Abstract
The phosphatidylinositide 3-kinase (PI3K) pathway is very commonly activated in a wide range of human cancers and is a major driving force in oncogenesis. One of the class I lipid kinase members of the PI3K family, p110α, is probably the most commonly mutated kinase in the human genome. Alongside genetic, molecular biological and biochemical studies, chemical inhibitors have been extremely helpful tools in understanding the role of PI3K enzymes in signal transduction and downstream physiological and pathological processes, and also in validating PI3Ks as therapeutic targets. Although they have been valuable in the past, the early and still frequently employed inhibitors, wortmannin and LY294002, have significant limitations as chemical tools. Here, we discuss the case history of the discovery and properties of an increasingly used chemical probe, the pan-class I PI3K and mTOR inhibitor PI-103 (a pyridofuropyrimidine) and its very recent evolution into the thienopyrimidine drug GDC-0941 that exhibits excellent oral anticancer activity in preclinical models and is now undergoing Phase I clinical trials in cancer patients. We also illustrate the impact of structural biology on the design of PI3K inhibitors and on the interpretation of their effects. The challenges and outlook for drugging the PI3 kinome are discussed in the more general context of the role of structural biology and chemical biology in innovative drug discovery.
Introduction: Points of activation and intervention
Phosphatidylinositide 3-kinases (PI3Ks) are critical elements in a signal transduction pathway that plays a key role in regulating many features of cell behaviour, including growth, survival, metabolism and various specialized functions. They belong to a family of lipid kinases that phosphorylate the 3′-hydroxy position of the inositol ring of phosphatidylinositides, yielding products of which the best characterized is phosphatidylinositol-3,4,5-trisphosphate (PIP3), the second messenger that recruits protein kinase B (AKT) to the cell membrane (1,2). PIP3 is generated by the class I PI3Ks, which comprise p110α, p110β and p110δ (class IA) and p110γ (class 1B), proteins that are activated to varying extents by receptor tyrosine kinases and G-protein coupled receptors. In addition to the class I PI3Ks, of note in the present context are the class II and III lipid kinases and also the class IV PI3K-related protein kinases (PIKKs), including mTOR, which is downstream on the PI3K pathway, and DNA-PK, ATM and ATR that are pivotal in DNA repair (1,3).
In terms of its importance in cancer, the PI3K signalling cascade is more appropriately referred to as a super-highway than a pathway. It is hijacked in multiple ways in many types of human malignancy (4,5). PIK3CA, which encodes the p110α catalytic subunit of PI3K, is probably the most commonly mutated kinase in the human genome (15% of all cancers) and is also amplified in some tumors, while PTEN, which encodes the opposing phosphatase to PI3K, is the second most commonly affected tumor suppressor gene after p53 (http://www.sanger.ac.uk/genetics/CGP/cosmic/; refs 6 and 7). Activation of PI3K signalling in cancer also occurs at the level of mutated or overexpressed receptor tyrosine kinases, AKT and RAS (4,5). The frequent genetic and epigenetic activation by a range of different molecular mechanisms strongly suggests that activation of the PI3K pathway is very likely to be a critical step in human oncogenesis. The overwhelming degree of genetic validation for PI3K signaling as a therapeutic target in cancer is supported by multiple lines of functional credentialing, including genetically engineered mouse models (5,8).
The optimal point of therapeutic intervention in the PI3K pathway remains unclear and will likely depend on the particular molecular pathology driving a given cancer (4, 5). Furthermore, emerging evidence shows that various abnormalities in the pathway can have different effects (4, 5). Nevertheless, all class I PI3Ks are compelling targets for therapeutic intervention because p110α is mutated and amplified in cancer and all four isoforms can generate PIP3 and are oncogenic in model systems (9). On the other hand, recent data indicate that the preferred class I PI3K target may be dependent on the molecular context – for example, mutation of p110α, loss of PTEN or overexpression of p110α, p110β or p110δ – thus fuelling the ongoing debate on the optimal selectivity profile of PI3K drugs for cancer treatment (5, 9). This is a point to which we will return later and upon which PI3K inhibitors are shedding invaluable light.
The emergence of chemical tools
Following on from the success with protein kinase inhibitors in cancer treatment (10) and in the general context of “drugging the cancer genome” (11), the subsequent therapeutic targeting of PI3K enzymes has been termed “drugging the PI3 kinome” (12).
Alongside genetic, molecular biological and biochemical studies, chemical inhibitors have been enormously helpful as tools in PI3K research (13-16). They have been used to help understand the role of PI3K enzymes in signal transduction and downstream physiological and pathological processes, as well as to assist validation of PI3Ks preclinically as therapeutic targets (13). The earliest and still widely employed inhibitors were wortmannin and LY294002 (Figure 1). Wortmannin is a fungal natural product originally identified in 1987 as a potent inhibitor of the respiratory burst in neutrophils and monocytes (17). It was subsequently found to inhibit PI3K (18) by covalent attack on the ATP site Lys 802 (human p110α numbering; ref 19). The synthetic flavone LY294002, based on the natural product, broad spectrum protein kinase inhibitor quercetin, was first reported as an inhibitor of PI3K in 1994 (20), at a time when relatively few selective inhibitors of any kinase had been discovered (21). Although these two agents have been extremely useful and are still widely employed as research probes (eg refs 13-15), they both have very significant limitations. Wortmannin is a reactive electrophilic and therefore unstable compound while LY294002 is a weak inhibitor with only micromolar potency, and both agents display significant off-target effects on other kinases (9-13, 15). Because of those properties, they are not ideal as probes for specific PI3Ks. Nevertheless, important proof of concept studies with these chemical tools and their analogs in animal models, together with the emerging biology of PI3Ks and their role in a range of disease pathologies – including not only cancer but also inflammation, autoimmune and cardiovascular disease – has led to an explosion of interest in the discovery of PI3K inhibitors (reviewed in refs 9, 18-21).
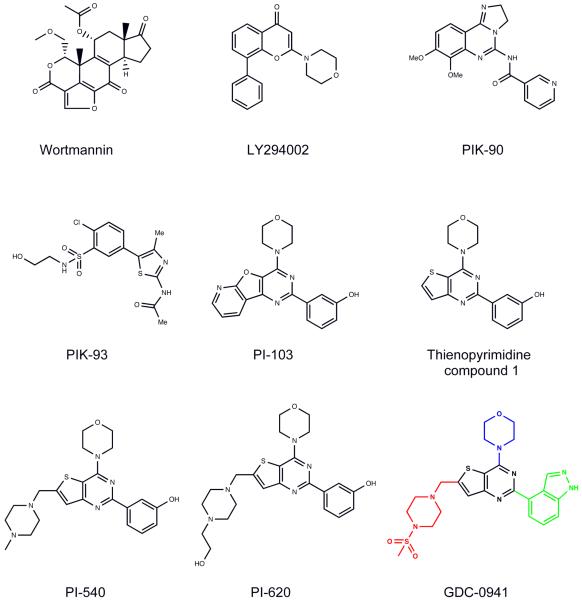
Figure 1.
Chemical structures of the PI3K inhibitors wortmannin, LY294002, PIK-90, PIK-93, PI-103, thienopyrimidine compound 1, PI-540, PI-620 and GDC-0941. In the chemical structure of GDC-0941 the indazole moiety is highlighted in green, the sulphonylpiperazine in red and the morpholino group in blue, while the thienopyrimidine core is shown in black.
