidin revealed filopodial bundles (arrowheads), lamellipodial veils at the leading edge (arrows) and T zone ruffles (R). C: Cortactin staining with 4F11. D: Overlay of F-actin and cortactin staining. Inset: zoom of the boxed region showing strong actin/cortactin colocalization in filopodia and the lamellipodia at the leading edge. Bar: 10 μm (5 μm for inset).
Cortactin localization after cytoskeletal perturbations
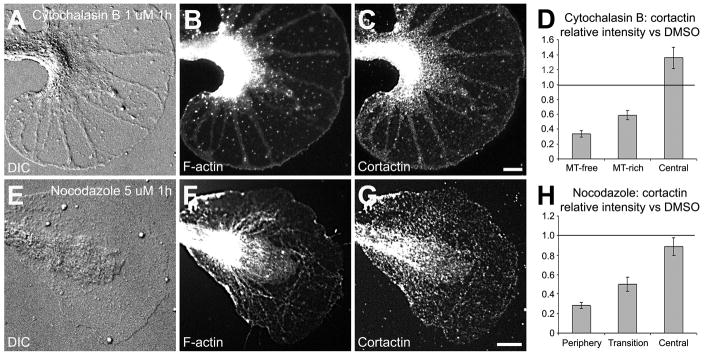
To further test whether cortactin distribution depends on F-actin, we perturbed the actin cytoskeleton by treating bag cell neurons with 1 μM cytochalasin B for 1h. This resulted in an almost complete disappearance of F-actin in the P and T regions (Fig. 3A and B), as described earlier (Forscher and Smith, 1988). The C domain was still strongly labeled with phalloidin and so was a thin margin along the leading edge (Fig. 3B). Cortactin mainly redistributed together with F-actin, although some labeling was still detected in the P domain (Fig. 3C). Cytochalasin B treatment also resulted in C domain (Fig. 3A) and microtubule extension (not shown) as previously reported (Forscher and Smith, 1988). Despite the colocalization with F-actin in unperturbed growth cones, here we detected a more intense cortactin staining in microtubule-rich than in microtubule-free regions in the P domain (Fig. 3C). To quantify the extent of cortactin redistribution, we compared cortactin signals in all growth cone regions between DMSO- and cytochalasin B-treated cells (Fig. 3D). Given the growth cone domain reorganization after cytochalasin B treatment, we decided to compare the microtubule-free and microtubule-rich regions to the peripheral and transition domains of DMSO-treated growth cones, respectively, based on the thickness and the fact that microtubules are more abundant in the T zone than in the P domain. We found that after cytochalasin B treatment cortactin intensity decreased by 65% and 40% in the microtubule-free and microtubule-rich regions, respectively (p<0.01, n = 31 growth cones), while increasing by 35% in the C domain (p<0.01). Thus, cortactin largely redistributes with F-actin upon perturbation of the actin cytoskeleton.
Figure 3. Cortactin redistributes with F-actin after cytoskeletal perturbation.
Bag cell cultures were treated with 1 μM cytochalasin B (A–D) or 5 μM nocodazole (EH) for 1h. After fixation, cells were co-stained for F-actin (B and F) and cortactin (C and G). DIC pictures (A and E) show the morphological changes occurring after cytoskeletal perturbations compared to control conditions (Fig. 2A). Cortactin staining in the different growth cone areas was quantified after cytochalasin B (n = 31 growth cones; see text for details) or nocodazole (n = 49) treatment and was normalized against cortactin staining of growth cones treated with DMSO only (D and H; n = 45). Histograms represent means ± SEM. Lines at the value 1 on the y axis indicate the level of staining in control DMSO-treated cells. MT: microtubule. Bars = 10 μm.
