Abstract
A trio of genome-wide association studies recently reported sequence variants at three loci to be significantly associated with schizophrenia. No sequence polymorphism had been unequivocally (P<5×10−8) associated with schizophrenia earlier. However, one variant, rs1344706[T], had come very close. This polymorphism, located in an intron of ZNF804A, was reported to associate with schizophrenia with a P-value of 1.6×10−7, and with psychosis (schizophrenia plus bipolar disorder) with a P-value of 1.0×10−8. In this study, using 5164 schizophrenia cases and 20 709 controls, we replicated the association with schizophrenia (odds ratio OR= 1.08, P= 0.0029) and, by adding bipolar disorder patients, we also confirmed the association with psychosis (added N= 609, OR= 1.09, P= 0.00065). Furthermore, as it has been proposed that variants such as rs1344706[T]—common and with low relative risk—may also serve to identify regions harboring less common, higher-risk susceptibility alleles, we searched ZNF804A for large copy number variants (CNVs) in 4235 psychosis patients, 1173 patients with other psychiatric disorders and 39 481 controls. We identified two CNVs including at least part of ZNF804A in psychosis patients and no ZNF804A CNVs in controls (P= 0.013 for association with psychosis). In addition, we found a ZNF804A CNV in an anxiety patient (P = 0.0016 for association with the larger set of psychiatric disorders).
Keywords: schizophrenia, bipolar disorder, ZNF804A, CNV, association
Introduction
Before the publication of recent genome-wide association (GWA) studies,1–3 the sequence variant having the strongest evidence for unconditional association with schizophrenia was rs1344706[T]. This variant was reported to be associated with schizophrenia with an odds ratio (OR) of 1.12 and a P-value of 1.6×10−7. Evidence for association was strengthened (OR = 1.12, P = 1.0×10−8) when a psychosis phenotype (schizophrenia plus bipolar disorder) was assessed.4 More recently, rs1344706 was shown to be associated with alterations in the functional connectivity of various regions of the brain.5 The single-nucleotide polymorphism (SNP) is located in an intron of ZNF804A, a gene encoding a protein predicted to be a transcription factor.
Current discussions of GWA studies have suggested that findings of common SNP association may be exploited by examining the region surrounding the initial variant for additional polymorphisms not tagged by the original variant.6–8 Of particular interest are low frequency (between 5 and 1%) and rare ( < 1%) susceptibility polymorphisms that may confer a higher risk than the originally described variant but are unlikely, because of their frequency, to be discovered by standard GWA study. An example of the successful use of this strategy is in nonalcoholic fatty liver disease where the finding of a common PNPLA3 susceptibility allele was followed by the discovery, using re-sequencing, of an excess of null sequence mutations in individuals with the highest hepatic fat levels as well as a protective allele that was rare in European Americans (about 0.3%) but common in African Americans (about 10%).9 To our knowledge, the discovery of rare copy number variants (CNVs) associated with disease in regions initially uncovered through common SNP susceptibility alleles has not been reported.
In this study, we confirmed the association of rs1344706[T] with schizophrenia and also corroborated the bolstering of the evidence when the phenotype was expanded to psychosis. In addition, we examined ZNF804A for large structural variants, and found CNVs in three patients with psychiatric disorders, but not in controls.
Materials and methods
Samples
Full information about each study group is presented in the Supplementary Methods. Individuals were diagnosed according to International Statistical Classification of Diseases and Related Health Problems 10th Revision (ICD-10) or Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) criteria. Because of variation in diagnostic criteria and protocols between the various centers, some phenotypic heterogeneity is expected. All individuals provided written, informed consent for participation and approval was obtained from the ethics committees at each location.
In the initial analysis of rs1344706 association with schizophrenia, genome-wide typed samples from seven European locations were included. After quality control, case/control samples from England (93/88), Finland/excluding Kuusamo (59/147), Finland/Kuusamo (123/50), Iceland (589/11 492), Italy (84/89), the Netherlands (693/3689) and Scotland (658/661) were included. With the exception of the samples from the Netherlands, these samples derived from the genome-wide data described in our primary GWA report.3 Additional samples from Bonn and Munich that were part of the earlier data set were not used here as they had been included in the initial ZNF804A study.4 Bipolar samples from Iceland (N= 404 after quality control) were also incorporated into the psychosis analysis.
