Abstract
Although the structural core of the ribosome is conserved in all kingdoms of life, eukaryotic ribosomes are significantly larger and more complex than their bacterial counterparts. The extent to which these differences influence the molecular mechanism of translation remains elusive. Multiparticle cryo-electron microscopy and single-molecule FRET investigations of the mammalian pre-translocation complex reveal spontaneous, large-scale conformational changes including an inter-subunit rotation of the ribosomal subunits. Through structurally related processes, tRNA substrates oscillate between classical and at least two distinct hybrid configurations facilitated by localized changes in their L-shaped fold. Hybrid states are favoured within the mammalian complex. However, classical tRNA positions can be restored by tRNA binding to the E site or by the eukaryotic-specific antibiotic and translocation inhibitor, cycloheximide. These findings reveal critical distinctions in the structural and energetic features of bacterial and mammalian ribosomes, providing a mechanistic basis for divergent translation regulation strategies and species-specific antibiotic action.
INTRODUCTION
Genetic information within messenger RNA (mRNA) is translated into protein by the ribosome, a highly conserved macromolecular RNA-protein assembly (70S in bacteria; 80S in eukaryotes) consisting of two asymmetric subunits (30S and 50S in bacteria; 40S and 60S in eukaryotes). The ribosome decodes the mRNA sequence in discrete codon steps during the elongation cycle of protein synthesis using transfer RNA (tRNA) substrates. In all domains of life, three classical tRNA binding sites exist within the ribosome created by the interface of the two ribosomal subunits: the Aminoacyl (A), Peptidyl (P) and Exit (E) sites. The L-shaped, 25kDa tRNA substrate serves as an adaptor molecule by bridging the functional centers of the small and large subunit responsible for decoding and peptidyl transferase (PTC) activities, respectively (Frank and Spahn, 2006; Ramakrishnan, 2002).
The ribosome actively synthesizes proteins during the elongation cycle of translation through three sequential reactions: (i) decoding, which leads to A-site occupation by an aminoacyl-tRNA (aa-tRNA); (ii) peptide bond formation, which generates a pre-translocation (PRE) complex bearing deacylated tRNA in the P site and peptidyl-tRNA in the A site; and (iii) translocation of A and P site tRNAs to the P and E sites, respectively –a multistep process resulting in a post-translocation (POST) state. The activation barriers for spontaneous PRE and POST complex formation are relatively high (Schilling-Bartetzko et al., 1992). In order to achieve rapid rates of protein synthesis in the cell, both PRE and POST ribosome complex formation are catalyzed by two conserved GTPases: elongation factors EF-Tu and EF-G, respectively (eEF1A and eEF2 in eukaryotes).
A tRNA molecule moves through the ribosome during sequential elongation cycles, first entering the A site and then translocating via the P site to the E site, where it is released. Understanding this multistep process has been expanded by the detection of hybrid states of tRNA binding (Moazed and Noller, 1989). Hybrid tRNA positions are defined by a spontaneous movement of A- and P-site tRNAs in the direction of translocation on the large ribosomal subunit, while the anticodon portions of tRNA remain fixed with respect to the small subunit (A/P and P/E hybrid sites, respectively). Hybrid tRNA configurations have been suggested to serve as bona fide intermediates in the translocation process by lowering the activation barrier for translocation (Dorner et al., 2006; Semenkov et al., 2000). Importantly, single-molecule fluorescence resonance energy transfer (smFRET) imaging has revealed the intrinsically dynamic nature of the bacterial PRE complex in which A- and P-site tRNAs reversibly exchange between classical and hybrid configurations by both coupled and independent motions on the sub-second time scale (Blanchard et al., 2004; Kim et al., 2007; Munro et al., 2007). tRNA translocation converts the dynamic PRE state to the POST state and re-establishes classical tRNA positioning in P/P and E/E sites by moving the adjacent anticodon elements of A- and P-site tRNAs, along with mRNA, with respect to the 30S subunit.
The process of translation is facilitated by large-scale conformational changes in the ribosome, including a dramatic, ratchet-like inter-subunit rearrangement (RSR) (Frank and Agrawal, 2000). Initially, such events were associated with the binding of translation factors, especially EF-G (Frank and Agrawal, 2000; Valle et al., 2003). More recently, evidence supporting the spontaneous nature of conformational modes within ribosomal complexes was provided by smFRET experiments and multiparticle cryo-EM investigations. These data have shown that A- and P-site tRNAs spontaneously fluctuate between classical and hybrid configurations in a manner that is loosely coupled to inter-subunit rotation (Cornish et al., 2008; Marshall et al., 2008; Munro et al., 2010a).
Structural characterizations of hybrid tRNA configurations have been hampered by their transient nature. However, recent multiparticle cryo-EM investigations of the bacterial PRE complex have revealed that non-rotated and rotated subunit arrangements of the ribosome as well as classical and hybrid tRNA positions (Agirrezabala et al., 2008; Fischer et al., 2010; Julian et al., 2008). Conformational heterogeneities are also present in bacterial ribosome•EF-G complexes, where a mixture of PRE and POST translocation intermediates containing inter-and intra-subunit hybrid state tRNA configurations have been identified (Ratje et al., 2010). Taken together, these findings suggest a metastable energy landscape view of ribosome function (Munro et al., 2009), where rather than displaying a single, “unique” structure, compositionally defined ribosomal complexes exhibit dynamic heterogeneities that reflect an exchange between distinct, functionally relevant structural configurations.
The eukaryotic (80S) ribosome is significantly larger and more complex than its bacterial counterpart. However, core elements of the particle and the substrate binding sites are evolutionarily conserved (Chandramouli et al., 2008; Spahn et al., 2001a; Taylor et al., 2009). Biochemical studies suggest that the elongation mechanism is similar in both domains of life (Budkevich et al., 2008). Despite recent advances towards the elucidation of eukaryotic ribosome structures (Ben-Shem et al., 2010; Rabl et al., 2010), it is not yet known to what extent the observed divergence in the ribosomal core and the additional, eukaryotic-specific components influence its interaction with ligands and factors and its dynamic behaviour. The existence of ribosome-targeting antibiotics that display domain specificity and evidence of essential, domain-specific translation factors (Andersen et al., 2006), implies that critical aspects of the translation mechanism and/or dynamics differ substantially. In order to further explore the nature of such distinctions and to understand how these features may be leveraged for potentially therapeutic interventions, comparative functional and structural investigations must be achieved.
Towards this goal, we report here a comprehensive study of the mammalian 80S ribosome PRE complex from rabbit liver using multiparticle cryo-EM and smFRET techniques. These studies, which include the elucidation of four, structurally distinct sub-states of the 80S particle, highlight the dynamic nature of the mammalian PRE complex and show how the energy landscape of the 80S ribosome is tuned by tRNA binding to the E site and by the antibiotic cycloheximide. Together, these findings provide important insights into conserved and divergent features of the protein synthesis mechanism among distinct domains of life and into the nature and role of dynamic processes intrinsic to the 80S ribosome that likely contribute directly to the mechanism of translocation.
RESULTS AND DISCUSSION
The mammalian PRE-translocation complex is dynamically heterogeneous
Insights into the dynamic heterogeneities intrinsic to the mammalian PRE complex were initially obtained through multiparticle cryo-EM reconstructions of an eEF1A·GDPNP decoding complex formed using 80S ribosomes from rabbit liver. Unexpectedly, a significant number of PRE complexes were formed in this reaction, in addition to the stalled decoding complex (data not shown), presumably due to inefficient stalling of ternary complex during the decoding process. Refinement of these data to 15–20 Å resolution revealed that about 33% of the sample contained PRE ribosome complexes of two distinct types. The larger PRE subpopulation (60% of 33%) was found to be in a non-rotated conformation containing classically configured tRNAs in the A, P, and E sites. The second subpopulation (40% of 33%) was characterized by a rotated ribosome conformation in which the P-site tRNA was in a hybrid configuration (Fig. S1). In order to validate these results, functional PRE complexes were directly prepared using tRNA molecules from bovine liver (Budkevich et al., 2008). Multiparticle, cryo-EM refinement again yielded PRE complexes of the same two types. Thus, analogous to the bacterial PRE complex (Agirrezabala et al., 2008; Julian et al., 2008), these data suggest that the 80S ribosome is intrinsically dynamic, where the process of inter-subunit rotation and the formation of hybrid tRNA configurations can occur spontaneously.