Given that about 60% of cortactin signal was still present in microtubule-rich regions after cytochalasin treatment, we wondered whether microtubules play a role in the distribution of cortactin in growth cones. To test this hypothesis, we treated the neurons with 5 μM of the microtubule-depolymerizing agent nocodazole. In agreement with earlier studies (Dent and Kalil, 2001; Goslin et al., 1989; Lafont et al., 1993), such a high dose of nocodazole not only disturbed microtubules (data not shown), but also F-actin organization in the P and C domain. One hour of nocodazole treatment resulted in reduced filopodia numbers and flattening of the T zone (Fig. 3E) when compared to control growth cones (Fig. 2A). These morphological changes were accompanied by a complete disorganization of F-actin networks and bundles in the P domain (Fig. 3F). Again, cortactin mostly relocated in a similar pattern to F-actin, besides a few dots in the P domain (Fig. 3G). The quantification of cortactin staining revealed a depletion of 70% and 50% in the P and T domains after nocodazole treatment, respectively (p<0.01, n=49 growth cones), while no significant change was observed in the C domain (Fig. 3H). Thus, both cytoskeleton perturbation protocols caused distinct actin reorganizations that were paralleled by cortactin redistribution, suggesting that cortactin localization in the growth cone periphery largely depends on F-actin, and not on microtubules. However, based on our current results we can not exclude a potential role for microtubules in cortactin localization in the C domain.
Cortactin colocalization with Src kinases in growth cones
In order to test whether cortactin could be a Src tyrosine kinase effector in growth cones, we investigated the potential colocalization of cortactin with Src1 and Src2, two Src tyrosine kinases recently identified in Aplysia (Wu et al., 2008). Both Src1 and Src2 exhibit a punctate staining throughout the growth cone and a partial colocalization with cortactin in the P domain (Fig. 4A and B). Src1 staining was generally stronger in the C than in the P domain, while Src2 was more evenly distributed. Activated Src2, visualized with pSrc2 antibody specific for autophosphorylated Src2, exhibits significant cortactin colocalization in filopodia (Fig. 4C and D). Compared to total Src2 labeling, pSrc2 signals were more enriched at the tip of filopodia, revealed by F-actin staining (Fig. 4D). However, a fraction of cortactin/Src colocalization could be purely coincidental due to plasma membrane-associated or cytoplasmic Src in the same areas as cortactin signals. Hence, to reduce signals from plasma membrane associated and cytosolic Src, and to better reveal cortactin/Src colocalization, we performed live cell extraction experiments under conditions of stabilized actin and microtubule cytoskeleton (Fig. 4E and F). Such a treatment resulted in a strong reduction of cortactin- (Fig. 4E) and of Src1- and Src2-labeling (data not shown) in the growth cone periphery, while pSrc2-staining was more resistant to live cell extraction, e.g. at the tip of filopodia that were visualized by F-actin labeling (Fig. 4F). Although this precluded us from assessing cortactin/Src colocalization using the live cell extraction method, these observations suggest that the binding of cortactin to F-actin is weak in the growth cone periphery, while stronger in the T zone ruffles.
Cortactin localizes to apCAM adhesion sites
Using the RBI assay, we have previously shown that apCAM-coated beads can induce growth cone guidance events when beads are physically restrained in the growth cone P domain (Suter et al., 1998). Both F-actin accumulation and Src activation were detected in the vicinity of the beads (Suter et al., 1998; Suter and Forscher, 2001). Since cortactin is a major Src substrate and involved in actin assembly, we asked whether cortactin would also accumulate around apCAM-coated beads. We carried out RBI assays followed by F-actin and cortactin staining, and quantified actin and cortactin amounts at restrained and unrestrained beads in ratiometric, volume-independent manner (Fig. 5). To achieve this, neuronal cell bodies were injected with Texas Red-dextran as a volume marker a few hours before carrying out the RBI assay (see methods).
After a typical latency period of 5–15 min with little morphological change, the C domain moved towards the beads and the leading edge advanced at similar rate (Fig. 5A–B). In agreement with previous findings (Suter et al., 1998), F-actin accumulated around the restrained beads (Fig. 5D). We also observed and measured an increase in cytoplasmic volume around the beads, as indicated by the dextran signal (Fig. 5C). The increased volume is likely due to the ruffling activity and actin assembly induced by the beads. Notably, cortactin strongly accumulated around the beads as well as in T zone ruffles close to the bead in a fashion similar to F-actin (Fig. 5E). In contrast, while amounts of both F-actin and cortactin were higher at the leading edge that protruded in front of the beads when compared to the adjacent regions, the density of the proteins remained unchanged as the volume increased as well (Fig. 5C–E). Thus, cortactin accumulates at sites of de novo actin assembly.