Early drug discovery efforts used wortmannin and LY294002 as chemical starting points for inhibitor design and sought to overcome their respective drawbacks in potency, selectivity and pharmaceutical properties (22-25). The inspiration that these early inhibitors provided is reflected in the fact that among the privileged chemical scaffolds commonly found in PI3K inhibitors, the most frequent is the aryl morpholine present in LY294002 (Figure 1). The oxygen atom of the morpholine moiety forms a key hydrogen bond interaction with the hinge region of the kinase, as well as making additional hydrophobic interactions in the ATP pocket (ref 26 and see later). In addition to inhibitors based on wortmannin and LY294002, high-throughput screening (HTS) of large diverse compound collections coupled to medicinal chemistry efforts, frequently supported by structural biology studies, has led to over fifteen chemical classes of PI3K inhibitors (13, 22-25) and has considerably enriched the available chemical equity.
The discovery of PI-103: An improved chemical tool
In 2006-7, Hayakawa et al published a series of four papers (27-30) that described the use of HTS, based on a scintillation proximity assay, to discover a range of chemically interesting PI3K hits with micromolar inhibitory potency that were then optimised in the same work by medicinal chemistry to yield single digit nanomolar leads (Figure 2). One of those lead compounds was the pyridofuropyrimidine PI-103 (Figures 1 and 2; ref 29) which in the initial and later work was shown to exhibit IC50 values of 2-3nM (at 1μM ATP) on the class IA α, β, and δ isoforms of p110 PI3K and a slightly higher though still very potent IC50 of 15nM on the class IB isoform p110γ (29, 31-33). Antitumor activity was initially reported against four human cancer cell lines in vitro and also against HeLa human cervical cancer xenografts in athymic mice (29).
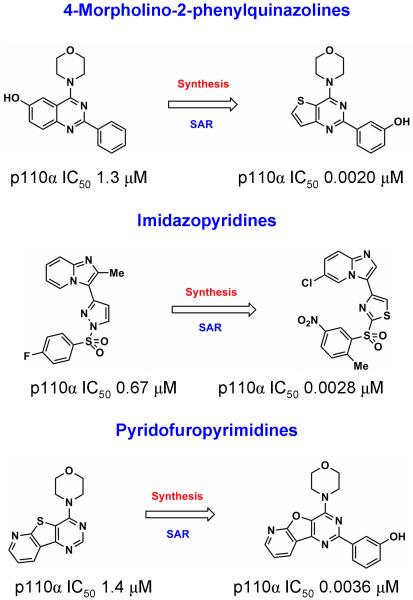
Figure 2.
Three PI3K inhibitory hit compounds identified by high-throughput screening against p110α and their optimization into nanomolar leads. From top to bottom, the 4-morpholino-2-phenylquinazolines optimised to the thienopyrimidine compound 1, the imidazopyridines, and the pyridofurapyrimidines resulting in PI-103. For further details see references 22-26.
Following the publication of the design, synthesis and structure-activity relationships for PI-103 (29), the same collaborative team provided a detailed pharmacological characterisation and mechanistic evaluation of the new agent (33). PI-103 exhibited advantages over wortmannin and LY294002, with excellent potency and selectivity for class I PI3Ks versus a panel of 70 other kinases, including class II and III PI3Ks and numerous representative protein kinases, as well as having improved stability. However, it was recognised that this agent still had some liabilities with respect to its use in animal models, specifically limited aqueous solubility and extensive metabolism (33). Nevertheless, the compound gave sufficient pharmacokinetic exposure in plasma and tumor tissue of treated mice to allow the demonstration of proof of concept for therapeutic activity against eight human tumor xenograft models representing various cancer types, including several with PI3KCA mutations and mutated or silenced PTEN – which are thus more likely to be “addicted” to the PI3K pathway (33). PI-103 showed evidence of effects on invasion, angiogenesis and metastasis, as well as direct antiproliferative activity, mainly resulting from a G1 arrest and involving reduced expression of cyclin D1 and increased p27 (33,34).
The antitumor activity of PI-103 was associated with effects on pharmacodynamic biomarkers that were fully consistent with the therapeutic mechanism being PI3K inhibition, including reduced phosphorylation of AKT and other pathway substrates; PI-103 also induced a gene expression profile consistent with PI3K pathway modulation (33-34).
PI-103 was included, along with a diverse array of other cell permeable, relatively drug-like PI3K inhibitors selected from the patent literature, in an innovative chemical biology study that revealed fascinating cryptic homologies across PI3K targets and chemotypes and demonstrated clear trends in selectivity across the class I PI3K and PIKKs that were not predictable from the amino acid sequence (35). In one important exemplification of the approach, a matrix of PI3K-inhibitory chemical tools – including PI-103 and other, more p110α-specific inhibitors – was used to identify p110α as the critical PI3K acting downstream of the insulin receptor (35), a result that was consistent with the mouse knock-in studies (36). These findings emphasized the concern at that time that p110α-specific PI3K inhibitors could potentially cause insulin resistance and associated diabetogenic side-effects.
A PubMed search (http://www.ncbi.nlm.nih.gov/sites/entrez; 1st Decemberr 2009) revealed that since its initial discovery (29, 31-33), PI-103 has been employed as a chemical tool in at least 30 published studies. In one of the earliest of these (37), PI-103 was shown to have potent activity against human glioma models, also seen in other reports (31-34). Through comparison with a small array of PI3K inhibitors with differing PI3K and PIKK selectivities, this was linked to the combinatorial inhibition of p110α and mTOR by PI-103 (37). Indeed the gene expression profile induced by PI-103 in glioma cells shared common elements with changes caused by the mTOR inhibitor rapamycin, amino acid deprivation and modulation of insulin/IGFR signalling (34). However, this combinatorial effect may not, in fact, be essential for antitumor activity and we will return later to the requirement for mTOR inhibition.
In a recent update (16) of the earlier compendium (14,15) of selectivity patterns of kinase inhibitors – revealed by profiling large, diverse kinase panels representative of the broader kinome – the suitability of these agents as chemical tools for use in cellular studies was determined. Based on this comparative assessment, the replacement of LY294002 by PI-103 was recommended alongside wortmannin as the two most suitable chemical probes to be used in tandem to determine the involvement of PI3K signalling in cellular phenotypes.
Despite its value a chemical tool (16, 35) and its utility in demonstrating the therapeutic potential of this series of PI3K inhibitors (33, 34, 37), PI-103 was recognized from the outset to exhibit serious drawbacks in terms of its drug-like properties and thus its suitability for clinical development (33). In particular, the planar tricyclic structure (Figure 3) results in limited aqueous solubility, and the phenolic hydroxyl group is rapidly glucuronidated. These adverse features needed to be fixed in order to produce a clinical development candidate with suitable formulation, pharmacokinetic and pharmacodynamic properties, while retaining potency and selectivity against PI3K.
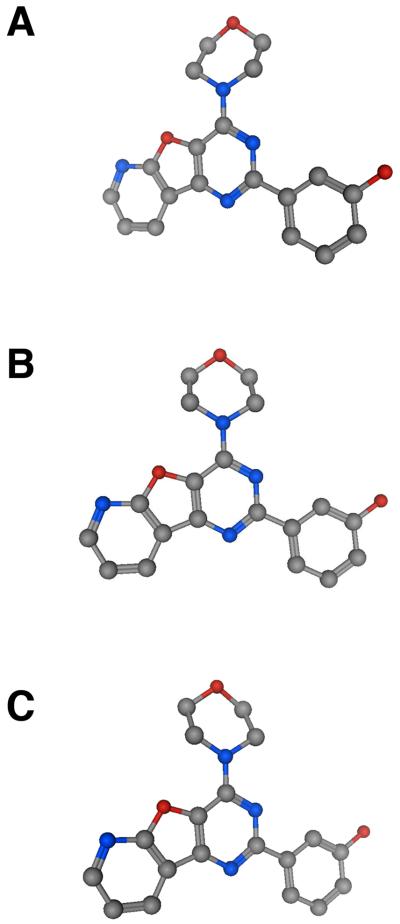
Figure 3.