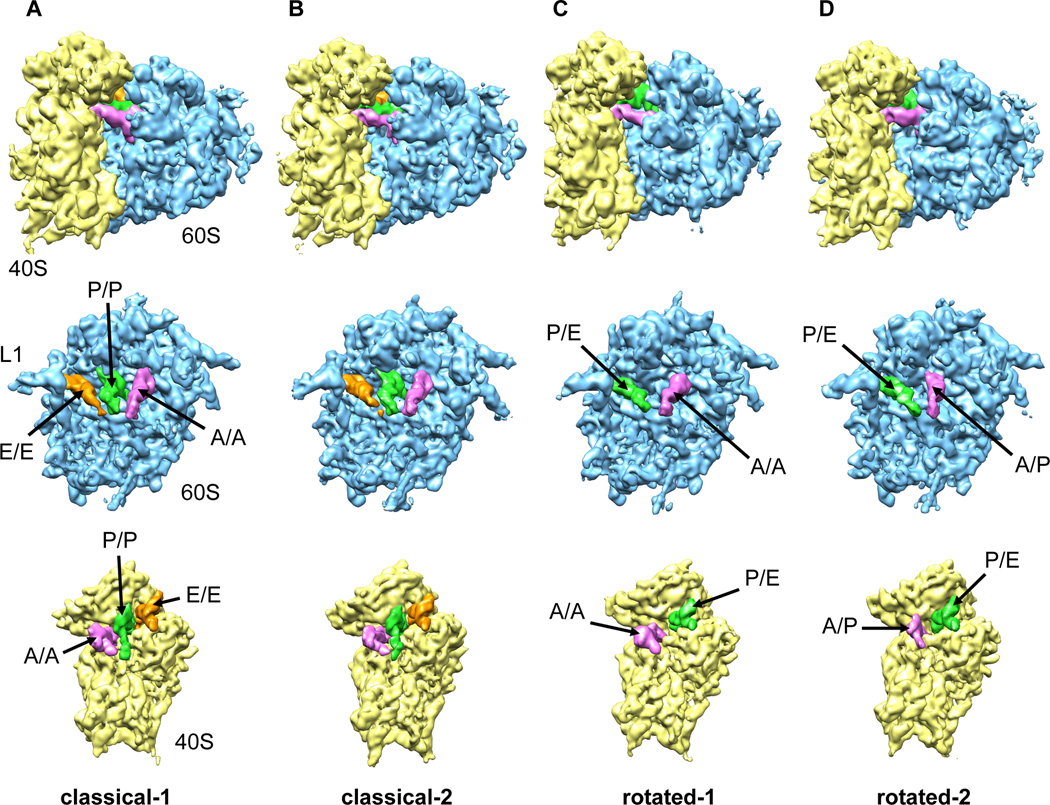
Image processing and multiparticle refinement of the combined data sets (298,800 particle images) resulted in further splitting of both the classical and rotated subpopulations yielding a total of four, structurally distinct reconstructions at resolutions ranging from 9 to 10.6 Å (Fig. 1, S1, S2). Accordingly, two structures were obtained of non-rotated PRE complexes containing three tRNAs in classical A, P and E sites (classical-1 and classical-2) and two structures were obtained of rotated PRE complex conformations with tRNAs in hybrid states (rotated-1 and rotated-2). Improvement of the resolution by processing the combined data set indicates that the two differently prepared PRE complexes are structurally equivalent. This was further verified by Fourier shell correlation curves comparing structures obtained from re-separated data (Fig. S2). By monitoring the distribution of the particle images during our sorting procedure (Fig. S1) it was also shown that the two PRE specimens exhibited similar ratios for all four subpopulations consistent with both data sets being comparable in nature.
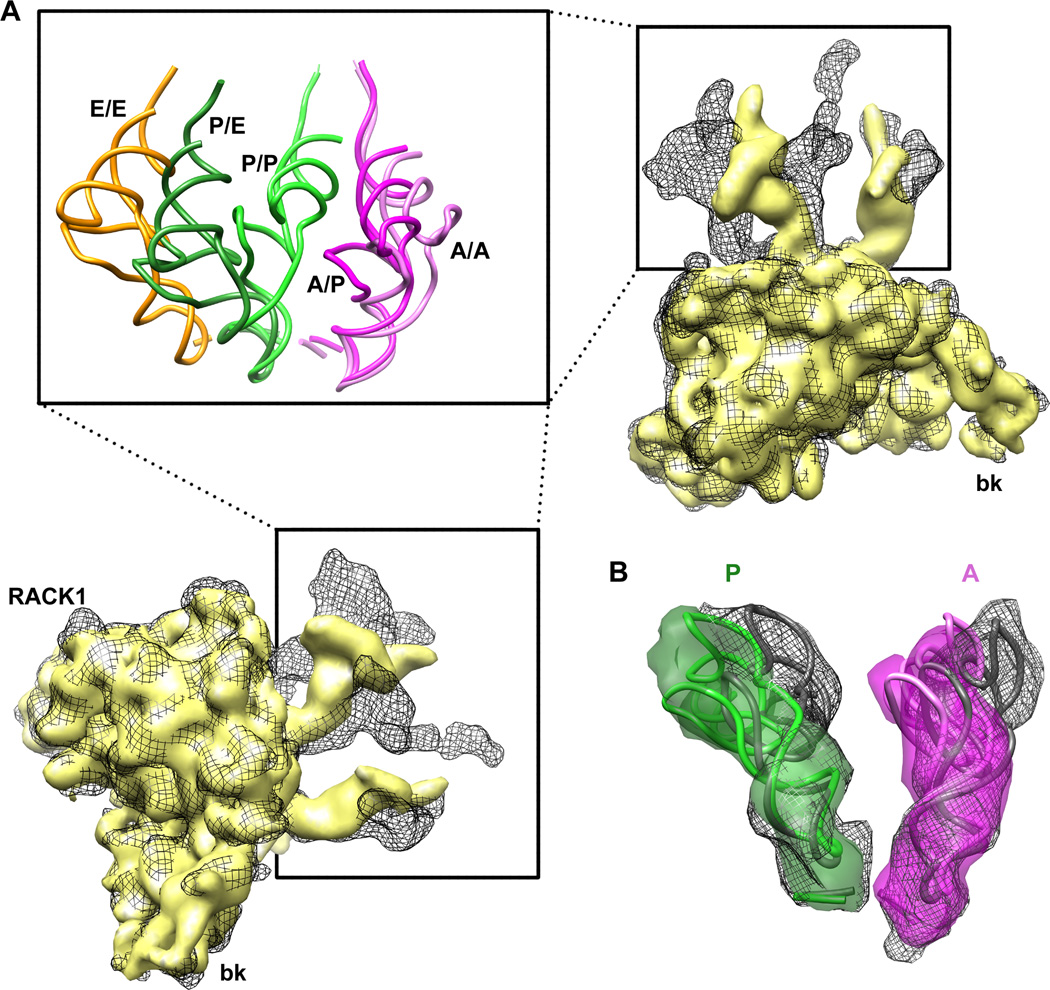
Figure 1. Multiparticle refinement reveals four structurally distinct sub-populations in the PRE complex.