Although unrestrained beads also induced increased cytoplasmic volumes around them (Fig. 5F), F-actin (Fig. 5G) and cortactin signals (Fig. 5H) were only slightly increased when compared to restrained beads. Quantification of volume-corrected F-actin and cortactin fluorescence intensities, hence staining density, around restrained and unrestrained beads normalized to adjacent regions without beads is shown in Fig. 5I. Dextran signals increased more around restrained (5.13 ± 0.72 fold) than unrestrained beads (2.76 ± 0.38 fold) when compared to adjacent regions (p<0.01). However, there was a larger difference in the fold increase of cortactin and actin signals between restrained (4.86 ± 0.71 and 4.97 ± 0.42, respectively) and unrestrained beads (1.35 ± 0.2 and 1.5 ± 0.14 for cortactin and actin, respectively; p<0.01). Thus, the staining density at restrained beads normalized against adjacent regions was increased by 80% (cortactin) and 90% (actin) when compared to unrestrained beads. While total protein amounts are increased at restrained beads, the density of cortactin and F-actin did not appear to be much different between restrained beads and adjacent P-domain areas, assuming that cortactin is equally distributed by volume (Fig. 5I). In summary, both cortactin and F-actin accumulate around restrained beads, implicating cortactin as a potential Src effector in apCAM-induced F-actin rearrangements and growth cone guidance.
Discussion
In the present study, we report that cortactin is highly expressed in the Aplysia nervous system and localizes to regions of dynamic F-actin assembly in growth cones, including filopodia tips and the leading edge of lamellipodia. Cortactin distribution largely depends on the F-actin cytoskeleton. Furthermore, cortactin exhibits significant colocalization with activated Src2, particularly along the leading edge. Finally, we demonstrate that cortactin is localizes to regions where apCAM receptors are clustered and immobilized during growth cone guidance events.
Aplysia cortactin sequence
Aplysia cortactin displays several unique characteristics in the different functional domains when compared to other cortactin sequences. The NTA domain in all the species listed in Fig. 1 have the Arp2/3 binding motif DDW, except Aplysia, which has the motif DEW, similar to the Wiskott-Aldrich syndrome protein (WASP) family (Weaver et al., 2003). Could Aplysia cortactin share WASP function? This idea is supported by the absence of WASP cDNA in the recently released Aplysia EST database (Moroz et al., 2006), although only approximately 70% of the Aplysia genome information is available to date. Aplysia cortactin is the only cortactin homolog known so far that has seven and a half tandem repeats, which may partly explain its higher molecular weight compared to mouse cortactin. There is no correlation between the position and function of the tandem repeats, except that in vertebrate cortactin the fourth repeat plus a portion of third or fifth repeat are an absolute requirement for F-actin binding (Weed et al., 2000). While little is known about the function of the helix, a recent study suggests that it could be involved in the regulation of actin binding (Cowieson et al., 2008). The proline-rich region is quite different among species. Interestingly, Aplysia cortactin is the only sequence that has six and a half repeats of the motif (PEPVREPS) in the proline-rich region. The presence of tyrosine residues in this domain suggests potential phosphorylation sites for Src kinases, but further investigation is needed to determine whether Aplysia Src1 and/or Src2 phosphorylate cortactin. The presence of the SH3 domain suggests interactions with additional regulatory proteins involved in actin assembly, as described in other species (Tehrani et al., 2007). However, while ESTs for Arp2/3 complex proteins are present in the Aplysia database, no EST matches were found for the cortactin-interacting molecules WASP, WIP, or Nck. Thus, additional molecular information is required to fully understand the machinery of actin polymerization and its potential regulation by cortactin in Aplysia growth cones.