Ball and stick representations of PI-103 to illustrate the planar nature of the tricyclic pyridofurapyrimidine, which limits the aqueous solubility of the compound.
From PI-103 to GDC-0941: A candidate for clinical development
Designing a drug candidate suitable for clinical evaluation is like solving Rubik’s Cube in that all of the essential elements need to be aligned simultaneously in the same molecule. Almost inevitably the eventual clinical agent is to some degree a compromise, since many of the different properties may be dependent on the same structural features within the drug molecule. Solubility, pharmacokinetic and pharmacodynamic behaviour are very important alongside target potency and selectivity (10). This is as true for PI3K inhibitors as it is for other drugs.
The multiparameter lead optimisation programme focused on improving pharmaceutical, pharmacokinetic and pharmacodynamic properties while maintaining the key inhibitory activity on class I PI3K enzymes and the related antiproliferative activity against cancer cells (38, 39). The two chemical starting points considered were: 1) the tricylic pyridofuropyrimidine PI-103 (29); and 2) the bicyclic thienopyrimidine ‘compound 1′, also optimized from an HTS hit (Figures 1 and 2; ref 27). The latter, although it exhibited even faster clearance than PI-103 in mice, offered greater potential for optimisation because of its lower molecular weight and capacity for key chemical substitutions to be made. From among the large number of chemical analogs designed, synthesized and tested in an iterative fashion (38), the more advanced lead compounds PI-520 and PI-620 illustrate the evolution to the clinical candidate GDC-0941 (Figure 1; ref 39).
Both PI-540 and PI-620 are bicyclic thienopyrimidines (Figure 1) with a solubilizing piperazine-based functionality at the 6-position of the thienopyrimidine ring (left-hand side), which was predicted by computer modelling to extend out of the ATP-pocket into the solvent. This modelling was done using PI-103, the bicyclic thienopyrimidine lead, and the p110γ structure (38). At the same time, PI-540 and PI-620 keep the phenol moiety (right-hand side) that was thought to be required for binding in the affinity pocket (refs 35, 38 and see later). PI-540 and PI-620 retained the <10nM potency against p110α and p110δ seen with PI-103; although potency against p110β and p110δ was lower by an order of magnitude these compounds are still potent pan-class I inhibitors (38, 39). Submicromolar activity was maintained against a panel of cancer cell lines, consistent with comparable inhibition of AKT phosphorylation (39). PI-540 and PI-620 exhibited much improved aqueous solubility and reduced in vitro microsomal metabolism. Clearance was still quite fast, due to remaining glucuronidation of the phenolic hydroxyl and extensive tissue uptake, but high tissue to plasma ratios led to improved concentrations and better pathway modulation in human tumor xenografts and resulted in correspondingly greater antitumor activity, as compared to PI-103 (39). Indazoles were investigated as phenol replacements that might markedly reduce glucuronidation while maintaining the necessary target interaction – specifically the hydrogen bound donor properties in the affinity pocket of the ATP site (see later). This strategy paid off and GDC-0941 was designed (38) by combining: 1) a 4-indazole substitution on the right-hand side of the thienopyrimidine ring; with 2) an optimised sulfonylpiperazine solubilizing group on the left-hand side; while at the same time 3) retaining the essential morpholine (top of structure; Figure 1).
Solving the Rubik’s Cube conundrum in this way resulted in a range of interesting and attractive features being exhibited by GDC-0941 (38, 39). The drug retains the 3nM IC50 against wild-type p110α and has equal potency against both of the common activated oncogenic hotspot oncogenic p110α mutants that were tested, namely the helical domain E545K mutant and the C-terminal kinase domain H1047R mutant. Inhibition of p110α was ATP-competitive with a Ki value of 10nM. The same 3nM IC50 seen for p110α was also exhibited against the class 1A PI3K p110δ, although the IC50 values for the class IA p110β and the class IB p110β are 10-fold and 25-fold lower at 33nM and 75nM respectively. Since all these IC50 values are <75nM, the overall profile is that of a potent pan-class I PI3K inhibitor with relatively greater selectivity for the p110α and p110δ isoforms. A very high degree of selectivity was observed against the class III PI3K C2β (IC50 670nM) and the class III PI3K Vps34 (1000nM). In addition, and in contrast to PI-103, GDC-0941 has a very weak potency against the class IV PIKKs DNA-PK (1230nM) and mTOR (580nM). Furthermore, when tested in a panel of 228 protein kinases at a concentration of 1μM, GDC-0941 was shown to inhibit only minor activity on Flt3 and TrkA A (50% and 61% inhibition respectively), further confirming it to be very selective class I PI3K inhibitor (38).
Despite some differences in selectivity compared to PI-103, especially the markedly reduced potency against DNA-PK and mTOR, GDC-0941 displayed comparable potency against a panel of human cancer cell lines and human vascular endothelial cells (HUVECs), suggesting that a high potency against the class IV PIKKs was not necessary to inhibit tumor cell and HUVEC proliferation (39). Furthermore, GDC-0941, PI-103, PI-540 and PI-620 showed comparable inhibition of phosphorylation of protein substrates on the PI3K pathway in cancer cells; for example, EC50 values were in the range 10-36nM for AKT phosphorylation (39), as measured by quantitative electrochemoluminescent ELISA assays (33, 34, 40). Results indicated that ~90% inhibition of AKT phosphorylation for several hours is needed to inhibit cancer cell proliferation, information that was valuable during lead optimisation and identification of GDC-0941 as the clinical candidate (38,39) and should also inform clinical studies. In addition, this group of compounds inhibited translocation of the FKHR forkhead transcription factor with EC50 values of 30-81nM, consistent with PI3K inhibition (38, 39).
In vivo animal model testing demonstrated that the major reduction in glucuronidation of GDC-0941 afforded by the indazole substitution resulted in markedly slower systemic clearance compared to PI-103, PI-540 and PI-640 and, moreover, led to 78% oral bioavailability in mice, a major advantage over the other analogs (38, 39). This in turn led to profound and prolonged inhibition of PI3 kinase pathway biomarkers in human cancer xenografts, consistent with the sustained tumor drug exposure (39). Thus tumor levels were well above antiproliferative GI50 concentrations for at least 6 hours and decreased phosphorylation of AKT, GSK3β and P70S6K was maintained for at least 8 hours (39). As a result of these enhanced pharmacokinetic and pharmacodynamic properties, GDC-0941 exhibited excellent dose-dependent therapeutic activity in the PTEN null, PI3K pathway-addicted U87MG human glioblastoma xenograft model in athymic mice, with up to 98% growth inhibition and significant regressions seen in this model (39).
Efficacy was confirmed in the PI3K-addicted IGROV-1 human ovarian cancer xenograft model, which harbors a hetT319F deletion and frameshift in PTEN as well as a p85 binding domain hetR38C mutation in p110α (39). Therapeutic activity was demonstrated in additional human tumor xenografts (41). The linkage of pharmacokinetic exposure, pharmacodynamic biomarker changes in the PI3K pathway and drug response provided a convincing pharmacologic audit trail (42-43) that forms a good platform for subsequent clinical studies. Based on its very attractive molecular, pharmacologic and therapeutic profile, including minimal effects on cytochrome P450s and the hERG channel (38), GDC-0941 was selected for clinical development.