(A, B) Representation of the two non-rotated populations with 3 classically positioned A-, P- and E-site tRNAs. (C, D) Representation of the two rotated sub-states, where deacylated tRNA occupies a P/E hybrid state while the peptidyl-tRNA body adopts configurations that are similar to classical (C) or hybrid (D) positions. Color code: 60S subunit (blue), 40S subunit (yellow), A-site tRNA (pink), P-tRNA (green), E-site tRNA (orange).
See alsoFigure S1. Heterogeneity in the 80S ribosome PRE complexes from rabbit liver.
Figure S2. Resolution curves for the four sub-populations of the mammalian PRE complex.
All four structures could be interpreted in molecular terms by docking atomic models of tRNAs (Voorhees et al., 2009) and the recent X-ray structure of eukaryotic ribosomal particles (Ben-Shem et al., 2010; Rabl et al., 2010) into the cryo-EM maps. As detailed below, each PRE complex subpopulation identified differs with respect to the positioning of peptidyl-tRNA. Thus, these data provide structural insights into the nature of classical and hybrid tRNA configurations within the mammalian PRE complex.
Structural features of the classically configured mammalian PRE complex
The classical-1 PRE complex showed strong density for tRNAs in A, P and E sites (Fig. S3). Global alignment of the classical-1 small subunit with that of the bacterial 70S PRE complex (Voorhees et al., 2009) revealed nearly identical positions of tRNA in the A site (Fig. 2). However, despite this overall similarity, two interactions were observed for the peptidyl-tRNA in the classical A site in the mammalian system that have not been previously reported in bacterial systems (Korostelev et al., 2006; Selmer et al., 2006; Voorhees et al., 2009; Yusupov et al., 2001). The first is an interaction between the tRNA acceptor stem and helix 89 (H89) of 28S rRNA within the large subunit; the second is an interaction between the tRNA anticodon stem loop in the region of positions 28/29 and the 40S subunit (Fig. 3A, B and Table S1, S2). The latter contact may involve the N-terminus of the eukaryotic-specific protein S30e.
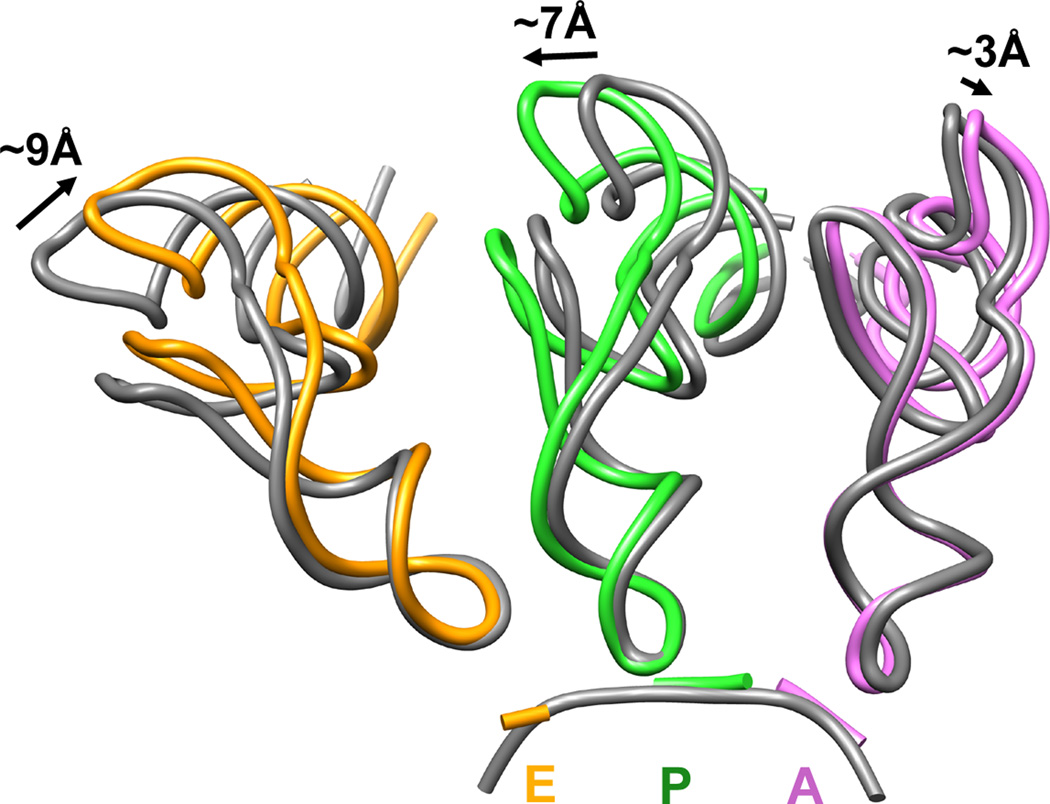
Figure 2. Analysis of classically positioned tRNAs observed in the classical-1 PRE complex from rabbit liver.
Deviation in the classical tRNA positions near the elbow regions of tRNA observed in the non-rotated 80S PRE complex populations (coloured ribbon) compared to those observed in T.thermophilus 70S structures after aligning the small subunits (Voorhees et al., 2009) (grey ribbon, pdb identifier 2WDG). Distances were measured between the phosphate backbone at position 56. Color code: A-site tRNA (pink), P-site tRNA (green), E-site tRNA (orange).
See also: Figure S3. tRNA in the classical A/A, P/P and E/E positions.
Table S1. tRNA – 40S contacts
Table S2. tRNA – 60S contacts.
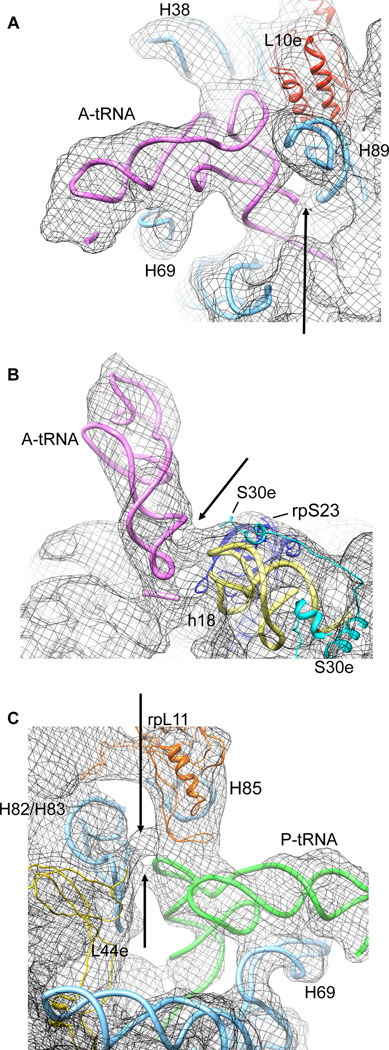
Figure 3. Eukaryotic-specific contacts between tRNAs and 80S ribosomes detected within the classical-1 80S PRE complex.
(A) Contact of the A-site tRNA acceptor stem with H89 of the 60S ribosomal subunit. (B) Contact of the A-site tRNA anticodon stem (positions 28/29) with the 40S ribosomal subunit. (C) Contacts between the T-loop of P-site tRNA and elements (H85, H82/83 and/or rpL44e) of the 60S ribosomal subunit. The models for rRNA helices and ribosomal proteins are derived from X-ray structures of the yeast 80S ribosome (Ben-Shem et al., 2010) for the 60S subunit (pdb identifier 3O58) and the 40S-eIF1 complex (Rabl et al., 2010) for the 40S subunit (pdb identifier 2XZN).