Colocalization with F-actin
We observed colocalization between cortactin and F-actin in Aplysia growth cones, particularly in areas of dynamic actin assembly such as filopodia tips, leading edge, T zone ruffles and around restrained beads. Previous reports have shown that cortactin localizes to sites of actin remodeling such as lamellipodia of non-neuronal cells (Kaksonen et al., 2000; Weed et al., 2000), and dendritic spines of hippocampal neurons (Hering and Sheng, 2003; Racz and Weinberg, 2004). A very recent study has shown that cortactin is present in lamellipodial veils of vertebrate growth cones, a localization that was correlated with the branching of actin filaments (Korobova and Svitkina, 2008). While these findings are in agreement with our lamellipodial cortactin localization in Aplysia growth cones, we also detected cortactin highly associated with F-actin bundles in filopodia. The lamellipodial localization is in agreement with the proposed role of cortactin to recruit the Apr2/3 complex to mother actin filaments and induce daughter filament nucleation, as well as stabilizing branched actin networks in lamellipodia (Weaver et al., 2001), for example by forming “actin sheets” (Cowieson et al., 2008). However, the presence of cortactin on filopodia, and especially at the tips of filopodia was unexpected. These are locations where typical filopodial proteins such as Ena/Vasp, formins, and fascins are found (Chhabra and Higgs, 2007; Faix and Grosse, 2006). Since Arp2/3 has been localized adjacent to filopodia in vertebrate growth cones and implicated in the formation of both lamellipodia and filopodia (Korobova and Svitkina, 2008), we speculate that in Aplysia growth cones cortactin could be an important regulator of Arp2/3-dependent actin assembly in both lamellipodia and filopodia. However, Arp2/3 localization in Aplysia growth cones and additional functional studies are needed to establish the roles of cortactin/Arp2/3- versus formin-triggered filopodial formation in Aplysia growth cones. Lastly, the abundance of cortactin in both Aplysia growth cones and total nervous system suggests that cortactin could have additional actin-related functions, such as stabilization of bundles and networks.
Colocalization with active Src at the tip of filopodia
Cortactin was first identified as a Src kinase substrate (Wu et al., 1991) and was further demonstrated to be phosphorylated by several other members of the Src kinase family, including Fer, Fyn and Syk (Cosen-Binker and Kapus, 2006; Lua and Low, 2005). However, the role of Src tyrosine phosphorylation in cortactin function is still controversial. While a few reports indicate that tyrosine phosphorylation decreases F-actin crosslinking activity of cortactin (Huang et al., 1997a) or is not required for cortactin-mediated actin nucleation (Illes et al., 2006a), several studies demonstrated a strong correlation between Src-mediated tyrosine phosphorylation of cortactin and actin-based processes, such as podosome formation in osteoclasts (Tehrani et al., 2006), strengthening of N-cadherin-mediated intercellular adhesions between fibroblasts (El Sayegh et al., 2005), and actin polymerization (Tehrani et al., 2007). Src-mediated tyrosine phosphorylation of cortactin also regulates cell motility but not cortactin localization in endothelial cells (Huang et al., 1998). Furthermore, cortactin and Src colocalization was observed in a number of non-neuronal cell types, including chicken embryonic fibroblasts transformed with v-Src (Okamura and Resh, 1995), osteoclasts (Matsubara et al., 2006) and human ovarian tumor cells (Bourguignon et al., 2001). In summary, Src-mediated tyrosine phosphorylation of cortactin appears to stimulate actin remodeling in non-neuronal cells. In agreement with our results, activated Src has been localized at the tip of filopodia in Xenopus neuronal growth cones (Robles et al., 2005). To our knowledge, the present study is the first to report colocalization of activated Src and cortactin at the tips of growth cone filopodia and the leading edge. Additional functional experiments are required to dissect whether Src-dependent tyrosine phosphorylation of cortactin regulates actin assembly at filopodia tips.