In the follow-up study of rs1344706 association with schizophrenia, samples from nine locations (after quality control)—China (460 cases, 466 controls), Denmark/Aarhus (236 cases, 500 controls), Denmark/Copenhagen (513 cases, 1338 controls), Germany/Bonn (275 cases, 510 controls), Germany/Munich (178 cases, 320 controls), Hungary (264 cases, 223 controls), Norway (201 cases, 357 controls), Russia (483 cases, 487 controls) and Sweden (255 cases, 292 controls)—were included. The European samples in this set originated from ‘follow-up set 2’ of our primary GWA study,3 although some samples that were used in the original ZNF804A study4 or that lacked sufficient DNA for typing were excluded. In addition, 205 bipolar samples from Norway were part of the psychosis analysis.
The CNV portion of the study included schizophrenia case/control samples from 10 European locations (following quality control): Denmark (547/541), England (85/80), Finland/excluding Kuusamo (61/144), Finland/Kuusamo (129/49), Germany/Bonn (485/848), Germany/Munich (585/586), Iceland (603/35 995), Italy (84/86), the Netherlands (604/489) and Scotland (645/663). These samples corresponded to the genome-wide typed samples used in our initial SNP association study,3 supplemented by material from Denmark and the Netherlands. Finally, 407 bipolar and 1173 depression, anxiety and anxiety-related disorder samples from Iceland were examined.
Genotyping
Genome-wide genotyping for all samples from England, Finland, Iceland and Italy was carried out at deCODE Genetics using the Illumina HumanHap300 chip (San Diego, CA). For the samples from Germany/Munich and Scotland, approximately one-third of both cases and controls were genome-wide typed at deCODE using the Illumina HumanHap300 chip, whereas the remaining two-thirds of cases and controls were genome-wide typed at Duke University using the Illumina HumanHap300 (Germany) and Illumina HumanHap550 (Scotland) chips. For the samples from the Netherlands, 715 cases and 643 controls were typed at UCLA using the Illumina HumanHap550 chip, whereas 3334 additional controls were typed at deCODE using the Illumina HumanCNV370 chip. The samples from Germany/Bonn were genome-wide typed at the University of Bonn using the Illumina HumanHap550 chip. The samples from Denmark were typed at deCODE Genetics using the Illumina HumanHap610 chip. For all the chips, yield for the two markers used for the surrogate (rs12477914 and rs1366840) was at least 98% and, in the controls of each group, neither marker deviated significantly from Hardy–Weinberg equilibrium (P > 0.05). For both the markers used, when the same study group was typed on more than one chip, there was no significant difference in allele frequency between the chips in either cases or controls (P > 0.05).
Of the follow-up groups, the Norwegian sample was genotyped at the University of Oslo using the Affymetrix 6.0 chip (Santa Clara, CA, USA). The remaining groups were single-marker genotyped at deCODE Genetics using Centaurus assays (Nanogen, San Diego, CA, USA). Yield for rs1344706 was at least 94% in both cases and controls of all study groups and, in the controls of each group, rs1344706 did not deviate significantly from the Hardy–Weinberg equilibrium (P > 0.01).
Sample quality control
For the genome-wide typed samples, duplicate samples, samples with a sex determined by X-chromosome homozygosity different from their reported sex and samples determined to be of non-European ancestry either by running STRUCTURE10 using the HapMap CEU, YRI and CHB/JPT individuals as reference samples or by examination of identity by state were removed. In addition, low-yield ( < 98% for the Illumina chips and <95% for the Affymetrix chip) samples were removed for the SNP association part of the study, whereas samples with an excess of large CNVs ( > 10 CNVs of at least 10 SNPs) were removed for the CNV part of the study. For the single-marker typed samples, samples that, based on additional genotyping, were duplicates of other samples in the data set or were low yield ( < 60%) were removed.