Interestingly, the P/P and E/E tRNA configurations in the bacterial and eukaryotic ribosome appear to deviate substantially relative to each other and to that of the A-site tRNA. These distinctions manifest most profoundly near the elbow regions of the L-shaped tRNA molecule (Fig. 2). Compared to the T. thermophilus 70S ribosome (Voorhees et al., 2009), the elbow region of the P-site tRNA is more distal from the A site, whereas the elbow region of E-site tRNA is closer to both the P and A sites. Correspondingly, the elbow domains of A-and P-site tRNAs are ~9Å further apart in the 80S particle than in the T. thermophilus ribosome, while P- and E-site tRNAs are ~14Å more proximal (Fig. 2). Disparities of this size are not found when X-ray structures of 70S complexes bearing classically configured tRNAs from T. thermophilus are compared to corresponding cryo-EM structures in the E. coli system (Valle et al., 2003; Voorhees et al., 2009, respectively). Thus, classical tRNA positions appear to possess a domain-specific character that likely reflects subtle changes in the nature of tRNA-ribosome interactions.
In addition to the conserved interactions of classically configured P-site tRNA (Tables S1 and S2), a tentative eukaryotic-specific interaction between the T-loop of P-site tRNA and the 60S subunit (Fig. 3C) may contribute to the observed positioning of the tRNA elbow. Despite its altered position, the classically configured E-site tRNA elbow remains in contact with the L1 stalk protuberance (Fig. 1A). Such interactions are likely maintained by an extended range of L1 stalk motions in the eukaryotic system afforded by the presence of an additional hinge element in the L1 stalk created by the presence of an asymmetric bulge within H76 of 28S rRNA (Ben-Shem et al., 2010; Spahn et al., 2004).
Dynamic behaviour of the A-site tRNA within the classically configured PRE complex
While classical-1 and classical-2 subpopulations of the mammalian PRE complex possess globally similar architectures and tRNA positions, closer inspection revealed distinctions in the contacts between A-site tRNA and the ribosome (Fig. 4). First, the point of contact between the D-stem of tRNA and H69, which is involved in a critical inter-subunit bridge, differs between classical-1 and -2 subpopulations (Fig. 4A, C). Second, contacts observed in the classical-1 subpopulation between positions 17-19/56 of the tRNA elbow and H38 of 28S rRNA, and the neighbouring positions 54, 55 of tRNA and rpL10e, are absent in the classical-2 subpopulation (Fig. 4B, D).
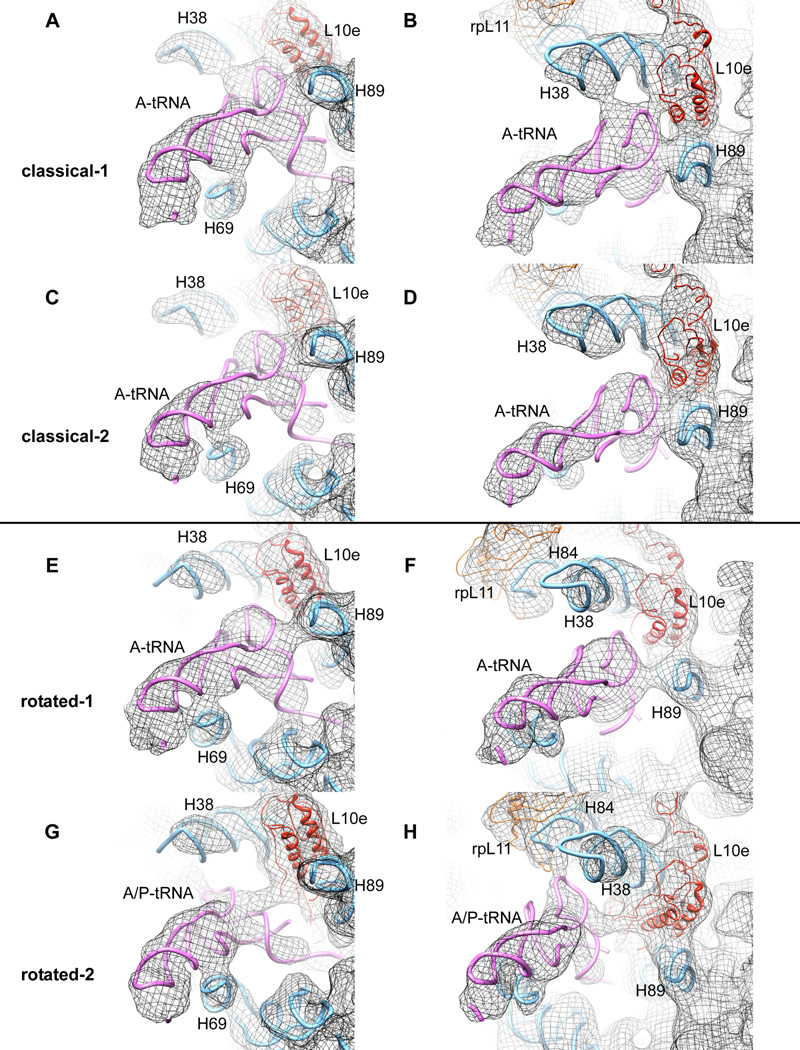
Figure 4. Comparison of A-site tRNA contacts with 60S ribosomal subunit for all four sub-populations described.
(from top to the bottom: A, B, classical-1; C, D, classical-2; E, F, rotated-1; G, H rotated-2). (A, C, E, G) Illustration of different contacts between A-site tRNA and H69. (B, D, F, H) Demonstration of the potentially dynamic interaction between the A-site tRNA elbow and components of the 60S subunit during the transition from classical-1 to rotated-2 intermediate states. Models for rRNA and separated proteins are derived from the X-ray structure of the yeast 80S ribosome ((Ben-Shem et al., 2010) PDB ID 3O58)
See alsoFigure S4. Distinct features of 80S ribosomes in the classical-1 and classical-2 subpopulations.
Movie S1. Spontaneous movement of tRNAs relative to the 60S subunit.
Movie S2. Spontaneous movement of tRNAs relative to the 40S subunit.
H38, also referred to as the A-site finger (ASF), forms the inter-subunit bridge B1a. Comparison of the ASF position in the two classical subpopulations reveals a conformational change of this important functional element. In the classical-1 subpopulation, the ASF contacts rpS15 (S19p), while in the classical-2 subpopulation, it contacts rpS18 (S13p) (Fig. S4A, B). This rearrangement likely contributes to a disruption of contacts between the ASF and the elbow domain of A-site tRNA in the classical-2 subpopulation. Conformational changes in this region are believed to be functionally relevant as similar rearrangements have been observed in the bacterial 70S ribosome upon EF-G binding (Valle et al., 2003) and ASF truncations have been shown to increase the rate of translocation (Komoda et al., 2006).
A ~2° rotation of the 40S subunit body centered on the long axis of h44 may also participate in the loss of contacts at the A/A tRNA elbow, as this leads to an opening of the A site (Fig. S4C). Thus, the rotation of the 40S subunit, the shift of the ASF and the loss of elbow contacts of the A/A tRNA in the classical-2 subpopulation appear to be interrelated. Interestingly, the A/A tRNA density is weaker in the classical-2 cryo-EM map. This finding may reflect increased structural fluctuations of the peptidyl-tRNA of the classical-2 subpopulation around the A/A position as a consequence of lost interactions with the 60S subunit.