Cortactin accumulation after apCAM clustering
Despite substantial findings regarding the molecular components of intracellular signaling pathways downstream of NCAM, very little is know about how these pathways affect the actin cytoskeleton in neuronal cells following an acute NCAM stimulation (Ditlevsen et al., 2008). Using the RBI growth cone guidance assay, we have demonstrated an accumulation of F-actin and an increase in Src kinase activity at apCAM adhesion sites (Suter et al., 1998; Suter and Forscher, 2001). Here, we observed an accumulation of cortactin around restrained beads that induce growth cone steering, but much less increase around unrestrained, flow-coupled beads. It is important to note that total cortactin amounts, but not density were increased around beads when compared to adjacent P domain areas, assuming a simple volume-like cortactin distribution. Collectively, our data provide evidence that cortactin could be one of the components bridging Src kinase activity and actin polymerization downstream of IgCAM receptors. Thus, cortactin recruitment appears to occur downstream of several receptor types (Cosen-Binker and Kapus, 2006), including integrins (Illes et al., 2006b), cadherins (El Sayegh et al., 2005), IgCAMs (Cavallaro et al., 2001), N-syndecan (Kinnunen et al., 1998) and acetylcholine receptors (Dai et al., 2000). Future functional studies are needed to dissect the precise role of cortactin in growth cone motility and guidance driven by IgCAMs and other cell adhesion receptors.
Acknowledgments
Contract grant sponsor: National Institutes of Health, U.S. Public Health Service; Contract grant number: NS049233 (to D.M.S.).
We would like to thank Dr. H. Chang, and Dr. Z.Q. Luo for their technical assistance. We are also grateful to Dr. P. Hollenbeck and Dr, C. Staiger for their valuable comments on the manuscript. This work was supported by grants from the NIH (R01 NS049233) and the Bindley Bioscience Center at Purdue University to D.M.S.
Abbreviations
- ApCAM
Aplysia cell adhesion molecule
- BSA
Bovine serum albumin
- C domain
Central domain
- CAM
Cell adhesion molecule
- DIC
Differential interference contrast
- IgCAM
Cell adhesion molecule of the immunoglobulin superfamily
- NCAM
Neural cell adhesion molecule
- NTA
N-terminal acidic region
- P domain
Peripheral domain
- PBS
Phosphate buffer saline
- RACE
Rapid amplification of cDNA ends
- RBI
Restrained bead interaction assay
- SDS-PAGE
Sodium-dodecyl sulphate polyacrylamide gel electrophoresis
- T zone
Transition zone
References
- Bourguignon LY, Zhu H, Shao L, Chen YW. CD44 interaction with c-Src kinase promotes cortactin-mediated cytoskeleton function and hyaluronic acid-dependent ovarian tumor cell migration. J Biol Chem. 2001;276(10):7327–7336. doi: 10.1074/jbc.M006498200. [DOI] [PubMed] [Google Scholar]
- Buday L, Downward J. Roles of cortactin in tumor pathogenesis. Biochim Biophys Acta. 2007;1775(2):263–273. doi: 10.1016/j.bbcan.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Campbell DH, Sutherland RL, Daly RJ. Signaling pathways and structural domains required for phosphorylation of EMS1/cortactin. Cancer Res. 1999;59(20):5376–5385. [PubMed] [Google Scholar]
- Cavallaro U, Niedermeyer J, Fuxa M, Christofori G. N-CAM modulates tumour-cell adhesion to matrix by inducing FGF-receptor signalling. Nat Cell Biol. 2001;3(7):650–657. doi: 10.1038/35083041. [DOI] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9(10):1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- Cosen-Binker LI, Kapus A. Cortactin: the gray eminence of the cytoskeleton. Physiology (Bethesda) 2006;21:352–361. doi: 10.1152/physiol.00012.2006. [DOI] [PubMed] [Google Scholar]