Association analysis
Association analysis of rs1344706 was carried out using a likelihood procedure described previously.11 For the genome-wide typed samples, a surrogate for rs1344706 made up of a linear combination of two-marker haplotypes, defined using the HapMap CEU, was used. This method, which we have used earlier,12 is an extension of the two-marker haplotype tagging described in Pe’er et al.13 and is similar in spirit to the methods described in Nicolae14 and Zaitlen et al.15 Genomic control16 was used to correct for relatedness and potential population stratification in each genome-wide typed study group. With the exception of Iceland, genomic control factors were < 1.1; in Iceland, some related individuals were included in the analysis and genomic control factors were 1.19, 1.12 and 1.18 for the schizophrenia, bipolar and psychosis analyses, respectively. The study groups within the Illumina genome-wide typed and follow-up sets were combined using the Mantel–Haenszel model.17 The combined Illumina genome-wide typed and combined follow-up sets were joined using summary statistics. P-values were calculated by summing z-scores with each data set’s z-score multiplied by the inverse of that data set’s s.e. divided by the square root of the sum of the squared inverse s.e., and combined ORs were calculated by summing log ORs with each log OR weighted by the inverse of its variance. The CNV association analysis was carried out using exact Mantel–Haenzel tests.
CNV detection
PennCNV,18 a free, open-source tool, was used for copy number variation detection. The input data for the program are log R ratio, a normalized measure of the total signal intensity for the two alleles of the SNP, and B-allele frequency, a normalized measure of the allelic intensity ratio of the two alleles. Values of these quantities are derived with the help of control genotype clusters (HapMap samples), using Illumina BeadStudio software. A hidden Markov model is then used to make CNV calls based on the probability of a given copy state at the current marker as well as the probability of observing a copy state change from the previous marker to the current one. A built-in correction model for GC content19 is included. As the samples were genotyped on several different types of Illumina BeadArray chips (HumanHap300, Human-Hap300-duo, HumanHap550, HumanHap610), we analyzed them with a twofold approach: first, using the full complement of markers on the chip and second, using only a subset of markers, present on most of the chip types, to ensure similar resolution of markers covering the genome. In this study, only CNVs including at least ten consecutive markers in the region chr2:185,171,338—185,512,459 Mb NCBI Build 36 were considered.
Results
In an attempt to replicate the association of rs1344706[T] with schizophrenia, we used two data sets: (1) an initial set of Illumina genome-wide typed samples from Europe nonoverlapping with the samples of the original report4 (2299 cases, 16 216 controls) and (2) a set of follow-up samples from Europe and China also without an overlap with those of the initial study4 (2865 cases, 4493 controls). Because rs1344706 was not included on the Illumina HumanHap300 or HumanHap550 BeadChips, a surrogate, composed of a linear combination of haplotypes, was used in the analysis of the genome-wide data.
The OR for rs1344706[T] (or the surrogate) was 1.09 in the initial data set and 1.08 in the follow-up data set; P-values were 0.037 in the initial sample and 0.033 in the follow-up sample (Table 1). In the combination of the two data sets, the OR was 1.08 and the P-value was 0.0029 (Table 1). The average control frequency in the combined sample was 0.59, identical to that of the original report.4 We found no evidence of heterogeneity between the study groups (P = 0.66, Supplementary Table 1) and no indication that we should reject the multiplicative model for the full model (P = 0.95 for the initial sample, P = 0.79 for the follow-up sample).
Table 1.
Association of rs1344706[T] with schizophrenia
| Data set (N cases/N controls) | Control frequency |
OR
(95% CI) |
P-value |
|---|---|---|---|
| Initial set (2280/16 099) | 0.58 | 1.09 (1.01, 1.18) | 0.037 |
| Follow-up (2797/4407) | 0.59 | 1.08 (1.01, 1.16) | 0.033 |
| Initial set and follow-up (5077/20 506) | 0.59 | 1.08 (1.03, 1.14) | 0.0029 |
Abbreviations: CI, confidence interval; OR, odds ratio.
Control frequency is the average, unweighted allele frequency over the various study groups of each data set, whereas ORs and P-values were estimated using the Mantel–Haenszel model. Analysis for each genome-wide typed group was adjusted using genomic control (see Materials and methods).
We also examined the association of rs1344706[T] with psychosis by including bipolar samples from Iceland (N= 404) and from Norway (N= 205). The strength of the evidence for association increased (P = 0.00065 from P = 0.0029, see Table 2 for psychosis analysis and Supplementary Table 2 for bipolar-only analysis), consistent with the results of the initial report.4
Table 2.