Inter-subunit rotation occurs spontaneously within the mammalian PRE complex
Compared to the classical ribosome structures, the rotated-1 and rotated-2 subpopulations exhibit a pronounced inter-subunit rotation: in both structures the 40S subunit is rotated ~8° counter-clockwise relative to the large subunit. This movement resembles, in part, the ratchet-like subunit rearrangement (RSR) described for the 70S ribosome in the presence of EF-G (Connell et al., 2007; Frank and Agrawal, 2000; Valle et al., 2003). Notably however, the ratcheted ribosome conformation observed in the presence of EF-G/eEF2 entails two rotational movements of the small subunit domains that are roughly orthogonal to each other: a 5–8° rotation of the small subunit body/platform relative to the large subunit as observed in the rotated-1 and -2 subpopulations and a ~12–15° swivel-like rotation of the small subunit head domain (Connell et al., 2007; Schuwirth et al., 2005; Spahn et al., 2004; Spahn et al., 2001b; Zhang et al., 2009). The motion of the 40S body observed in the rotated-1 and rotated-2 subpopulations of the mammalian PRE complex is not accompanied by the 40S head rotation (Fig. S5). These observations suggest that the spontaneous inter-subunit rotation observed here is distinct from that observed when factors are bound. Accordingly, inter-subunit rotation is a spontaneous conformational mode in both bacterial and mammalian PRE complexes (Cornish et al., 2008; Marshal et al., 2008), while swivelling motions of the small subunit head may be driven, or stabilized, by EF-G/eEF2 binding (Ratje et al., 2010).
Peptidyl-tRNA adopts two distinct configurations within the rotated PRE complex
While P-site tRNA adopts indistinguishable P/E hybrid positions in both rotated-1 and -2 subpopulations, two distinct binding modes of A-site peptidyl-tRNA are observed within the rotated sub-states of the mammalian PRE complex. In the rotated-1 subpopulation, the A-site tRNA body occupies a classical-like (A/A) configuration. In contrast, the A-site tRNA elbow in the rotated-2 subpopulation is displaced by almost 18 Å towards the P site where it contacts the central protuberance (Fig. 4F, H). Thus, according to the elbow domain the peptidyl-tRNA in the rotated-2 subpopulation occupies a hybrid (A/P) position.
Cryo-EM investigations of the bacterial PRE complex have revealed only a single hybrid tRNA configuration where A- and P-site tRNAs were proposed to occupy hybrid A/P and P/E positions, respectively (Agirrezabala et al., 2008; Julian et al., 2008). Notably, the tRNA configurations observed in both structures most closely resemble those found in the rotated-1 complex. As described above, this state more closely approximates an A/A, P/E hybrid state as the A-site tRNA body appears classically configured. Because the 3’-CCA end of peptidyl-tRNA could not be resolved in either rotated-1 or rotated-2 subpopulations it cannot be presently distinguished whether the A-site tRNA position in the rotated-1 structure formally represents a hybrid (A/P) configuration as defined by localization of the 3’-CCA end of peptidyl-tRNA in the P site of the large subunit PTC. Thus, the position observed may represent a classical (A/A) configuration or a partially formed hybrid state (A/ap) wherein the anticodon and elbow domains occupy the classical A site, while the 3’-CCA-end occupies the P site. Indeed, both inter- and intra-subunit P-site tRNA hybrid configurations have been observed in the EF-G-bound ribosome (Ratje et al., 2010). Ambiguity is less likely in this regard in the rotated-2 structure due to the large displacement of the peptidyl-tRNA elbow towards the P site. In line with bulk and smFRET observations made on bacterial PRE complexes (Munro et al., 2007; Pan et al., 2007), these data suggest that A- and P-site tRNAs can move independently within the mammalian PRE complex leading to at least two distinct hybrid tRNA configurations distinguished by the position of the peptidyl-tRNA elbow. As both A- and P-site tRNA bodies have moved in the direction of translocation within the rotated-2 complex, this state likely represents a configuration transited during eEF2-mediated translocation. Indeed, similar states have been observed in the bacterial system in the context of a spontaneous retro-translocation (Fischer et al., 2010).
Conformational changes in both the ribosome and peptidyl-tRNA underpin formation of the A/P hybrid state
The distinct A-site tRNA positions observed in rotated-1 and -2 subpopulations is accompanied by a substantial remodelling of tRNA-large subunit interactions: (i) The A-site tRNA T-stem appears to change its interaction site from H89 in the rotated-1 subpopulation to ribosomal protein L10e in the rotated-2 structure (compare Fig. 4F and H), (ii) A/P hybrid state formation in the rotated-2 subpopulation results in a ~7Å movement of the D-stem relative to the tip of H69 in the direction of the P site (Fig. 4E, G) and (iii) the A-site tRNA elbow region achieves a new interaction that appears to include rpL11 (L5p) (compare Fig. 4F, H). In the classical-1 and -2 subpopulations, this protein directly contacts the classically configured P-site tRNA (Fig. 3C). As the elbow domain of peptidyl-tRNA does not fully reach the P site, this A/P hybrid state configuration may require alterations in the position of rpL11 (L5p) or the central protuberance.
Despite substantial displacement of the acceptor arm of peptidyl-tRNA towards the P site in the rotated-2 structure, the position and orientation of the anticodon stem-loop is similar to what is observed in the rotated-1 subpopulation (Fig. 5A). Thus, movements of the A-site tRNA elbow towards the P site appear to require flexion/bending of the tRNA body rather than a pivoting movement of a rigid body, implying that conformational changes within the tRNA contribute to hybrid state formation (Fig. 5B). In line with previous suggestions for the process of tRNA selection (Cochella and Green, 2005; Schmeing et al., 2009; Schuette et al., 2009; Villa et al., 2009) and translocation (Ratje et al., 2010; Dunkle et al., 2011), these observations reinforce the notion that tRNA plays an active role in the mechanisms of protein synthesis.
Figure 5. Comparison of tRNA positions in rotated-1 and rotated-2 sub-populations of 80S PRE complex.
(A) Superposition of 40S subunits separated from classical-1 (grey mesh) and rotated-2 (yellow solid) sub-populations tracks A/A and P/P tRNAs movements inside the inter-subunit cavity (inset shows the same scene with usage of crystallographic models derived from (Voorhees et al., 2009), PDB ID 2WDG) (B) Superposition of ASL regions of classical (A/A) tRNA (grey mesh and X-ray model) with hybrid (A/P) tRNA (pink, map and X-ray model) indicates a conformational difference between the two structures. A similar distortion (kink) is even more pronounced for P/P (grey mesh, X-ray model) and P/E (green solid map and X-ray model) tRNAs.
See alsoFigure S5. Rotation of the 40S subunit in the non-rotated and rotated subpopulations.