- Cowieson NP, King G, Cookson D, Ross I, Huber T, Hume DA, Kobe B, Martin JL. Cortactin adopts a globular conformation and bundles actin into sheets. J Biol Chem. 2008 doi: 10.1074/jbc.M708917200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Luo X, Xie H, Peng HB. The actin-driven movement and formation of acetylcholine receptor clusters. J Cell Biol. 2000;150(6):1321–1334. doi: 10.1083/jcb.150.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly RJ. Cortactin signalling and dynamic actin networks. Biochem J. 2004;382(Pt 1):13–25. doi: 10.1042/BJ20040737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decourt B, Bouleau Y, Dulon D, Hafidi A. Expression analysis of neuroleukin, calmodulin, cortactin, and Rho7/Rnd2 in the intact and injured mouse brain. Brain Res Dev Brain Res. 2005;159(1):36–54. doi: 10.1016/j.devbrainres.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Dent EW, Kalil K. Axon branching requires interactions between dynamic microtubules and actin filaments. J Neurosci. 2001;21(24):9757–9769. doi: 10.1523/JNEUROSCI.21-24-09757.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditlevsen DK, Povlsen GK, Berezin V, Bock E. NCAM-induced intracellular signaling revisited. J Neurosci Res. 2008;86(4):727–743. doi: 10.1002/jnr.21551. [DOI] [PubMed] [Google Scholar]
- Du Y, Weed SA, Xiong WC, Marshall TD, Parsons JT. Identification of a novel cortactin SH3 domain-binding protein and its localization to growth cones of cultured neurons. Mol Cell Biol. 1998;18(10):5838–5851. doi: 10.1128/mcb.18.10.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Sayegh TY, Arora PD, Fan L, Laschinger CA, Greer PA, McCulloch CA, Kapus A. Phosphorylation of N-cadherin-associated cortactin by Fer kinase regulates N-cadherin mobility and intercellular adhesion strength. Mol Biol Cell. 2005;16(12):5514–5527. doi: 10.1091/mbc.E05-05-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Grosse R. Staying in shape with formins. Dev Cell. 2006;10(6):693–706. doi: 10.1016/j.devcel.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Forscher P, Smith SJ. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J Cell Biol. 1988;107(4):1505–1516. doi: 10.1083/jcb.107.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goslin K, Birgbauer E, Banker G, Solomon F. The role of cytoskeleton in organizing growth cones: a microfilament-associated growth cone component depends upon microtubules for its localization. J Cell Biol. 1989;109(4 Pt 1):1621–1631. doi: 10.1083/jcb.109.4.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering H, Sheng M. Activity-dependent redistribution and essential role of cortactin in dendritic spine morphogenesis. J Neurosci. 2003;23(37):11759–11769. doi: 10.1523/JNEUROSCI.23-37-11759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Liu J, Haudenschild CC, Zhan X. The role of tyrosine phosphorylation of cortactin in the locomotion of endothelial cells. J Biol Chem. 1998;273(40):25770–25776. doi: 10.1074/jbc.273.40.25770. [DOI] [PubMed] [Google Scholar]
- Huang C, Ni Y, Wang T, Gao Y, Haudenschild CC, Zhan X. Down-regulation of the filamentous actin cross-linking activity of cortactin by Src-mediated tyrosine phosphorylation. J Biol Chem. 1997a;272(21):13911–13915. doi: 10.1074/jbc.272.21.13911. [DOI] [PubMed] [Google Scholar]
- Huang C, Tandon NN, Greco NJ, Ni Y, Wang T, Zhan X. Proteolysis of platelet cortactin by calpain. J Biol Chem. 1997b;272(31):19248–19252. doi: 10.1074/jbc.272.31.19248. [DOI] [PubMed] [Google Scholar]
- Huber AB, Kolodkin AL, Ginty DD, Cloutier JF. Signaling at the growth cone: ligand-receptor complexes and the control of axon growth and guidance. Annu Rev Neurosci. 2003;26:509–563. doi: 10.1146/annurev.neuro.26.010302.081139. [DOI] [PubMed] [Google Scholar]