Association of rs1344706[T] with psychosis
| Data set (N cases/N controls) | Control frequency |
OR
(95% CI) |
P-value |
|---|---|---|---|
| Initial set (2683/16 099) | 0.58 | 1.10 (1.02, 1.18) | 0.012 |
| Follow-up (2991/4407) | 0.59 | 1.08 (1.01, 1.16) | 0.020 |
| Initial set and follow-up (5674/20 506) | 0.59 | 1.09 (1.04, 1.14) | 0.00065 |
Abbreviations: CI, confidence interval; OR, odds ratio.
Control frequency is the average, unweighted allele frequency over the various study groups of each data set, whereas ORs and P-values were estimated using the Mantel–Haenszel model. Analysis for each genome-wide typed group was adjusted using genomic control (see Materials and methods).
To explore the suggestion that loci initially identified through GWA studies may also harbor independent susceptibility variants, perhaps rarer and of greater effect, we examined the region overlapping ZNF804A (chr2:185,171,338—185,512,459 in NCBI Build 36) for large (containing at least 10 consecutive SNPs) CNVs in 3828 schizophrenia patients, 407 bipolar disorder patients and 39 481 controls not known to have any psychiatric disorder (see Materials and methods). In addition, 1173 patients with other psychiatric disorders (945 anxiety or anxiety-related disorder patients and 662 depression cases, 434 of whom were also diagnosed with anxiety or anxiety-related disorders) were available.
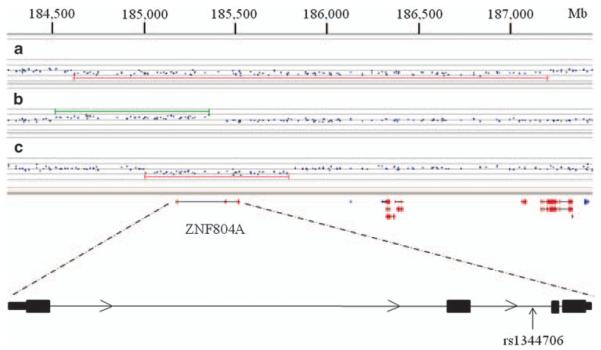
We carried out tests for association with schizophrenia and psychosis and also with the broader phenotype of combined psychiatric disorders. A schizophrenia patient harbored a deletion of the entire ZNF804A gene (Figure 1), but no ZNF804A structural variants were identified in the control set, leading to a P-value of 0.49 for association with schizophrenia. A bipolar disorder patient had a duplication of the first exon of the gene (Figure 1), resulting in a P-value of 0.013 for association with psychosis. A ZNF804A deletion was also identified in a patient with anxiety (Figure 1), leading to a P-value of 0.0016 for association with the larger set of psychiatric disorders.
Figure 1.
Large structural variants in ZNF804A.(a) A deletion in an Icelandic anxiety patient (patient 3), (b) a duplication in an Icelandic bipolar patient (patient 2), (c) a deletion in a Scottish schizophrenia patient (patient 1). The coordinates are based on NCBI Build 36.
Carriers of ZNF804A CNVs were of both sexes and were generally unremarkable with respect to age of onset and other features of their disorder (Supplementary Table 3). For the duplication carrier, genotypes from the patient’s father and from a sibling of the patient’s mother were available. Because the mother’s sibling carried the haplotype background the duplication was located on, but not the duplication, it could be inferred that the duplication event had most likely taken place either in the germ line of the patient’s mother or during gametogenesis in one of the maternal grandparents. For the deletion carriers, genotypes were not available for either the parents or the parents’ relatives; thus, it was not possible to estimate the timing of the mutation.
