Abstract
Purpose
Histamine N-methyltransferase (HNMT) catalyzes one of two major histamine metabolic pathways. Histamine is a mediator of pruritus in atopic dermatitis (AD). The aim of this study was to evaluate the association between HNMT polymorphisms and AD in children.
Methods
We genotyped 763 Korean children for allelic determinants at four polymorphic sites in the HNMT gene: -465T>C, -413C>T, 314C>T, and 939A>G. Genotyping was performed using a TaqMan fluorogenic 5' nuclease assay. The functional effect of the 939A>G polymorphism was analyzed.
Results
Of the 763 children, 520 had eczema and 542 had atopy. Distributions of the genotype and allele frequencies of the HNMT 314C>T polymorphism were significantly associated with non-atopic eczema (P=0.004), and those of HNMT 939A>G were significantly associated with eczema in the atopy groups (P=0.048). Frequency distributions of HNMT -465T>C and -413C>T were not associated with eczema. Subjects who were AA homozygous or AG heterozygous for 939A>G showed significantly higher immunoglobulin E levels than subjects who were GG homozygous (P=0.009). In U937 cells, the variant genotype reporter construct had significantly higher mRNA stability (P<0.001) and HNMT enzyme activity (P<0.001) than the common genotype.
Conclusions
Polymorphisms in HNMT appear to confer susceptibility to AD in Korean children.
Keywords: Atopic dermatitis; children; histamine, N-Methyltransferase; polymorphism
INTRODUCTION
Atopic dermatitis (AD) is a chronic pruritic inflammatory skin disease caused by complex interactions among various susceptibility genes, host environment, infectious agents, skin barrier defects, and immunological responses.1 The prevalence of AD has doubled or tripled in industrialized countries during the past three decades.2 In Korea, an increased prevalence of symptoms, diagnosis, and treatment of AD has been noted.3 AD often represents the first clinical manifestation of atopy in childhood and suggests a high risk for the development of asthma.4 The underlying pathology of AD is complex, with interactions among genes and environmental factors contributing to disease manifestation.5
Histamine is released from mast cells and basophils, and is an important mediator in the development of allergic diseases. Histamine is degraded by two major metabolic pathways involving the enzymes histamine N-methyltransferase (HNMT) and diamine oxidase (DAO; ABP1), respectively.6 DAO may scavenge extracellular histamine, whereas HNMT catalyzes the inactivation of intracellular histamine. The HNMT gene, located on the long arm of chromosome 2 (q22.1), is approximately 34 kb in length, with six exons and a second intron of approximately 15 kb.7
Of the eight reported polymorphisms in HNMT, only three are common single nucleotide polymorphisms (SNPs): -463T>C, 314C>T, and 939A>G.8 For 314C>T, a nonsynonymous polymorphism which results in a change from threonine to isoleucine at amino acid 105, the 314T allele is associated with decreased enzyme activity of HNMT.8 In a study of Chinese females, -1,637T>C or -463T>C tended to be associated with decreased HNMT activity, and 939A>G or 1097A>T was associated with increased enzyme activity.9 In another study, the variant HNMT allele frequencies were slightly higher in patients with asthma and rhinitis as compared with healthy subjects.10 In our previous study,11 the 939A>G polymorphism was significantly associated with the acetylsalicylic acid-intolerant chronic urticaria phenotype in adult patients, and the variant genotype of the 939A>G polymorphism showed higher mRNA stability and increased enzyme activity. However, to our knowledge, there is only one study of HNMT polymorphisms in AD patients, which demonstrated a significant association between AD and 314C>T in HNMT.12
The purpose of this study was to evaluate the role of HNMT gene in susceptibility to AD by investigating the polymorphisms of this gene in Korean children with AD
MATERIALS AND METHODS
Subjects
A total of 763 children were recruited from Severance Children's Hospital of Yonsei University for this study. These subjects included 396 children with atopic eczema, 124 children with non-atopic eczema, 146 atopic controls, and 97 non-atopic controls.