Structural studies on the PI3 kinome: Invaluable insights and unfinished business
The design of GDC-0941 was guided by structural data on PI3Ks and their interaction with small molecule inhibitors, a situation that is almost taken for granted in many current drug discovery programmes on soluble drug targets. However, it was only a decade ago that the first light was shed on the three-dimensional structure of PI3Ks with the elucidation of the apo- and ATP-bound crystal structures of porcine p110γ (44).
These studies revealed a five-domain structure, which includes an N-terminal adaptor binding domain (ABD-domain), a Ras-Binding Domain (RBD-domain), a C2 domain that has been proposed to be involved in membrane binding, a helical domain and a catalytic domain (Figure 4a). The fold of the catalytic domain of the lipid kinase turned out to be very similar to the archetypical protein kinase fold, and comprises an N-terminal and a C-terminal lobe connected by a flexible peptide linker called the hinge region. Similar to the protein kinases, the PI3K ATP binding site is located in the groove in between the two components of the catalytic domain.
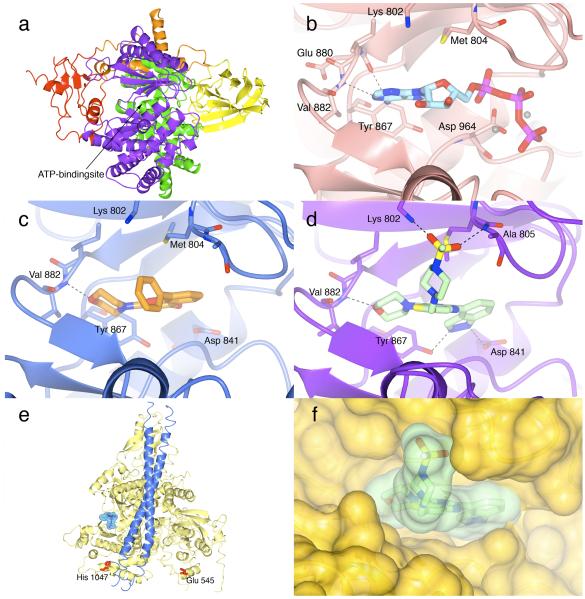
Figure 4.
a Ribbon diagram of human p110γ (PDB code 3dbs) illustrating the five-domain structure of the enzyme. The Ras-binding (RBD) domain is shown in red, the C2 domain in yellow, the helical domain in green and the bilobal catalytic domain in purple. The N-terminal ABD domain preceding the RBD-domain is shown in orange. The ATP-binding site is located in between the N- and C-terminal lobes of the catalytic domain, as indicated by the arrow. All figures were made with CCP4MG (66).
b Binding of ATP to porcine p110γ (PDB code 1e8x), showing the hydrogen bond interactions of ATP with the hinge region. The protein is shown in pink, ATP is displayed in light blue. The hydrogen bond interactions with the amide group of Val 882 and with the carbonyl group of Glu 880 are shown as dotted lines. Shown in grey are the two metal ions coordinating the phosphate groups of ATP.
c Binding of LY294002 to porcine p110γ (PDB code 1e7v) illustrates the crucial hydrogen bonding interaction between the oxygen of morpholino group of the weakly potent and relatively unselective inhibitor and Val 882 of the PI3K hinge region (left-hand side). The protein is shown in blue with key PI3K residues displayed and labelled. LY294002 is shown in orange. The critical hydrogen bond is shown as a dotted line. Although LY294002 extends into the affinity pocket (right-hand side) it does not fill the pocket as efficiently as the indazole moiety of GDC-0941 (see panel d).
d Binding of GDC-0941 to human p110γ (PDB code 3dbs) showing the hydrogen bond interactions that anchor the inhibitor in the ATP-site. p110γ is shown in purple and GDC-0941 is shown in light green. The oxygen of the morpholino group in GDC-0941 forms a hydrogen bond with the amide of the hinge residue Val 882, (left-hand side) while the indazole moiety (right-hand side) binds deep in the affinity pocket with its two nitrogen atoms forming hydrogen bonds with the side chains of Tyr 867 and Asp 841, respectively. The oxygen atoms of the sulfonyl group (projecting out of the plane, centre) interact with the side chain of Lys 802 and the amide group of Ala 805 at the mouth of the ATP pocket.
e Ribbon diagram of the p110α/p85α structure, showing p110α in yellow and the p85 niSH2 domain blue (PDB code 2rd0). To indicate the location of the ATP site, the ATP from the ATP-bound p110γ structure (PDB code 1e8x) superimposed on the p110α/p85α complex is shown. The ATP is displayed in cylinder representation with its semitransparent molecular surface superimposed in blue. Highlighted in red are the oncogenic mutation hotspots Glu 545 and His1047.
f Superposition of the GDC-0941-bound p110γ structure and the human p110α/p85α structure (PDB code 2rd0). The superposition was done using the CCP4 program suite (67). p110α is shown with its solvent-accessible surface in yellow and GDC-0941 with its (semi-transparent) surface in light green. Based on this superposition, the overall fit of GDC-0941 in the p110α ATP site is very good with only a minor clash of the indazole ring with the wall of the ATP site (right-hand side). It seems likely that GDC-0941 will be able to form hydrogen bond interactions with the hinge valine (Val 851 human p110α numbering left-hand side) and with the tyrosine and aspartate side chains (Tyr 836 and Asp 810, p110α numbering; right hand side) in the affinity pocket, as observed in the GDC-0941 p110γ structure. Differences could occur in the interaction of the sulfonyl oxygens of GDC-0941 (centre) with the protein, because the equivalent of Lys 802 in p110γ is an arginine residue in p110α and the equivalent residue of Ala 805 in p110γ has a different backbone conformation in p110α.
However, in contrast to the abundance of protein kinase structures, the structural studies on PI3Ks have long been restricted to porcine and human p110γ. Nevertheless, crystal structures of porcine p110γ complexed with ATP and early non-specific PI3K inhibitors such as LY294002, wortmannin and the broad spectrum protein kinase inhibitors quercetin, myricetin and staurosporine have yielded exciting insight into the molecular determinants of ligand binding in the ATP site of PI3Ks (22). For example, the structures show that the inhibitors fill the space occupied by the adenine moiety of ATP (Figure 4b) and demonstrate that the overlapping ring systems bind in the same plane as the adenine group. They further mimic protein-ATP interactions through the formation of a hydrogen bond with the backbone nitrogen of the hinge residue Val 882 (porcine p110γ numbering), which also interacts with the N1 atom in ATP, analogous to the hinge-binding observed for protein kinase inhibitors (Figure 4b,c). In addition, the five inhibitors studied extend to some degree into the affinity pocket, located at the back of the ATP site, which is not occupied by ATP in the ATP-bound structure (26). This pocket is defined by Lys 833, Asp 836, Leu 838, Asp 841, Asp 861, Tyr 867, Ile 879 and Asp 964 and is crucial in controlling inhibitor potency (35)
Indeed, the crystal structures of p110γ bound to two potent PI3K inhibitors, the imidazoquinazoline PIK-90 and the phenylthiazole PIK-93, show that both exploit the affinity pocket, PIK-90 through its pyridine moiety and PIK-93 using a chlorine atom (35). In addition, PIK-90 forms the hinge hydrogen bond with the backbone nitrogen of Val 882, whereas PIK-93 forms two hydrogen bonds with this residue, one with its backbone amide and one with its carbonyl group. Based on these protein-inhibitor structures, a model of the binding of PI-103 suggests that the morpholino group of PI-103 forms the hydrogen bond with the hinge and that its phenol moiety binds in the affinity pocket (35).