Intrinsic dynamics within the mammalian PRE complex
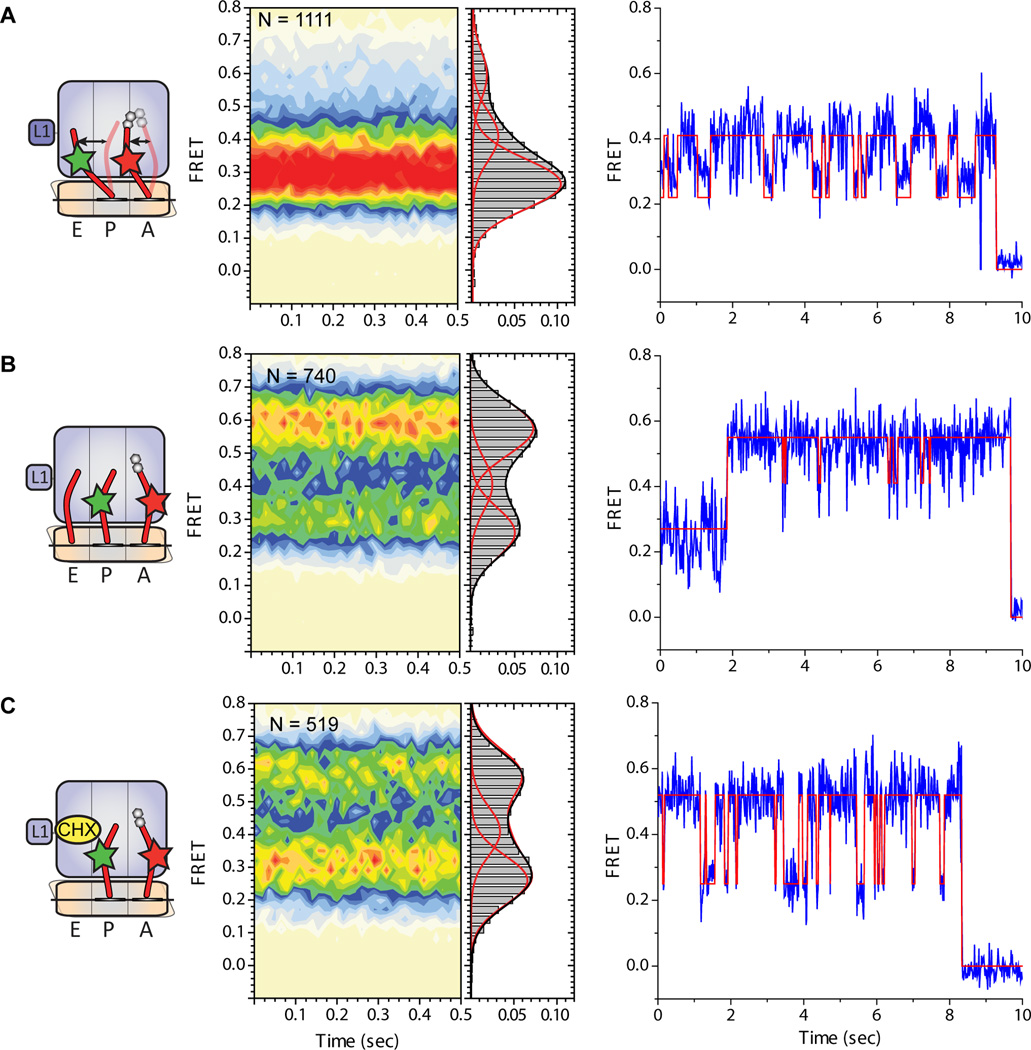
In order to investigate the hypothesis that A- and P-site tRNAs spontaneously exchange between classical and hybrid configurations within the mammalian 80S PRE complex, smFRET studies were performed in which dye-labeled E. coli tRNAs were incorporated into the 80S particle. Here, the P-site was non-enzymatically filled with Cy3-labeled NAcPhe-tRNAPhe and the A-site was enzymatically filled with Cy5-labeled Lys-tRNALys (Experimental Procedures). Surface-immobilized 80S PRE complexes were imaged at 15ms time resolution and data were analyzed using hidden Markov Modelling procedures analogous to those previously described (Munro et al. 2007). Using this approach, 80S PRE complexes were observed to predominantly occupy a broadly distributed, low-to-intermediate (~0.26–0.41) FRET configuration (Fig. 6A) corresponding to an inter-elbow, A- to P-site tRNA distance of ~50–70Å. By analogy to the bacterial ribosome system, where the FRET states have been rigorously defined through rRNA mutagenesis and imaging from multiple structural perspectives (Munro et al., 2007; Munro et al., 2010a; Munro et al., 2010b), as well as the structural features of rotated-1 and -2 subpopulations, such findings are consistent with P-site tRNA predominantly residing in a P/E hybrid configuration within the mammalian PRE complex.
Figure 6. tRNA dynamics on the mammalian PRE complex bearing fluorescently labelled tRNAs in the A and P sites.
(A) 80S PRE complexes containing deacylated Cy3-labeled tRNAPhe in the P site and Cy5-labeled NAc-Phe-Lys-tRNALys in the A site prepared as described in Experimental Methods. (B) 80S PRE complexes imaged in the presence of 5uM deacylated E.coli tRNAfMet in solution. (C) 80S PRE complexes imaged in the presence of 200uM cycloheximide in solution. For each panel, the nature of the complex investigated and the putative motions of tRNA between classical and hybrid positions is schematized (left panel) along with population histograms showing the distribution of FRET values observed (center panel) as well as representative single-molecule FRET trajectories for individual molecules from within each experiment overlaid by the idealization (red) obtained through hidden Markov modelling (Experimental Procedures).
See alsoFigure S6. Inclusion of deacylated tRNA in the imaging buffer increased E-site tRNA occupancy in a concentration dependent fashion.
Inspection of individual FRET trajectories further revealed that tRNAs dynamically exchange between at least three structurally and kinetically distinct configurations within the mammalian PRE complex. As expected from the population distribution of FRET states observed, such fluctuations predominantly occurred between low- (~0.26) and intermediate-(~0.41) FRET states (Fig. 6A). These data suggest that the principle dynamic processes in the system correspond to a ~10–15Å change in the intermolecular distance between the elbow domains of A- and P-site tRNA. By analogy to observations reported for the bacterial PRE complex (Munro et al., 2007), and the present finding that a P/E hybrid state is observed in both rotated-1 and rotated-2 subpopulations, such dynamics likely arise from the motions of the A-site tRNA elbow domain between classical and hybrid positions. Correspondingly, the conversion between low- and intermediate-FRET states may be interpreted as reflecting transitions between rotated-1 and rotated-2 subpopulations, respectively. This model is supported by both structural and kinetic information. First, the change in FRET observed between low- and intermediate FRET is consistent with the amplitude of peptidyl-tRNA motions into and out of the P site observed in rotated-1 to rotated-2 subpopulations. Second, consistent with bacterial PRE complex investigations (Munro et al., 2007), the lifetime of the intermediate-FRET state, wherein both A- and P-site tRNAs are in hybrid configurations, was observed to be the most transient in nature (approximately 200 ms vs. 650 ms for the low-FRET state).
Transient excursions to high-FRET (~0.59) configurations were also rarely observed in this system. As reflected in this state’s relatively low occupancy (~11% on average) (Fig. 6A), the high-FRET state lifetime was estimated at ~275 ms. Structurally, the mean value of the high-FRET state is consistent with the anticipated proximity of A- and P-site tRNAs in their classical positions (A/A; P/P) as observed in classical-1 and classical-2 subpopulations. Thus, the smFRET experiments are in good overall agreement with the multiparticle cryo-EM structures and suggest that the mammalian PRE complex undergoes spontaneous remodelling, observed in the smFRET experiment as a dynamic exchange between at least three distinct tRNA positions.
Multiparticle analysis revealed classical tRNA configurations predominate in the mammalian PRE complex (Fig. S1) whereas the smFRET data suggest a preference for hybrid states of tRNA binding. However, the E-site was found to be occupied with tRNA in both classical-1 and -2 sub-populations (Fig. 1), most likely due to the excess of deacylated tRNA over ribosomes used to efficiently fill the P-site when preparing the cryo-EM sample. Reasoning that the binding of deacylated tRNA to the E site may shift the equilibrium from a rotated, hybrid-tRNA configuration to one in which tRNAs were bound classically, smFRET experiments were performed in the presence of unlabeled, deacylated E. coli tRNAfMet (1–10µM). In line with this hypothesis, the assignment of FRET states made, and as anticipated from both classical-1 and -2 structures, tRNA inclusion in the imaging buffer increased high-FRET state occupancy in a concentration-dependent manner (Fig. S6; Fig. 6B). At the highest tRNA concentrations examined (10µM), ~60% of the PRE complexes occupied a relatively stable, high-FRET state (lifetime ~3 seconds). These data suggest that tRNA occupancy in the E site efficiently promotes both classical A- and P-site tRNA positions and a non-rotated configuration of the mammalian PRE complex. Residual dynamics between low- and high-FRET states observed even at the highest solution concentrations of tRNA, may either reflect dynamic processes within the classically configured ribosome containing A-, P- and E-site tRNAs and/or the kinetic features of E site tRNA binding.