- Illes A, Enyedi B, Tamas P, Balazs A, Bogel G, Buday L. Inducible phosphorylation of cortactin is not necessary for cortactin-mediated actin polymerisation. Cell Signal. 2006a;18(6):830–840. doi: 10.1016/j.cellsig.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Illes A, Enyedi B, Tamas P, Balazs A, Bogel G, Melinda, Lukacs, Buday L. Cortactin is required for integrin-mediated cell spreading. Immunol Lett. 2006b;104(1–2):124–130. doi: 10.1016/j.imlet.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Peng HB, Rauvala H. Association of cortactin with dynamic actin in lamellipodia and on endosomal vesicles. J Cell Sci. 2000;113(Pt 24):4421–4426. doi: 10.1242/jcs.113.24.4421. [DOI] [PubMed] [Google Scholar]
- Kinnunen T, Kaksonen M, Saarinen J, Kalkkinen N, Peng HB, Rauvala H. Cortactin-Src kinase signaling pathway is involved in N-syndecan-dependent neurite outgrowth. J Biol Chem. 1998;273(17):10702–10708. doi: 10.1074/jbc.273.17.10702. [DOI] [PubMed] [Google Scholar]
- Korobova F, Svitkina T. Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol Biol Cell. 2008;19(4):1561–1574. doi: 10.1091/mbc.E07-09-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafont F, Rouget M, Rousselet A, Valenza C, Prochiantz A. Specific responses of axons and dendrites to cytoskeleton perturbations: an in vitro study. J Cell Sci. 1993;104 ( Pt 2):433–443. doi: 10.1242/jcs.104.2.433. [DOI] [PubMed] [Google Scholar]
- Lee AC, Decourt B, Suter DM. Neuronal cell cultures from Aplysia for high-resolution imaging of growth cones. J Vis Experim. 2008;12 doi: 10.3791/662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lua BL, Low BC. Cortactin phosphorylation as a switch for actin cytoskeletal network and cell dynamics control. FEBS Lett. 2005;579(3):577–585. doi: 10.1016/j.febslet.2004.12.055. [DOI] [PubMed] [Google Scholar]
- Matsubara T, Myoui A, Ikeda F, Hata K, Yoshikawa H, Nishimura R, Yoneda T. Critical role of cortactin in actin ring formation and osteoclastic bone resorption. J Bone Miner Metab. 2006;24(5):368–372. doi: 10.1007/s00774-006-0701-4. [DOI] [PubMed] [Google Scholar]
- Mayford M, Barzilai A, Keller F, Schacher S, Kandel ER. Modulation of an NCAM-related adhesion molecule with long-term synaptic plasticity in Aplysia. Science. 1992;256(5057):638–644. doi: 10.1126/science.1585176. [DOI] [PubMed] [Google Scholar]
- Moroz LL, Edwards JR, Puthanveettil SV, Kohn AB, Ha T, Heyland A, Knudsen B, Sahni A, Yu F, Liu L, Jezzini S, Lovell P, Iannucculli W, Chen M, Nguyen T, Sheng H, Shaw R, Kalachikov S, Panchin YV, Farmerie W, Russo JJ, Ju J, Kandel ER. Neuronal transcriptome of aplysia: neuronal compartments and circuitry. Cell. 2006;127(7):1453–1467. doi: 10.1016/j.cell.2006.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura H, Resh MD. p80/85 cortactin associates with the Src SH2 domain and colocalizes with v-Src in transformed cells. J Biol Chem. 1995;270(44):26613–26618. doi: 10.1074/jbc.270.44.26613. [DOI] [PubMed] [Google Scholar]
- Racz B, Weinberg RJ. The subcellular organization of cortactin in hippocampus. J Neurosci. 2004;24(46):10310–10317. doi: 10.1523/JNEUROSCI.2080-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles E, Woo S, Gomez TM. Src-dependent tyrosine phosphorylation at the tips of growth cone filopodia promotes extension. J Neurosci. 2005;25(33):7669–7681. doi: 10.1523/JNEUROSCI.2680-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochlin MW, Dailey ME, Bridgman PC. Polymerizing microtubules activate site-directed F-actin assembly in nerve growth cones. Mol Biol Cell. 1999;10(7):2309–2327. doi: 10.1091/mbc.10.7.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AW, Kabir N, Forscher P. Filopodia and actin arcs guide the assembly and transport of two populations of microtubules with unique dynamic parameters in neuronal growth cones. J Cell Biol. 2002;158(1):139–152. doi: 10.1083/jcb.200203038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Backert S. Cortactin: an Achilles’ heel of the actin cytoskeleton targeted by pathogens. Trends Microbiol. 