A crystal structure of the tridentate GDC-0941 bound to human p110γ shows that it is very efficiently anchored in the ATP-binding site which may explain its high potency (Figure 4d and Supplementary movie: ref 38). Similar to the early PI3K inhibitor LY294002, GDC-0941 uses an essential morpholino group to form the key hydrogen bond with the hinge. In addition, the indazole moiety fits snugly deep in the affinity pocket with the two indazole nitrogen atoms forming hydrogen bonds with the hydroxyl group of Tyr 867 and the carboxylate group of Asp 841, further strengthening the interactions in this pocket. Furthermore, the 4-methanesulfonyl-piperazin-1-ylmethyl group of GDC-0941 points towards the solvent, with the piperazine ring lying against the side chain of Met 804 and the oxygen atoms of the sulfonyl group forming hydrogen bonds with the backbone amide of Ala 805 and the side-chain nitrogen atom of Lys 802. Finally, the thienopyrimidine scaffold of GDC-0941 is effectively sandwiched between on the one hand the side chains of Met 953 and Ile 963, which form the floor of the ATP-binding site, and on the other hand the side chains of Met 804, Trp 812 and Ile 831 which form its ceiling.
As mentioned, so far all crystallographic investigations into the binding interactions of PI3K inhibitors, from the first inhibitors to advanced clinical candidates, have been carried out using porcine and human p110γ. However, as discussed earlier, it is p110α which is most frequently mutated and amplified in human cancers (6,7). The recent elucidation of the crystal structure of the human p110α/p85α complex (45) yielded a detailed structural understanding of the effects of oncogenic mutations in PI3Kα. For example, the p110α/p85α structure with the poorly visible nSH2 domain of p85 modelled in is consistent with a biochemically identified interaction between the frequently mutated Glu 542, Glu 545 and Gln 546 with residues in the p85 nSH2 domain and with the conclusion that mutation of these residues disrupts the inhibition of the catalytic subunit by this interaction (45). In addition, the structure shows that another oncogenic mutation hotspot His 1047 is located close to the C-terminal end of the activation loop, suggesting that His 1047 mutations could influence the conformation of the activation loop and therefore affect the binding of phosphoinositide substrates (45). However, both Glu 545 and His 1047 are quite far away from the ATP-binding site in PI3K (Figure 4e) and it is therefore not surprising that the oncogenic E545K and H1047R mutations have no effect on the potency of GDC-0941.
Although the structure elucidation of the p110α/p85α complex is an important milestone in the structural characterisation of the PI3K family, the quest for potent and isoform-specific next generation PI3K inhibitors would greatly benefit from the availability of protein-inhibitor structures of this and other isoforms. Unfortunately, the p110α/p85α structure shows that the crystals used to obtain it are not suitable for crystallographic soaking experiments, as a loop from the RBD domain of a neighbouring molecule protrudes into the ATP-binding site, thereby blocking the binding of nucleotides or small molecule PI3K inhibitors in these crystals. Nevertheless, some insight into inhibitor binding to PI3Kα can be obtained from superposition of the p110α/p85α structure with that of p110γ-inhibitor complexes. Such overlays show that inhibitors like the potent pan-class I-selective imidazoquinazoline PIK-90, phenylthiazole PIK-93 and GDC-0941 itself (Figure 4f) would fit into the p110α ATP site with only relatively minor clashes, consistent with their pan-specific PI3K inhibitor profile. However, the sequence and structural similarities of the p110α and p110γ ATP sites illustrate the challenges faced in the design of highly specific PI3K inhibitors.
Despite these challenges, inhibitors are emerging with surprising selectivity profiles, even differentiating between the class I PI3K isoforms (35), and as a result we are beginning to uncover the structural determinants that govern selectivity across the PI3Ks. For example, the crystal structure of p110γ bound to the quinazolinone purine PIK-39 (IC87114), one of the most isoform-specific (p110δ-selective) PI3K inhibitors to date, suggested that differences in plasticity in PI3Ks coupled to a conformational rearrangement of Met 804 (porcine p110γ numbering) could provide a mechanism to obtain isoform selectivity (35). In addition, recent results have illustrated the role of non-conserved residues at the entrance of the ATP-pocket with respect to inhibitors that are surprisingly p110β-selective (46). Future design efforts could greatly benefit from the availability of structures of the other class I isoforms PI3Kβ, PI3Kδ and also class IV protein kinases such as mTOR, ATM and ATR. It is therefore exciting to see the first structural insights provided by the crystal structures of p110δ in complex with a series of pan- and PI3Kδ-specific inhibitors that were published at the time of writing this review (47). These structures reveal a slightly different binding of GDC-0941 to p110δ compared to its binding to P110γ. Importantly, in contrast to GDC-0941 and other predominantly flat pan-specific inhibitors, the new structures also show that the PI3Kδ-specific inhibitors prefer a propellor shape, which allows them to efficiently exploit the greater plasticity of PI3Kδ, by accessing the so-called specificity pocket that was first observed in the p110γ-PIK-39 complex.
In addition to the increased understanding of selectivity of inhibitors within the PI3K family, a recent attempt to identify hotspots for resistance mutations revealed that, in contrast to the protein kinases, non-conservative mutations in the PI3K gatekeeper residue Ile 848 (p110α numbering) are not well tolerated, suggesting that this residue is an unlikely hotspot for resistance mutations (48). Indeed, the study showed that resistance mutants overall are likely to be less common than with many protein kinases. Intriguingly, mutations of Ile 800 to leucine and methionine were shown to confer some resistance to a variety of PI3K inhibitors including PIK-90 and PIK-93, but the I800L mutation was sensitized to the inhibitors PW-12 and BEZ-235. From the available crystal structures it can be seen that Ile 800 and its equivalent in p110γ are located in the ceiling of the ATP-binding site and interact with a variety of inhibitors through hydrophobic contacts. It therefore seems reasonable to speculate that mutations of this residue, in particular to a large methionine residue, could sterically hinder efficient binding of various classes of inhibitors. It will be exciting to see the emergence of crystal structures of these resistance mutants complexed with a variety of inhibitors, which will undoubtedly aid the design of new PI3K agents.
Context, Progress, Issues and Outlook
The case history discussed in this article highlights a number of issues that that are of general more general significance and interest.
Tools to drugs
Since the availability of the human genome sequence (http://www.ornl.gov/sci/techresources/Human_Genome/home.shtml), there has been an increasingly urgent need to develop not only genetic tools (such as RNA interference), but also small molecule probes to interrogate the function of all genes in normal biology and disease states, and to identify and exploit druggable targets for clinical benefit. Chemical probes can often give different phenotypes from RNAi knockdown, for example providing an immediate inhibitory effect that is usually confined to the catalytic activity whereas RNAi gives a slower effect and removes the entire protein from the cell. Developments in the biology of PI3Ks and the discovery of inhibitors have illustrated the value of the combined small molecule and genetic approach very well and the field has moved forward aggressively over the last few years. The progression of PI3K inhibitors from bioactive chemical biology tools – used to probe mechanisms and demonstrate feasibility and consequences of pathway inhibition – into inhibitors with improved, more drug-like properties that provide preclinical proof of concept and candidates for clinical evaluation is a pattern that is seen with other molecular targets (10, 49).