All patients with AD were diagnosed according to the criteria of Hanifin and Rajka.13 We excluded children with AD who also had respiratory allergies. In this study, we divided patients with AD patients into atopic eczema and non-atopic eczema.14 Tests for specific immunoglobulin E (IgE) against Dermatophagoides pteronyssinus, Dermatophagoides farinae, egg whites, cow's milk, peanuts, and soybeans were performed, as these represent six common allergens in Korea. Children with atopy were sensitive to one or more of the six common allergens, and their total IgE level was >150 IU/mL. The total serum IgE level of children without atopy was <150 IU/mL, with no detectable specific IgE antibodies to any of the six common allergens. Controls were children who visited the hospital for general health checkups and did not have any history of allergic or inflammatory disease. Written informed consent was obtained from all participants before enrollment in the study, which was approved by the Severance Hospital Institutional Review Board.
Measurement of blood eosinophils and serum total and specific IgE levels
Peripheral blood samples were obtained from the subjects, and the levels of total IgE and specific IgE against each of the six common antigens were measured using a fluorenzymeimmunoassay (CAP FEIA; Pharmacia & Upjohn Diagnostic AB, Uppsala, Sweden) according to the manufacturer's instructions. Specific IgE levels >0.35 kUA/L were considered positive. A hematology analyzer (NE-8000; Sysmax, Kobe, Japan) was used to automatically count peripheral blood eosinophils.
Genotyping and PCR amplification
Whole blood was obtained from each subject, and genomic DNA was extracted using a FlexiGene DNA kit (Qiagen, Valencia, CA, USA). We genotyped the allelic determinants of the 763 children at four known polymorphic sites in the HNMT gene: -465T>C, -413C>T, 314C>T, and 939A>G.8,9,15 Genotyping was performed using a TaqMan fluorogenic 5' nuclease assay (ABI, Foster City, CA, USA). The polymerase chain reaction (PCR) contained 10 ng of genomic DNA, 2.5 µL of TaqMan Universal PCR Master Mix, and 0.13 µL of 40× assay mix in a final volume of 5 µL. Thermal cycle conditions were: 50℃ for 2 minutes to activate uracil N-glycosylase and prevent carry-over contamination, and 95℃ for 10 minutes to activate DNA polymerase, followed by 45 cycles of 95℃ for 15 seconds and 60℃ for 1 minute. All PCRs were performed in 384-well plates with a Dual 384-Well GeneAmp PCR System 9700 (ABI). Endpoint fluorescence readings were performed using a Prism 7900 HT sequence detection system (ABI). Duplicate samples and negative controls were included to ensure the accuracy of genotyping.
mRNA stability according to the 939A>G polymorphism
Two luciferase reporter constructs for the common and variant genotypes of 939A>G, which comprised a 535-bp fragment (from nt 880 to 1,415), were transiently transfected into a myelomonocytic cell line (U937) by electroporation. Briefly, 1×106 cells were seeded in six-well plates, transfected with 2 µg of an enhanced green fluorescent protein (EGFP) reporter plasmid (pEGFP-HNMT 3'-UTR), and incubated for 18 hours. Then, actinomycin D (10 µg/mL; Sigma, St. Louis, MO, USA) was added, and the cells were collected at 0, 0.5, 1, 2, 4, and 6 hours.
EGFP mRNA levels were assessed by real-time PCR using a thermocycler and Prism 7500HT sequence detection system (ABI)11. The EGFP mRNA expression level was normalized to that of β-actin.
EGFP protein levels were assessed by measuring fluorescence intensity with a Synergy HT multi-detection microplate reader (BioTek Instruments, Winooski, VT, USA). Fluorescence intensities are given in relative fluorescence units calculated relative to the values of mock-treated cells. Autofluorescence of non-transfected cells was measured as a negative control. EGFP protein expression levels were normalized to total protein concentration.
HNMT enzyme activity according to the 939A>G polymorphism
Transfection of pHNMT CDS-3'-UTR constructs into U937 cells and the measurement of HNMT activity in cell lysates were performed as previously described.11 Radioactivity was measured using a liquid scintillation counter (LS-3801; Beckman Instruments, Irvine, CA, USA). Enzyme activity was measured as the formation of Nτ-methylhistamine per hour of incubation at 37℃.