2005;13(4):181–189. doi: 10.1016/j.tim.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Suter DM, Errante LD, Belotserkovsky V, Forscher P. The Ig superfamily cell adhesion molecule, apCAM, mediates growth cone steering by substrate-cytoskeletal coupling. J Cell Biol. 1998;141(1):227–240. doi: 10.1083/jcb.141.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter DM, Forscher P. Transmission of growth cone traction force through apCAM-cytoskeletal linkages is regulated by Src family tyrosine kinase activity. J Cell Biol. 2001;155(3):427–438. doi: 10.1083/jcb.200107063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter DM, Schaefer AW, Forscher P. Microtubule dynamics are necessary for SRC family kinase-dependent growth cone steering. Curr Biol. 2004;14(13):1194–1199. doi: 10.1016/j.cub.2004.06.049. [DOI] [PubMed] [Google Scholar]
- Tehrani S, Faccio R, Chandrasekar I, Ross FP, Cooper JA. Cortactin has an essential and specific role in osteoclast actin assembly. Mol Biol Cell. 2006;17(7):2882–2895. doi: 10.1091/mbc.E06-03-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehrani S, Tomasevic N, Weed S, Sakowicz R, Cooper JA. Src phosphorylation of cortactin enhances actin assembly. Proc Natl Acad Sci U S A. 2007;104(29):11933–11938. doi: 10.1073/pnas.0701077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uruno T, Liu J, Zhang P, Fan Y, Egile C, Li R, Mueller SC, Zhan X. Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat Cell Biol. 2001;3(3):259–266. doi: 10.1038/35060051. [DOI] [PubMed] [Google Scholar]
- van Rossum AG, de Graaf JH, Schuuring-Scholtes E, Kluin PM, Fan YX, Zhan X, Moolenaar WH, Schuuring E. Alternative splicing of the actin binding domain of human cortactin affects cell migration. J Biol Chem. 2003;278(46):45672–45679. doi: 10.1074/jbc.M306688200. [DOI] [PubMed] [Google Scholar]
- Weaver AM. Cortactin in tumor invasiveness. Cancer Lett. 2008;265(2):157–166. doi: 10.1016/j.canlet.2008.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver AM, Karginov AV, Kinley AW, Weed SA, Li Y, Parsons JT, Cooper JA. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr Biol. 2001;11(5):370–374. doi: 10.1016/s0960-9822(01)00098-7. [DOI] [PubMed] [Google Scholar]
- Weaver AM, Young ME, Lee WL, Cooper JA. Integration of signals to the Arp2/3 complex. Curr Opin Cell Biol. 2003;15(1):23–30. doi: 10.1016/s0955-0674(02)00015-7. [DOI] [PubMed] [Google Scholar]
- Weed SA, Karginov AV, Schafer DA, Weaver AM, Kinley AW, Cooper JA, Parsons JT. Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J Cell Biol. 2000;151(1):29–40. doi: 10.1083/jcb.151.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weed SA, Parsons JT. Cortactin: coupling membrane dynamics to cortical actin assembly. Oncogene. 2001;20(44):6418–6434. doi: 10.1038/sj.onc.1204783. [DOI] [PubMed] [Google Scholar]
- Wu H, Reynolds AB, Kanner SB, Vines RR, Parsons JT. Identification and characterization of a novel cytoskeleton-associated pp60src substrate. Mol Cell Biol. 1991;11(10):5113–5124. doi: 10.1128/mcb.11.10.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Decourt B, Zabidi MA, Wuethrich LT, Kim WH, Zhou Z, MacIsaac K, Suter DM. Microtubule-mediated Src tyrosine kinase trafficking in neuronal growth cones. Mol Biol Cell. 2008 doi: 10.1091/mbc.E08-06-0603. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XF, Schaefer AW, Burnette DT, Schoonderwoert VT, Forscher P. Rho-dependent contractile responses in the neuronal growth cone are independent of classical peripheral retrograde actin flow. Neuron. 2003;40(5):931–944. doi: 10.1016/s0896-6273(03)00754-2. [DOI] [PubMed] [Google Scholar]