To be useful as chemical tools, small molecule inhibitors need to have at least a minimum level of performance. Based on continuously increasing experience, useful criteria are emerging for chemical tools. For example in the initial pilot phase of the US National Institutes of Health Molecular Libraries and Imaging Initiative (MLI), the chemical probes to be generated were envisaged as having “adequate potency and solubility for in vitro (cell-based) experimentation” but requiring further “chemical modifications ... to confer the selectivity, pharmacokinetric, and metabolic properties required for in vivo use” (50). Increasingly, the focus is not only on potency (for MLI <100nM), but also on selectivity (for MLI >10-fold over related targets) and on aqueous solubility, together with demonstrable improvements over existing probes for the designated target (see ref 51 and http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-08-005.html). There is now an urgent need for chemical tools to be usable in vivo, for example in animal models of cancer, and for this the pharmaceutical, pharmacokinetic and pharmacodynamic demands are usually much greater, requiring extensive medicinal chemistry efforts. There is in reality a continuum in terms of the somewhat overlapping criteria for initial chemical hits; improved chemical tools, some of which may evolve into lead compounds for drug discovery; and eventual clinical candidates. The latter are usually differentiated by virtue of fully optimized drug-like properties such as oral bioavailability and freedom from effects on anti-targets such as hERG channels and cytochrome P450s, as exemplified by GDC-0941.
The early chemical tools for PI3K inhibition, particularly wortmannin and LY294002 were extremely valuable but clearly had limitations. The case history reviewed herein illustrates how various micromolar hits were improved to a potent and selective chemical tool inhibiting class I PI3Ks/mTOR (PI-103), then into more advanced leads and potential preclinical drug candidates (PI-540 and P-620) and finally into the eventual pan-class I PI3K drug GDC-0941 that is now in Phase I clinical trials. In addition to the lessons learned when morphing from a chemical biology tool to a clinical candidate, another take-home message is the value of structure-based strategies now used in all stages in the design of molecular cancer therapeutics against a wide variety of targets (52). A structure-based approach was also used in another example, the imidazoquinoline clinical agent BEZ-235 which is, like PI-103, a dual class I PI3K/mTOR inhibitor; BEZ-235 evolved by target-hopping from a PDK1 inhibitor lead (53). There are now al least nine PI3K inhibitors in the clinic with various isoform selectivity profiles (see refs 23, 24, 54; http://clinicaltrials.gov).
Isoform selectivity, biomarkers, efficacy and tolerability
A key challenge for the development of inhibitors of p110α isoforms and indeed other PI3K pathway inhibitors in cancer is to determine the optimal selectivity profile, or more likely profiles, and to identify those patients in which a particular profile will be most effective (4, 5, 9, 23, 54). Intriguing new results suggest that genetic context will be key. For example, whereas tumor cells harboring activating p110α are dependent on this isoform, consistent with addiction to it, PTEN-deficient cancer cells are, in contrast, dependent on p110β (55). Further confusing the issue is the finding that p110β appears to have a non-kinase dependent role (9, 56). In addition, AKT-independent signalling downstream of oncogenic p110α has been discovered, involving PDK1/SGK3 (57). The p110δ isoform is mainly restricted to hemopoietic cells and may be a target in leukaemia and lymphoma as well as immune and inflammatory disease (9). Results with chemical inhibitors do not currently give a clear picture regarding the impact of PI3K pathway and RAS mutations on sensitivity to agents that generally hit the class I PI3Ks, with or without mTOR (reviewed in 39, 54).
In the case of GDC-0941, findings to date suggest that although cancer cells lines with mutations in PIK3CA or loss of PTEN are often sensitive to this agent, while some with RAS mutations may be resistant, the drug nevertheless shows broad activity across malignant cell panels and human tumor xenografts (39, 41). One contributory factor is that cancer cells with activated upstream receptor tyrosine kinases may also be sensitive. The situation in vivo may be further complicated by the clear antiangiogenic effects of class I/mTOR inhibitors (33, 53) that may relate to the role of p110α in endothelial cell migration and vascular development shown by mouse genetic studies (58). In addition, PI3K inhibitors may exert other effects on the tumor microenvironment and immune cells (4). In some ways the above results argue for the broad therapeutic utility of pan-class I inhibitors, assuming that such agents are well tolerated, which seems to be the case from animal model studies and early clinical trials (5, 23). Prediction of sensitivity may be require more complex signatures rather than single mutational events. Nevertheless, it is not unreasonable to look for initial clinical activity by enriching trials with patients with 1) oncogenic mutations in PIK3CA, 2) loss of PTEN expression or 3) activated receptor tyrosine kinases, before broadening the studies to a wider patient population. The alternative is to treat a more diverse cohort of patients up front and to collect information on their molecular status.
Although inhibition of p110α was predicted to block insulin signaling and potentially to induce diabetes (35, 36) this does not seem to be a problem with chronic dosing. Nevertheless, experience with protein kinase inhibitors in the clinic, where inhibition of multiple kinases can be advantageous (10), would support the view that at least limited polypharmacology with PI3K inhibitors may be a good thing. Inhibitors of specific isoforms may be valuable in particular settings (see above). Drugs targeting particular isoforms could avoid particular side-effects (5). For example, p110α/β inhibitors that do not hit p110γ and p110δ would likely spare immune cells. P110δ-selective inhibitors may be advantageous in some leukaemias and lymphomas and avoid possible metabolic effects on p110α/β. Preferential activity on p110α may be desirable in cancers with mutant p110α whereas p110β selectivity may be valuable in PTEN-deficient malignancies. There are no reports of inhibitors selective to mutant versus wild type p110α, which could have advantages in relevant cancers addicted to that mutant kinase. Ongoing use of chemical biology and structure-based design approaches should continue to deliver a variety of isoform-specific or pan-selective profiles. Unexpected cancer dependencies and synthetic lethalities may emerge, as shown by the preferential activity of PI3K pathway inhibitors in mismatch repair deficient colorectal cell lines (59).
Combinations
Experience with modern targeted molecular cancer therapeutics (10) suggests that the presence of multiple genetic abnormalities and the induction of feedback loops and other resistance mechanisms is likely to mean that PI3K inhibitors will need to be given in combination with other agents (4,5). In support of this approach, recent studies with GDC-0941 have shown promising results by combining this agent with the HER2-directed antibody trastuzumab, including treatment of trastuzumab-resistant cancer models (60, 61). The combination of GDC-0941 with a BRAF inhibitor was beneficial in cancer models with oncogenic BRAF and PTEN deficiency (61) and dual blockade involving GDC-0941 with a MEK inhibitor was advantageous in PTEN wild type or PTEN null basal-like breast cancer models (63). ‘Horizontal’ inhibition of parallel pathways as well as ‘vertical’ blockade at more than one point within the PI3K pathway are of great interest, while also recognizing that these pathways likely operate in complex networks where the outcomes of pharmacologic modulation may be difficult to predict.
Concluding remarks
The twenty plus-year journey from chemical tools to PI3K drugs has been an exciting and productive one. We have interesting times ahead as the first generation of PI3K inhibitors progress through early clinical trials. Initial reports from Phase I clinical trials show GDC-0941 to be well tolerated with appropriate pharmacokinetics, biomarker evidence of target modulation, and some signs of biologic activity (64, 65). A number of other inhibitors of PI3K are now in the clinic (22, 23). With suitable pharmacodynamic biomarkers in hand to show proof of mechanism, the key challenges are now to identify predictive biomarkers so that PI3K inhibitors can be directed to the most appropriate patients and to develop rational mechanism-based combinatorial treatments for optimal therapeutic efficacy.
Supplementary Material
Acknowledgements
Grant support: Cancer Research UK program grant number C309/A8274 and National Health Service funding to the National Institute for Health Biomedical Research Centre. Paul Workman is a Cancer Research UK Life Fellow
We would like to thank our many colleagues and collaborators who have worked with us on PI3K inhibitors. We thank Dr Swen Hoelder for suggesting the Rubik’s Cube analogy. We are grateful to Val Cornwell for help with preparation of the manuscript. We apologize to the authors of many excellent papers that could not be cited because of space restrictions.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors are employees of The Institute of Cancer Research, which has a commercial interest in the development of PI3K inhibitors, including GDC-941, and operates a rewards-to-inventors scheme. They have been involved in a commercial collaboration with Yamanouchi (now Astellas Pharma) and with Piramed Pharma, and intellectual property arising from the programme has been licensed to Genentech. Paul Workman was a founder of, consultant to, and Scientific Advisory Board member of Piramed Pharma and was formerly an employee of AstraZeneca. Florence I Raynaud is a consultant for ELARA Pharmaceuticals. Rob Van Montfort is a former employee of Astex Therapeutics.