Statistical analysis
Statistical analyses were performed using SPSS ver. 11.5 (SPSS Inc., Chicago, IL, USA). The Chi-squared test was used to detect a significant departure from Hardy-Weinberg equilibrium for the genotype frequency of each SNP and to assess differences in genotype frequencies between patients and controls. SNPs were analyzed by logistic regression with three alternative models (co-dominant, dominant, and recessive), controlling for age and gender as covariates. Fisher's exact test was used when expected cell frequencies were less than five. Differences in mean phenotypic values were compared using Student's t-test. Significance was indicated by P<0.05. Bonferroni's correction was used for multiple comparisons.
RESULTS
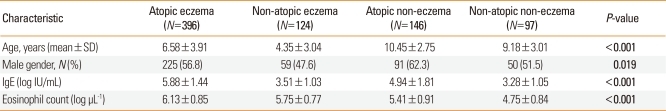
Characteristics of subjects
The characteristics of the study subjects are summarized in Table 1. There were significant differences in age and gender among the four groups (P<0.05). Total IgE levels were significantly higher in the atopic eczema group than in the other groups, and were higher in the atopic control group than in the non-atopic eczema group or non-atopic controls (P<0.05). Blood eosinophils were higher in the order of: atopic eczema, non-atopic eczema, atopic control, and non-atopic control groups (P<0.05).
Table 1.
Characteristics of study subjects
Values in bold indicate statistical significance.
N, number of patients; IgE, immunoglobulin E.
Genotype and allele frequencies of HNMT polymorphisms
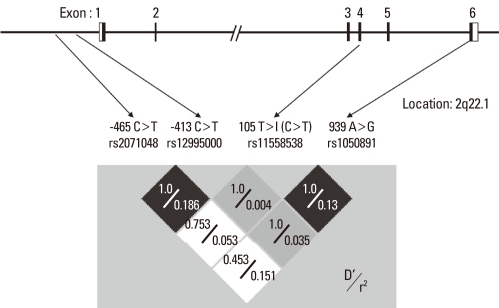
We performed a genetic association study of four genetic polymorphisms of HNMT, -465T>C, -413C>T, 314C>T, and 939A>G, among the four subject groups. The genomic structure and linkage disequilibrium of HNMT are shown in Fig. 1.
Fig. 1.
Linkage-disequilibrium structure of histamine N-methyltransferase (HNMT) in Koreans. The upper diagram shows the genomic structure of HNMT. The lower diagram shows the linkage disequilibrium (D'/r2) among the SNPs in this study.
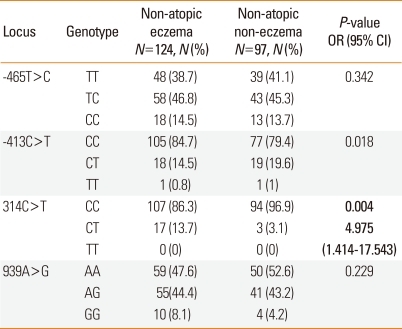
The genotype distributions of the polymorphisms in the eczema and control groups of children without atopy are showed in Table 2. The heterozygous CT frequency of 314C>T was significantly higher in the non-atopic eczema group than in the non-atopic control group (P=0.004). The genotype and allele frequencies of the -465T>C, -413C>T and 939A>G polymorphisms did not differ significantly between these two groups.
Table 2.
Genotype and allele frequencies of histamine N-methyltransferase (HNMT) polymorphisms in children without atopy
P values were calculated using dominant models. Values in bold indicate statistical significance. For multiple comparisons of genotype and allele frequencies, Bonferroni's multiple adjustment was applied to the level of significance, which was set at P<0.0125 (0.05/4). Logistic regression analysis was applied to control for age and gender as covariables.
N, number of patients.
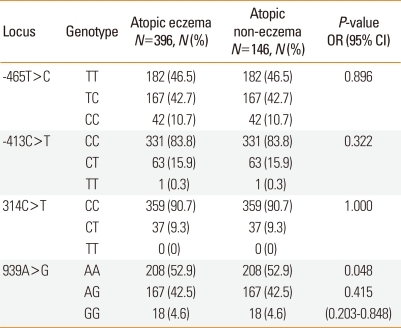
Table 3 shows the genotype distributions of the polymorphisms in the eczema and control groups of children with atopy. The homozygous GG frequency of 939A>G was lower than the combined homozygous AA and heterozygous AG frequencies in the eczema group (P=0.048). There were no significant differences in the genotype and allele frequencies of -465T>C, -413C>T, and 314C>T between these two groups.