Discussion
In the first part of this study, we confirmed the association of rs1344706[T] with schizophrenia (P = 0.0029), using genotypes from 5164 schizophrenia cases and 20 709 controls. Although, in this study, only a modest number of bipolar disorder cases was available (N= 609), adding these cases to the analysis increased the evidence for association, in line with the original report. Thus, rs1344706[T] may confer risk of both schizophrenia and bipolar disorder, a finding consistent with epidemiological studies suggesting shared genetic risk factors for the two diseases.20,21
Meta-analysis P-values for rs1344706[T], based on the results from this study and the original report, were 2.8×10−9 (OR= 1.10) for schizophrenia and 3.8×10−11 (OR= 1.11) for psychosis, making association with both of these phenotypes unequivocal. In addition, three schizophrenia GWA studies1,2,22 recently reported association results for rs1344706[T], although the samples used by each study at least partially overlap the material included here or in the original study. The International Schizophrenia Consortium (ISC) achieved a one-tailed P-value of 0.029 (OR= 1.08) based on 2519 cases and 2110 controls, of which about 650 cases and 650 controls were included in this study.1 The Molecular Genetics of Schizophrenia (MGS) consortium reported a two-tailed P-value of 0.026 (OR= 1.10) using about 5300 samples, of which ~1900 were part of the original ZNF804A study.2 A third GWA study gave results for a rs1344706 surrogate;22 however, the Aberdeen samples used in that study are entirely incorporated into this study. In addition, while this work was under review, a replication study using the Irish Case–Control Study of Schizophrenia (ICCSS) sample reported a one-tailed P-value of 0.011 (OR= 1.20) based on a unique group of 993 cases and 570 controls.23 Taken together, the ISC, MGS and ICCSS reports are consistent with an association between rs1344706[T] and schizophrenia, and they also provide additional support for that conclusion.
In the second part of the study reported in this paper, we examined the ZNF804A region for gain or loss of copy number, using 4235 psychosis patients, 1173 patients with other psychiatric disorders and 39 481 controls. Two psychosis patients harbored CNVs affecting at least part of ZNF804A, resulting in a P-value of 0.013 for association with psychosis. In addition, a patient with anxiety had a deletion, leading to a P-value of 0.0016 for association with psychiatric disorders.
The CNV portion of this study involved a large data set, but additional information about CNVs is also publicly available. In approximately 12 000 controls from studies included in the Database of Genomic Variants24 and three other reports,25–27 two CNVs involving ZNF804A exons, both from the same study,28 are found. One event is a deletion of the entire gene in an African-American parent–offspring pair, and the second is a deletion affecting the 3′ exons of the gene in a European-American child. Although these CNV carriers are classified as healthy controls, the children, aged 0–18 years, may subsequently develop psychiatric disorders, and the parent, though free of major medical problems, does not seem to have been extensively interviewed for psychiatric disorders. In about 3800 schizophrenia cases not overlapping with those in this report25,27,29,30 and about 1000 bipolar cases,26 no additional ZNF804A events are found (note that the ZNF804A deletion observed in the ISC data set1 is from an individual also included in this study), but in approximately 1700 autism families and 400 unrelated autism cases,31–36 there is a duplication of the entire gene in two affected siblings and a partial ZNF804A duplication in another patient. Because of the incompleteness of the available phenotypic information, drawing strong conclusions about the association of ZNF804A CNVs and mental disorders from the publicly accessible data is difficult. However, considered together with the association between ZNF804A CNVs and psychiatric disorders observed in this report, the existence of two ZNF804A deletion events in controls, several young, and all without detailed phenotype information, and the presence of two ZNF804A duplication events in autism patients, suggests that the link between ZNF804A CNVs and mental disorders should be investigated further.