References
- 1.Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD. Synthesis and function of 3-phosphorylated inositol lipids. Ann Rev Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 2.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–7. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 3.Abraham RT. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amst) 2004;3:883–7. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu P, Cheng H, Roberts TM, Zhao JJ. Disc Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–44. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ligresti G, Militello L, Steelman LS, Cavallaro A, Basile F, Nicoletti F, Stivala F, McCubrey JA, Libra M. PIK3CA mutations in human solid tumors. Cell Cycle. 2009;8:1352–58. doi: 10.4161/cc.8.9.8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the Pik3CA in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 8.Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–6. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogt PK, Gymnopoulos M, Hart JR. PI 3-kinase and cancer: changing accents. Curr Opin Genet Dev. 2009;19:12–7. doi: 10.1016/j.gde.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins I, Workman P. New approaches to molecular cancer therapeutics. Nat Chem Biol. 2006;2:689–700. doi: 10.1038/nchembio840. [DOI] [PubMed] [Google Scholar]
- 11.Workman P. Drugging the cancer genome: new challenges of infrequent and combinatorial targets. Curr Opin Investig Drugs. 2007;8:445–6. [PubMed] [Google Scholar]
- 12.Workman P, Clarke PA, Gulliard S, Raynaud FI. Drugging the PI3 kinome. Nat Biotechnol. 2006;24:1033. doi: 10.1038/nbt0706-794. [DOI] [PubMed] [Google Scholar]
- 13.Knight ZA, Shokat KM. Chemically targeting the PI3K family. Biochem Soc Trans. 2007;35:245–9. doi: 10.1042/BST0350245. [DOI] [PubMed] [Google Scholar]
- 14.Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bain J, McLauchlan H, Elliott M, Cohen P. The specificities of protein kinase inhibitors. Biochem J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baggiolini M, Dewald B, Schnyder J, Ruch W, Cooper PH, Payne TG. Inhibition of the phagocytosis-induced respiratory burst by the fungal metabolite wortmannin and some analogues. Exp Cell Res. 1987:408–18. doi: 10.1016/0014-4827(87)90201-1. [DOI] [PubMed] [Google Scholar]
- 18.Arcaro A, Wymann MP. Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: the role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil response. Biochem J. 1993;296:297–301. doi: 10.1042/bj2960297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wymann MP, Bulgarelli-Leva G, Zvelebil MJ, Pirola L, Vanhaesebroeck B, Waterfield MD, Panayotou G. Wortmannin inactivates phosphoinositide 3-kinase by covalent modification of Lys-802, a residue involved in the phosphate transfer reaction. Mol Cell Biol. 1996;16:1722–33. doi: 10.1128/mcb.16.4.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–8. [PubMed] [Google Scholar]
- 21.Cohen P. Protein kinase – the major targets of the twenty-first century. Nat Rev Drug Discov. 2002;1:309–15. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Echevria C, Sellers WR. Drug discovery approaches targeting the PI3K/Akt pathway in cancer. Oncogene. 2008;27:5511–26. doi: 10.1038/onc.2008.246. [DOI] [PubMed] [Google Scholar]
- 23.Yap T, Workman P. Targeting the PI3K-AKT-mTOR pathway: progress, pitfalls and promises. Curr Opin Pharmacol. 2008;8:393–412. doi: 10.1016/j.coph.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Drees BE, Mills GB, Rommel C, Prestwich GD. Therapeutic potential of phosphoinositide 3-kinase inhibitors. Exp Opin Ther. 2004;14:1354–3776. [Google Scholar]
- 25.Crabbe T, Welham MJ, Ward SG. The PI3K inhibitor arsenal: choose your weapon! Trends Biochem Sci. 2007;32:450–6. 22. doi: 10.1016/j.tibs.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 26.Walker EH, Pacold ME, Perisic O, Stephen L, Hawkins PT, Wymann MP, Williams RL. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol Cell. 2000;6:909–19. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- 27.Hayakawa M, Kaizawa H, Moritomo H, et al. Synthesis and biological evaluation of 4-morpholino-2-phenylquinazolines and related derivatives as novel PI3 kinase p110alpha inhibitors. Bioorg Med Chem. 2006;14:6847–58. doi: 10.1016/j.bmc.2006.06.046. [DOI] [PubMed] [Google Scholar]
- 28.Hayakawa M, Kaizawa H, Kawaguchi K, et al. Synthesis and biological evaluation of imidazo[1,2-a]pyridine derivatives as novel PI3 kinase p110alpha inhibitors. Bioorg Med Chem. 2007;15:403–12. doi: 10.1016/j.bmc.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 29.Hayakawa M, Kaizawa H, Moritomo H, et al. Synthesis and biological evaluation of pyrido[3′,2′:4,5]furo[3,2-d]pyrimidine derivatives as novel PI3 kinase p110alpha inhibitors. Bioorg Med Chem Lett. 2007;17:2438–42. doi: 10.1016/j.bmcl.2007.02.032. [DOI] [PubMed] [Google Scholar]
- 30.Hayakawa M, Kawaguchi K, Kaizawa H, et al. Synthesis and biological evaluation of sulfonylhydrazone-substituted imidazo[1,2-a]pyridines as novel PI3 kinase p110alpha inhibitors. Bioorg Med Chem. 2007;15:5837–44. doi: 10.1016/j.bmc.2007.05.070. [DOI] [PubMed] [Google Scholar]
- 31.Workman P. Drugging the cancer kinome: progress and challenges in developing personalized molecular cancer therapeutics. Cold Spring Harb Quant Biol. 2005;70:499–515. doi: 10.1101/sqb.2005.70.020. [DOI] [PubMed] [Google Scholar]
- 32.Workman P, Raynaud FI, Clarke PA, Te Poele R, Eccles S, Kelland L, Di Stefano F, Ahmadi K, Parker P, Waterfield M. Pharmacological properties and in vitro and in vivo antitumor activity of the potent and selective PI3 kinase inhibitor PI103. Eur J Cancer (Suppl) 2004;40:414A. [Google Scholar]
- 33.Raynaud FI, Eccles S, Clarke PA, et al. Pharmacologic characterization of a potent inhibitor of class I phosphatidylinositide 3-kinases. Cancer Res. 2007;67:5840–50. doi: 10.1158/0008-5472.CAN-06-4615. [DOI] [PubMed] [Google Scholar]
- 34.Guillard S, Clarke PA, Te Poele R, Mohri Z, Bjerke L, Valenti M, Raynaud F, Eccles SA, Workman P. Molecular pharmacology of phosphatidylinositol 3-kinase inhibition in human glioma. Cell Cycle. 2009;8:443–53. doi: 10.4161/cc.8.3.7643. [DOI] [PubMed] [Google Scholar]
- 35.Knight ZA, Gonzalez B, Feldman ME, et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125:733–47. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foukas LC, Claret M, Pearce W, Okkenhaug K, Meek S, Peskett E, Sancho S, Smith AJ, Withers DJ, Vanhaesebroeck B. Critical role for the p110α phosphoinositide-3-OH kinase in growth and metabolic regulation. Nature. 2006;441:366–70. doi: 10.1038/nature04694. [DOI] [PubMed] [Google Scholar]
- 37.Fan QW, Knight ZA, Goldenberg DD, Yu W, Mostov KE, Stokoe D, Shokat KM, Weiss WA. A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell. 2006;9:341–9. doi: 10.1016/j.ccr.2006.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Folkes AJ. Ahmadi K, Alderton WK, et al. The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I P13 kinase for the treatment of cancer. J Med Chem. 2008;51:5522–32. doi: 10.1021/jm800295d. [DOI] [PubMed] [Google Scholar]