Table 3.
Genotype and allele frequencies of histamine N-methyltransferase (HNMT) polymorphisms in children with atopy
P values were calculated using dominant models. Values in bold indicate statistical significance. For multiple comparisons of genotype and allele frequencies, Bonferroni's multiple adjustment was applied to the level of significance, which was set at P<0.0125 (0.05/4). Logistic regression analysis was applied to control for age and gender as covariables.
N, number of patients.
IgE level classified by genotype distribution of the HNMT 939A>G polymorphism
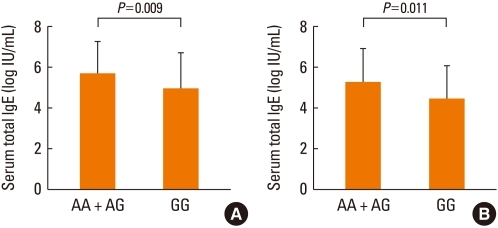
Total IgE was examined according to the genotype distribution of the HNMT 939A>G polymorphism. In children with atopy (Fig. 2A), the homozygous GG genotype of 939A>G was associated with significantly lower serum total IgE, compared with the combined homozygous AA and heterozygous AG genotypes (P=0.009). In children with eczema, a significantly lower total IgE level was also associated with the G allele. Patients who were homozygous GG for the 939A>G polymorphism tended to have significantly lower serum total IgE (P=0.011; Fig. 2B), and genotypes AG and GG were associated with lower serum total IgE levels compared with the AA genotype (P=0.007; data not shown).
Fig. 2.
Immunoglobulin E (IgE) level in children with atopy classified by distribution of the histamine N-methyltransferase (HNMT) 939A>G polymorphism in (A) children with atopy and (B) children with eczema. After Bonferroni's correction for multiple comparisons, P<0.0166 (0.05/3) was considered to be statistically significant. Logistic regression analysis was applied to control for age and gender as covariables.
Effect of the HNMT 939A>G polymorphism on mRNA stability
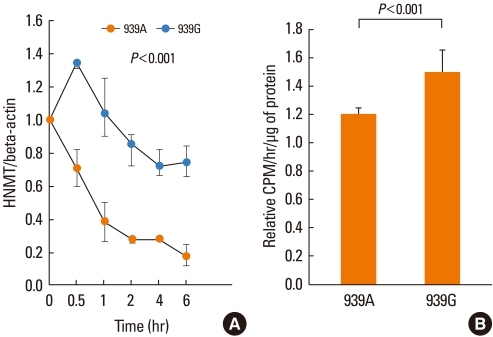
We examined the association of the 939A>G polymorphism with HNMT mRNA stability to identify the effect of the polymorphism on HNMT enzymatic activity. The mRNA stabilities of the 939A and 939G alleles were analyzed by real-time PCR in U937 cells transfected with each genotype (939A or 939G) in the pEGFP-HNMT 3'-UTR construct, after actinomycin D (10 µg/mL) treatment. The mRNA of the 939G-fused reporter gene was significantly more stable than that of the 939A-fused reporter gene (P<0.001). As seen in Fig. 3A, after actinomycin D treatment, the mRNA level of the 939A-fused reporter gene rapidly decreased, whereas that of the 939G-fused reporter gene decreased only slightly. We also investigated EGFP protein expression according to the 939A>G polymorphism by measuring the fluorescence signal from transfected U937 cells. The fluorescence intensity induced by the 939G-fused reporter gene was significantly higher than that induced by the 939A-fused reporter gene (P=0.001; data not shown).
Fig. 3.