To examine the functional significance of the observed dynamics within the mammalian PRE complex containing A- and P-site tRNAs, experiments were performed in the presence of the potent translocation inhibitor, cycloheximide. While the mechanism of cycloheximide action is not presently known, translation inhibition has been recently hypothesized to arise from its direct binding to the large subunit E site (Schneider-Poetsch et al., 2010). Inclusion of cycloheximide in the imaging buffer increased high-FRET state occupancy in a concentration-dependent manner, reaching ~45–50% high-FRET state occupancy at a concentration of 200µM (Fig. 6C). This demonstration of the metastable nature of the mammalian PRE complex suggests that cycloheximide binding specifically promotes classical configurations to inhibit translocation by directly interfering with P/E hybrid state formation. These observations are reminiscent of the action of unrelated aminoglycoside antibiotics on the bacterial PRE complex, where classical state stabilization was shown to directly correlate with translocation inhibition (Feldman et al., 2010).
A metastable energy landscape view of the mammalian 80S PRE complex
The combination of multiparticle cryo-EM at sub-nanometer resolution and smFRET imaging has provided important insights into the structural and dynamic features of a mammalian PRE complex. Clear evidence of two distinct hybrid states was observed in the mammalian PRE complex that appear to be differentiated by A-site tRNA motions. While these findings are globally consistent with the intrinsically dynamic nature of bacterial ribosomes, the relative stabilities of classical and hybrid states within the mammalian ribosome appear altered. The 70S PRE complex prefers classical tRNA configurations (~60% occupancy) (Munro et al., 2010a; Munro et al., 2010b), while 80S prefers hybrid tRNA configurations (~89% occupancy) (Fig. 6A). Identical conditions and the same (bacterial) tRNAs were used for smFRET imaging leaving the nature of the ribosome (bacterial or mammalian) as the differentiating variable in these experiments. The findings are consistent with the observation that the vacant eukaryotic ribosome favours a rotated 40S subunit conformation (Ben-Shem et al., 2010; Spahn et al., 2004). Accordingly, the energy landscape governing intrinsic motions within the mammalian PRE complex appears distinct from that of the bacterial PRE complex. Future investigations will be needed to further explore this comparison and to test the influence of the nature and identity of the tRNA.
When the four cryo-EM subpopulations are considered in the order classical-1, classical-2, rotated-1 and rotated-2, the respective A- and P-site tRNA positions lie along a plausible reaction coordinate for tRNA movements from classical A/A and P/P states to hybrid A/P and P/E states, respectively. Gradual movements, as well as a remarkable plasticity of tRNA elbow interactions with the 80S ribosome, are especially apparent for peptidyl-tRNA in the A site (Fig. 4). The first step in this trajectory would be a slight rotation of the 40S subunit around its long axis and a movement of the ASF (H38) of the 60S subunit from the classical-1 to classical-2 state. Both conformational changes in the ribosome contribute to an opening up of the A site, which looses interactions between the A-site tRNA elbow and the ASF. Increased mobility of the A-site tRNA elbow may facilitate a transition between the classical-2 and rotated-1 configurations, where interactions between the A-site tRNA and ASF are also absent. Small subunit rotation could then lead to P/E hybrid state formation and additional adjustments in the A-site tRNA elbow towards the P site. This set of movements would reflect a transition between classical-2 and rotated-1 configurations. Finally, formation of the A/P hybrid state, where the A-site tRNA elbow contacts rpL11 (L5p) within the P site, could then be achieved by a transition between rotated-1 and rotated-2 configurations.
Although additional intermediate states and alternative pathways may also exist, the proposed trajectory describes a partial translocation of the tRNAs from the perspective of the 60S subunit. As shown in Movie S1, the combined series of transitions results in tRNA 3’-CCA ends moving from the 60S A and P sites to the 60S P and E sites, respectively, as well as an approximately 10 Å motion of both tRNA anticodon stem-loops in the direction of translocation.
Notably, similar intermediate states have been recently observed for E. coli ribosomes during a back-translocation reaction (Fischer et al., 2010). However, the present observations were obtained in the absence of back-translocation or translocation. Indeed, from the perspective of the 40S subunit, significant movements of tRNA anticodon stem loops in the direction of translocation are not observed (Movie S2), because the 40S subunit and the tRNA anticodon stem loops move together relative to the 60S subunit. Thus, the large-scale fluctuations of tRNA positions observed do not lead to a conversion of the PRE to the POST complex. We therefore consider the classical-1, classical-2, rotated-1 and rotated-2 substates as intermediates along a reaction coordinate that precede substrate translocation. In line with emerging models of the translocation mechanism (Ratje et al., 2010; Schuwirth et al., 2005; Spahn et al., 2004; Zhang et al., 2009; Munro et al., 2010b), tRNA translocation on the small ribosomal subunit likely requires swivel-like motions of the head domain in the direction of substrate movement, which are not observed in any of the structures described. Although head swivel can occur spontaneously in the vacant bacterial 70S particle in the absence of a full 30S body rotation (Schuwirth et al., 2005), such motions have not been observed in multiparticle cryo-EM analysis of mammalian (this study) or bacterial PRE complexes (Agirrezabala et al., 2008; Ratje et al., 2010). These data lead to the speculation that spontaneous head swivelling in rotated PRE complexes carrying A- and P- site tRNAs either does not occur, or only rarely occurs, and that conformational events of this nature are strongly enhanced by EF-G/eEF2 binding.
Collectively, our data suggests that metastability is an evolutionarily conserved feature of the pre-translocation ribosome complex and that similar configurations of the system are transited along the translocation reaction coordinate. Nevertheless, significant energetic differences appear to exist between bacterial and mammalian systems that alter how tRNAs are positioned with respect to each other, and how they interact with the ribosome. Here, the observed propensity of the eukaryotic ribosome to adopt a rotated configuration, perhaps afforded by the existence of additional inter-subunit bridging contacts (Ben-Shem et al., 2010; Spahn et al., 2004), may play an important, differentiating role. Furthermore, such distinctions, along with local features of the ribosome’s functional centers, may be important, and perhaps underappreciated features, that contribute to the species specificities of antibiotic action. Further comparative studies will be needed to derive precise conclusions about the relative energies of functionally relevant states and the potential regulatory significance of these differences.
EXPERIMENTAL PROCEDURES
Cryo-EM of mammalian PRE complexes
Re-associated 80S ribosomes from rabbit liver, free of endogenous tRNAs and mRNAs, were prepared according to (Bommer et al., 1997) with slight modifications. The PRE-translocation and decoding complexes were prepared in polyamine buffer as described in (Budkevich et al., 2008). Briefly, 80S ribosomes were programmed with MFK-mRNA or MFVK-mRNA for PRE-translocation complex and decoding complex, respectively. Ribosomes contained deacylated tRNAPhe (or N-acetyl-Phe-tRNAPhe for decoding complex) in the P site and (or Val-tRNAVal for decoding complex) in the A site. The ribosome preparation used for the cryo-EM experiments had been tested before and found to be ~70% active (Budkevich et al., 2008). Complexes were imaged using an FEI Tecnai G2 Polara. Image processing was carried out using SPIDER (Frank et al., 1996). Multiparticle refinement was carried out as described previously (Loerke et al., 2010; Penczek et al., 2006; Ratje et al., 2010). Crystal structures of prokaryotic tRNAs (Voorhees et al., 2009) (PDB ID 2WDG) and X-ray structures of Saccharomyces cerevisiae 80S (Ben-Shem et al., 2010) (PDB IDs 3O58 and 3O2Z) and Tetrahymena thermophila 40S-eIF1 complex (Rabl et al., 2010) (PDB ID 2XZN) subunits were fit into the cryo-EM maps using Chimera (Pettersen et al., 2004). For further details see Supplemental Experimental Procedures.
smFRET Experiments
Ribosome complexes used for single-molecule imaging studies were prepared as previously described for analogous bacterial PRE complexes (Munro et al., 2007) with the exception that isolated eukaryotic ribosomal subunits were substituted in the reaction mixture and initiation factors were not utilized. Initiation 80S complexes with Cy3-labeled, NAc-Phe-tRNAPhe(Cy3-s4U8) in the P site were surface immobilized within passivated microfluidic reaction chambers, followed by enzymatic delivery of Cy5-labeled Lys-tRNALys(Cy5-acp3U47) to the A site in the presence of 1mM GTP.