- 39.Raynaud FI, Eccles SA, Patel S, et al. Biological properties of potent inhibitors of class I phosphatidylinositide 3-kinases: from PI-103 through PI-540, PI-620 to the oral agent GDC-0941. Mol Cancer Ther. 2009;8:1725–38. doi: 10.1158/1535-7163.MCT-08-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gowan SM, Hardcastle A, Hallsworth AE, et al. Application of meso scale technology for the measurement of phosphoproteins in human tumor xenografts. Assay Drug Dev Technol. 2007;5:391–401. doi: 10.1089/adt.2006.044. [DOI] [PubMed] [Google Scholar]
- 41.Friedman L. GDC-0941, a potent selective orally bioavailable inhibitor of class I PI3K. Proc American Assoc Cancer Res. 2008:LB–110. [Google Scholar]
- 42.Workman P. Auditing the pharmacological accounts for Hsp90 molecular chaperone inhibitors: unfolding the relationship between pharmacokinetics and pharmacodynamics. Mol Cancer Ther. 2003;2:131–8. [PubMed] [Google Scholar]
- 43.Sarker D, Workman P. Pharmacodynamic biomarkers for molecular cancer therapeutics. Adv Cancer Res. 2007;96:213–268. doi: 10.1016/S0065-230X(06)96008-4. [DOI] [PubMed] [Google Scholar]
- 44.Walker EH, Perisic O, Ried C, Stephens l, Williams RL. Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature. 1999;402:313–320. doi: 10.1038/46319. [DOI] [PubMed] [Google Scholar]
- 45.Huang C-H, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, Vogelstein B, Gabelli SB, Amzel M. The structure of a human p110a/p85a complex elucidates the effects of oncogenic PI3Ka mutations. Science. 2007;318:1744–8. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 46.Frazzetto M, Suphioglu C, Zhu J, Schmidt-Kittler O, Jennings IG, Cranmer SL, Jackson SP, Kinzler KW, Vogelstein B, Thompson PE. Dissecting isoform selectivity of PI3K inhibitors: the role of non-conserved residues in the catalytic pocket. Biochem J. 2008;414:383–90. doi: 10.1042/BJ20080512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berndt A, Miller S, Williams O, Le DD, et al. The p110δ crystal structure uncovers mechanisms for selectivity and potency of novel PI3K inhibitors. Nature Chem. Biol. In press. [Google Scholar]
- 48.Zunder ER, Knight ZA, Houseman BT, Apsel B, Shokat KM. Discovery of drug-resistant and drug-sensitizing mutations in the oncogenic PI3K isoform p110 alpha. Cancer Cell. 2008;14:180–92. doi: 10.1016/j.ccr.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perfecting probes. Nat Chem Biol. 2009;5:435. doi: 10.1038/nchembio0709-435. Editorial. [DOI] [PubMed] [Google Scholar]
- 50.Austin CP, Brady LS, Insel TR, Collins FS. NIH Molecular Libraries Initiative. Science. 2004;306:1138–9. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 51.Oprea TI, Bologa CG, Boyer S, et al. A crowdsourcing evaluation of the NIH chemical probes. Nat Chem Biol. 2009;5:441–7. doi: 10.1038/nchembio0709-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Montfort RL, Workman P. Structure-based design of molecular cancer therapeutics. Trends Biotechnol. 2009;27:315–28. doi: 10.1016/j.tibtech.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 53.Liu TJ, Koul D, LaFortune T, Tiao N, Shen RJ, Maira SM, Garcia-Echevrria C, Yung WK. NVP-BEZ235, a novel dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor, elicits multifaceted antitumor activities in human gliomas. Mol Cancer Ther. 2009;8:2204–10. doi: 10.1158/1535-7163.MCT-09-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ihle NT, Powis G. Take your PIK: phosphatidylinositol 3-kinase inhibitors race through the clinic and toward cancer therapy. Mol Cancer Ther. 2009;8:1–9. doi: 10.1158/1535-7163.MCT-08-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wee S, Wiederschain D, Maira SM, Loo A, Miller C, deBeaumont R, Stegmeier F, Yao YM, Lengauer C. PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci USA. 2008;105:13057–62. doi: 10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jia S, Liu Z, Zhang S, et al. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature. 2008;454:776–9. 55. doi: 10.1038/nature07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vasudevan KM, Barbie DA, Davies MA, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Graupera M, Guillermet-Guibert J, Foukas LC, et al. Angiogenesis selectively requires the p110alpha isoform of PI3K to control endothelial cell migration. Nature. 2008;453:662–6. doi: 10.1038/nature06892. [DOI] [PubMed] [Google Scholar]
- 59.Vilar E, Mukherjee B, Kuick R, et al. Gene expression patterns in mismatch repair-deficient colorectal cancers highlight the potential therapeutic role of inhibitors of the phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin pathway. Clin Cancer Res. 2009;15:2829–39. doi: 10.1158/1078-0432.CCR-08-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Junttila TT, Akita RW, Parsons K, Fields C, Lewis Phillips GD, Friedman LS, Sampath D, Sliwkowski MX. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell. 2009;15:429–40. doi: 10.1016/j.ccr.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 61.Yao E, Zhou W, Lee-Hoeflich ST, et al. Suppression of HER2/HER3-Mediated Growth of Breast Cancer Cells with Combinations of GDC-0941 PI3K Inhibitor, Trastuzumab, and Pertuzumab. Clin Cancer Res. 2009;15:4147–56. doi: 10.1158/1078-0432.CCR-08-2814. [DOI] [PubMed] [Google Scholar]
- 62.Hoeflich KP, Herter S, Tien J, et al. Antitumor efficacy of the novel RAF inhibitor GDC-0879 is predicted by BRAFV600E mutational status and sustained extracellular signal-regulated kinase/mitogen-activated protein kinase pathway suppression. Cancer Res. 2009;69:3042–51. doi: 10.1158/0008-5472.CAN-08-3563. [DOI] [PubMed] [Google Scholar]
- 63.Hoeflich KP, O’Brien C, Boyd Z, et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin Cancer Res. 2009;15:4649–64. doi: 10.1158/1078-0432.CCR-09-0317. [DOI] [PubMed] [Google Scholar]
- 64.Sarker D, Kristeleit R, Mazina KE, Ware JA, Yan Y, Dresser M, Derynck MK, De-Bono J. A phase I study evaluating the pharmacokinetics (PK) and pharmacodynamics (PD) of the oral pan-phosphoinositide-3 kinase (PI3K) inhibitor GDC-0941. Journal of Clinical Oncology (Suppl) 2009;27:3538. [Google Scholar]
- 65.Wagner AJ, Von Hoff DH, LoRusso PM, Tibes R, Mazina KE, Ware JA, Yan Y, Derynck MK, Demetri GD. A first-in-human phase I study to evaluate the pan-PI3K inhibitor GDC-0941 administered QD or BID in patients with advanced solid tumors. Journal of Clinical Oncology (Suppl) 2009;27:3501. [Google Scholar]
- 66.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta Cryst. 2004;D60:2288–94. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 67.Collabrative Computing Project 4 The CCP4 suite: programs for protein crystallography. Acta Cryst. 1994;D50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.