This is, to our knowledge, the first report of the identification of rare, disease-associated CNVs in a region initially discovered through common SNP association. Genes implicated in Mendelian disorders, however, have frequently been found to display a broad spectrum of mutations. In the cystic fibrous gene, CFTR, more than 1600 mutations have been identified (Cystic Fibrosis Mutation Database; http://www.genet.sickkids.on.ca/cftr/app), including large structural events. Rare, risk-conferring sequence mutations have also been identified in genes such as PNPLA3 that were initially uncovered through common SNP association.9 Similarly, some genes such as TCF2 first connected to disease through rare, highly penetrant risk alleles were later found, through GWA studies, to harbor common susceptibility alleles of modest effect.37 At CNTNAP2, the earliest reports associated rare SNPs with epilepsy and autism;38 more recently, evidence of the association of common SNPs with autism, although not at the genome-wide significant level, has been described,39,40 and rare structural variants in individuals with autism have also been identified.41–43
Association with disease is more difficult to establish for very rare genetic variants. In this study, we pooled deletions and duplications, considering them together. This approach allowed us to present statistical evidence for the connection of ZNF804A CNVs to psychiatric disorders. In addition, the large-scale nature of the structural alterations reported here makes phenotypic effects more likely, compared with most sequence changes. Both the deletions identified here remove the entire sequence of ZNF804A (Figure 1), which is likely to result in differences in mRNA abundance that may have downstream consequences, especially because ZNF804A is a putative transcription factor, a class of protein that has often been implicated in haploinsufficiency disorders.44 The duplication that includes one or two exons of ZNF804A (Figure 1) may lead to a protein that acts in a dominant-negative manner, interfering with the actions of the wild-type ZNF804A, or, because of the manner in which the duplicated sequence is inserted, the event may result in alterations in the transcription of the original copy of ZNF804A.
Mouse models may be useful in helping to understand the functional consequences of these variants. The reverse may also be true: these rare structural variants may turn out to be more valuable than the originally identified common variant in establishing a link between ZNF804A and a psychosis-related phenotype in an animal model. This is because gene knockout or overexpressing mice can be engineered relatively easily, the penetrance of pathogenic CNVs is generally much higher than common SNP variants, and temporal or cell specific manipulation of expression can be carried out with conditional mutants.
The ZNF804A variants examined in this study confer risk of more than one category of disease. Rs1344706[T] is associated with both schizophrenia and bipolar disorder, and the CNVs identified here suggest a connection with anxiety as well, although we do not find evidence for the association of rs1344706[T] with anxiety (data not shown). The functional connectivity alterations identified in the brains of rs1344706[T] carriers5 provide a possible mechanism for the link between ZNF804A variants and a diversity of disorders. Thus, examination of brain functional connectivity in ZNF804A CNV carriers would be of interest.
In this study, we have replicated the previously reported association of rs1344706[T] with schizophrenia and psychosis. In addition, we have further explored the locus and have identified rare CNVs in patients with psychiatric disorders, but not in controls.
Supplementary Material
Acknowledgments
We thank the subjects, their families and the recruitment center staff. This work was supported by the European Union (LSHM-CT-2006-037761 (Project SGENE), PIAP-GA-2008-218251 (Project PsychGene) and HEALTH-F2-2009-223423 (Project PsychCNVs)), the National Genomic Network (NGFN-2) of the German Federal Ministry of Education and Research (BMBF), the National Institute of Mental Health (R01 MH078075), the Center of Excellence for Complex Disease Genetics of the Academy of Finland (Grants 213506, 129680) and the Biocentrum Helsinki Foundation and Research Program for Molecular Medicine, Faculty of Medicine, University of Helsinki.
Appendix
Members of Genetic Risk and Outcome in Psychosis (GROUP) are as follows:
René S Kahn, MD, PhD; Wiepke Cahn, MD PhD; Department of Psychiatry, Rudolf Magnus Institute of Neuroscience, University Medical Center Utrecht, Postbus 85060, Utrecht, The Netherlands.
Don H Linszen, MD, PhD; Lieuwe de Haan, MD PhD; Academic Medical Centre University of Amsterdam, Department of Psychiatry, Amsterdam, NL326 Groot-Amsterdam, The Netherlands.
Jim van Os, MD, PhD; Lydia Krabbendam, MD PhD; Inez Myin-Germeys, MD PhD; Maastricht University Medical Centre, South Limburg Mental Health Research and Teaching Network, 6229 HX Maastricht, The Netherlands.
Durk Wiersma, MD, PhD; Richard Bruggeman, MD PhD; University Medical Center Groningen, Department of Psychiatry, University of Groningen, PO Box 30.001, 9700 RB Groningen, The Netherlands.
Footnotes
Conflict of interest Some of the authors, including Kari Stefansson (CEO of deCODE Genetics) and Augustine Kong (VP Statistics of deCODE Genetics), are shareholders in deCODE Genetics.