Fluorescence traces were extracted from the recorded movies using an automated protocol implemented in Matlab. FRET was calculated according to the equation FRET = ICy5/(ICy3+ICy5). The underlying FRET values for each system were obtained by fitting the raw experimental data as shown in Figure 6 (center panels), where only the widths of each FRET state were constrained according to what is observed for DNA oligonucleotide standards imaged under identical imaging conditions (Geggier et al. JMB 2010). In so doing, the raw data was fit to three Gaussian distributions whose mean FRET values were 0.59, 0.41 and 0.26 (± 0.01). Correspondingly, individual FRET trajectories were idealized to a three-state hidden Markov kinetic model using a segmental k-means algorithm implemented in QuB as previously described (Munro et al. 2007). Based on these idealizations, the average lifetimes in the observed FRET states were determined by fitting dwell-time histograms to a single-exponential function. For further details see Supplemental Experimental Procedures.
Highlights.
The mammalian PRE complex undergoes large-scale dynamic processes
tRNAs fluctuate between classical and at least two distinct hybrid configurations
The mammalian PRE complex favours tRNA positioning in hybrid states
Cycloheximide inhibits translocation by favouring classical tRNA positions
Supplementary Material
Acknowledgments
The present work was supported by grants from the Deutsche Forschungsgemeinschaft DFG (SFB 740 TP A3 and TP Z1 to CMTS; 436 UKR 113/64/1-1 to TB,) the European Union and Senatsverwaltung für Wissenschaft, Forschung und Kultur Berlin (UltraStructureNetwork, Anwenderzentrum), US NIH Grant GM079238 (to SCB) and US NSF Grant MCB-0747230 (to SCB). The electron density maps of the four sub-populations of the mammalian PRE complex have been deposited in the 3D-EM database (European Molecular Biology Laboratory–European Bioinformatics Institute, Cambridge, UK) with the accession codes EMD-5326, 5237, 5238 and 5239.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agirrezabala X, Lei J, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen CB, Becker T, Blau M, Anand M, Halic M, Balar B, Mielke T, Boesen T, Pedersen JS, Spahn CM, et al. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature. 2006;443:663–668. doi: 10.1038/nature05126. [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci U S A. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommer U, Burkhardt N, Junemann R, Spahn CMT, Triana-Alonso F, Nierhaus KH. Ribosomes and polysomes. In: Graham J, Rickwood D, editors. Subcellular Fractionation: A practical Approach. Washington, DC: IRL Press; 1997. pp. 271–301. [Google Scholar]
- Budkevich TV, El'skaya AV, Nierhaus KH. Features of 80S mammalian ribosome and its subunits. Nucleic Acids Res. 2008;36:4736–4744. doi: 10.1093/nar/gkn424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramouli P, Topf M, Menetret JF, Eswar N, Cannone JJ, Gutell RR, Sali A, Akey CW. Structure of the mammalian 80S ribosome at 8.7 A resolution. Structure. 2008;16:535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell SR, Takemoto C, Wilson DN, Wang H, Murayama K, Terada T, Shirouzu M, Rost M, Schuler M, Giesebrecht J, et al. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell. 2007;25:751–764. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave R, Terry DS, Munro JB, Blanchard SC. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys J. 2009;96:2371–2381. doi: 10.1016/j.bpj.2008.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman MB, Terry DS, Altman RB, Blanchard SC. Aminoglycoside activity observed on single pre-translocation ribosome complexes. Nat Chem Biol. 2010;6:244. doi: 10.1038/nchembio0310-244c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. Journal of Structural Biology. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Frank J, Spahn CM. The ribosome and the mechanism of protein synthesis. Rep Prog Phys. 2006;69:1383–1417. [Google Scholar]
- Geggier P, Dave R, Feldman MB, Terry DS, Altman RB, Munro JB, Blanchard SC. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J. Mol. Biol. 2010;399:576–595. doi: 10.1016/j.jmb.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julian P, Konevega AL, Scheres SH, Lazaro M, Gil D, Wintermeyer W, Rodnina MV, Valle M. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci U S A. 2008;105:16924–16927. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HD, Puglisi JD, Chu S. Fluctuations of transfer RNAs between classical and hybrid states. Biophys J. 2007;93:3575–3582. doi: 10.1529/biophysj.107.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoda T, Sato NS, Phelps SS, Namba N, Joseph S, Suzuki T. The A-site finger in 23 S rRNA acts as a functional attenuator for translocation. J Biol Chem. 2006;281:32303–32309. doi: 10.1074/jbc.M607058200. [DOI] [PubMed] [Google Scholar]
- Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Loerke J, Giesebrecht J, Spahn CM. Multiparticle cryo-EM of ribosomes. Methods Enzymol. 2010;483:161–177. doi: 10.1016/S0076-6879(10)83008-3. [DOI] [PubMed] [Google Scholar]
- Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci U S A. 2008;105:15364–15369. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- Munro JB, Altman RB, O'Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Altman RB, Tung CS, Cate JH, Sanbonmatsu KY, Blanchard SC. Spontaneous formation of the unlocked state of the ribosome is a multistep process. Proc Natl Acad Sci U S A. 2010a;107:709–714. doi: 10.1073/pnas.0908597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Altman RB, Tung CS, Sanbonmatsu KY, Blanchard SC. A fast dynamic mode of the EF-G-bound ribosome. EMBO J. 2010b;29:770–781. doi: 10.1038/emboj.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Sanbonmatsu KY, Spahn CM, Blanchard SC. Navigating the ribosome's metastable energy landscape. Trends Biochem Sci. 2009;34:390–400. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penczek PA, Frank J, Spahn CM. A method of focused classification, based on the bootstrap 3D variance analysis, and its application to EF-G-dependent translocation. J Struct Biol. 2006;154:184–194. doi: 10.1016/j.jsb.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal Structure of the Eukaryotic 40S Ribosomal Subunit in Complex with Initiation Factor 1. Science. 2010 doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling-Bartetzko S, Bartetzko A, Nierhaus KH. Kinetic and thermodynamic parameters for tRNA binding to the ribosome and for the translocation reaction. Journal of Biological Chemistry. 1992;267:4703–4712. [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FVt, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuette JC, Murphy FVt, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 2009;28:755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Semenkov YP, Rodnina MV, Wintermeyer W. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat Struct Biol. 2000;7:1027–1031. doi: 10.1038/80938. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CMT, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S Ribosome from Saccharomyces cerevisiae - tRNA-Ribosome and Subunit-Subunit Interactions. Cell. 2001a;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- Spahn CMT, Blaha G, Agrawal RK, Penczek P, Grassucci RA, Trieber CA, Connell SR, Taylor DE, Nierhaus KH, Frank J. Localization of the tetrecycline resistance protein Tet(O) on the ribosome and the inhibition mechanism of tetracycline. MolCell. 2001b;7:1037–1045. doi: 10.1016/s1097-2765(01)00238-6. [DOI] [PubMed] [Google Scholar]
- Taylor DJ, Devkota B, Huang AD, Topf M, Narayanan E, Sali A, Harvey SC, Frank J. Comprehensive molecular structure of the eukaryotic ribosome. Structure. 2009;17:1591–1604. doi: 10.1016/j.str.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc Natl Acad Sci U S A. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.