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
References
- 1.International Schizophrenia Consortium. Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–748. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 5.Esslinger C, Walter H, Kirsch P, Erk S, Schnell K, Arnold C, et al. Neural mechanisms of a genome-wide supported psychosis variant. Science. 2009;324:605. doi: 10.1126/science.1167768. [DOI] [PubMed] [Google Scholar]
- 6.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- 8.McCarthy MI, Hirschhorn JN. Genome-wide association studies: potential next steps on a genetic journey. Hum Mol Genet. 2008;17(R2):R156–R165. doi: 10.1093/hmg/ddn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gretarsdottir S, Thorleifsson G, Reynisdottir ST, Manolescu A, Jonsdottir S, Jonsdottir T, et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat Genet. 2003;35:131–138. doi: 10.1038/ng1245. [DOI] [PubMed] [Google Scholar]
- 12.Rafnar T, Sulem P, Stacey SN, Geller F, Gudmundsson J, Sigurdsson A, et al. Sequence variants at the TERT-CLPTM1 L locus associate with many cancer types. Nat Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pe’er I, de Bakker PI, Maller J, Yelensky R, Altshuler D, Daly MJ. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nat Genet. 2006;38:663–667. doi: 10.1038/ng1816. [DOI] [PubMed] [Google Scholar]
- 14.Nicolae DL. Testing untyped alleles (TUNA)-applications to genome-wide association studies. Genet Epidemiol. 2006;30:718–727. doi: 10.1002/gepi.20182. [DOI] [PubMed] [Google Scholar]
- 15.Zaitlen N, Kang HM, Eskin E, Halperin E. Leveraging the HapMap correlation structure in association studies. Am J Hum Genet. 2007;80:683–691. doi: 10.1086/513109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 17.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 18.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diskin SJ, Li M, Hou C, Yang S, Glessner J, Hakonarson H, et al. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res. 2008;36:e126. doi: 10.1093/nar/gkn556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cardno AG, Rijsdijk FV, Sham PC, Murray RM, McGuffin P. A twin study of genetic relationships between psychotic symptoms. Am J Psychiatry. 2002;159:539–545. doi: 10.1176/appi.ajp.159.4.539. [DOI] [PubMed] [Google Scholar]
- 21.Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Need AC, Ge D, Weale ME, Maia J, Feng S, Heinzen EL, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riley B, Thiselton D, Maher BS, Bigdeli T, Wormley B, McMichael GO, et al. Replication of association between schizophrenia and ZNF804A in the Irish Case-Control Study of Schizophrenia sample. Mol Psychiatry. 2010;15:29–37. doi: 10.1038/mp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 25.International Schizophrenia Consortium Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D, Cheng L, Qian Y, Alliey-Rodriguez N, Kelsoe JR, Greenwood T, et al. Singleton deletions throughout the genome increase risk of bipolar disorder. Mol Psychiatry. 2009;14:376–380. doi: 10.1038/mp.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaikh TH, Gai X, Perin JC, Glessner JT, Xie H, Murphy K, et al. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 30.Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 31.Christian SL, Brune CW, Sudi J, Kumar RA, Liu S, Karamohamed S, et al. Novel submicroscopic chromosomal abnormalities detected in autism spectrum disorder. Biol Psychiatry. 2008;63:1111–1117. doi: 10.1016/j.biopsych.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 36.Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 38.Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 39.Alarcon M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bakkaloglu B, O’Roak BJ, Louvi A, Gupta AR, Abelson JF, Morgan TM, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poot M, Beyer V, Schwaab I, Damatova N, Van’t Slot R, Prothero J, et al. Disruption of CNTNAP2 and additional structural genome changes in a boy with speech delay and autism spectrum disorder. Neurogenetics. 2009 doi: 10.1007/s10048-009-0205-1. e-pub ahead of print. [DOI] [PubMed] [Google Scholar]
- 43.Rossi E, Verri AP, Patricelli MG, Destefani V, Ricca I, Vetro A, et al. A 12Mb deletion at 7q33-q35 associated with autism spectrum disorders and primary amenorrhea. Eur J Med Genet. 2008;51:631–638. doi: 10.1016/j.ejmg.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 44.Seidman JG, Seidman C. Transcription factor haploinsufficiency: when half a loaf is not enough. J Clin Invest. 2002;109:451–455. doi: 10.1172/JCI15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.