Effect of the histamine N-methyltransferase (HNMT) 939A>G polymorphism on mRNA stability and enzyme activity. The mRNA levels of the pEGFP-HNMT 3'-UTR reporter construct in U937 cells were assessed by real-time PCR after actinomycin D (10 µg/mL) treatment. (A) EGFP mRNA stability according to the 939A>G polymorphism. The P value was determined using repeated measures ANOVA. (B) HNMT enzyme activity was examined by measuring the formation of Nτ-methylhistamine in pHNMT CDS-3'-UTR-transfected U937 cells. The relative enzyme activity (CPM: counts per minute) is represented as the ratio of the activity to the enzyme activity in cells transfected with the empty control vector pcDNA3. The P value was determined by a Mann-Whitney test. Three independent determinations were performed on each sample.
In vitro functional study of the HNMT 939A>G polymorphism
We examined the effect of the 939A>G polymorphism on HNMT enzyme activity and histamine release. Enzyme activity was assayed by measuring the formation of Nτ-methylhistamine in pHNMT CDS-3'-UTR-transfected U937 cells. Cells transfected with the HNMT 939G construct showed significantly higher enzyme activity than cells transfected with the HNMT 939A construct (P<0.001; Fig. 3B).
DISCUSSION
In this study, the effects of the HNMT -465T>C, -413C>T, 314C>T, and 939A>G polymorphisms on AD susceptibility in Korean children were investigated. The genotype and allele frequencies of 314C>T in HNMT were significantly associated with non-atopic eczema, and the genotype and allele frequencies of 939A>G were associated with atopic eczema. Subjects with the GG genotype for 939A>G had significantly lower IgE levels than homozygous AA or heterozygous AG subjects. In a functional analysis, the variant genotype reporter construct in U937 cells showed significantly higher mRNA stability and HNMT enzyme activity than the common genotype.
The 314C>T polymorphism, previously identified as a common nonsynonymous coding SNP located in HNMT exon 4, results in a Thr105Ile amino acid change.15 The function of the 314T allele has been determined.6,8,15,16 Genotype-phenotype correlation analyses in HNMT pharmacogenetic studies8,15 have revealed a significant association of the 314T allele with reduced thermal stability and decreased activity of HNMT. Based on these results, a role for the 314C>T polymorphism in the pathogenesis of asthma and allergic diseases has been investigated. The 314C>T HNMT polymorphism was significantly associated with the asthma phenotype in Caucasian patients,16 but no significant association of this polymorphism with asthma, bronchial hyperresponsiveness, or other asthma-related phenotypes was detected in German children.17 Further studies also failed to identify an association of this polymorphism with asthma.10,18 In our previous study of adult patients with chronic urticaria, there was no significant association with the 314C>T HNMT polymorphism.11 The only study of this polymorphism in AD patients suggested that the 314C>T polymorphism may confer an increased risk for AD.12 In the present study, the 314C>T polymorphism was significantly associated with non-atopic eczema. The importance of histamine as a pruritic mediator in eczema19 may explain the association between the 314C>T polymorphism and non-atopic eczema.
The 939A>G polymorphism is a common SNP in HNMT.15 A study in a European population reported no association between 939A>G and food allergy,20 and a study in an Indian population found no association of this polymorphism with asthma.18 In our previous study, a significant association was demonstrated between the 939A>G polymorphism and the acetylsalicylic acid-intolerant chronic urticaria phenotype.11 In the present study, 939A>G was associated with atopic eczema. The GG genotype frequency of 939A>G was lower in the atopic eczema group compared with the atopic control group.
Interactions of IgE with its receptors are central to the phenomenon of allergy, and an elevated serum IgE level is an important diagnostic feature of AD.21 To our knowledge, no published study has suggested an association between 939A>G in HNMT and the serum IgE level. Here, we detected a significant association between the 939A>G genotype distribution and serum IgE levels. In children with atopy and in those with eczema regardless of atopy, the homozygous GG genotype was significantly associated with lower serum IgE levels, compared with the other 939A>G genotypes. These results suggest a significant relationship between the 939G allele and lower IgE levels.
Although the 939A>G polymorphism in the 3'-UTR of HNMT has not been significantly correlated with HNMT phenotype, a study of a healthy Chinese population demonstrated that HNMT activity is modulated by the HNMT 939A>G polymorphism.9,15 In the present study, the 939G genotype was significantly related to higher mRNA stability, increased protein expression, and higher enzyme activity. This replicated a previous study, which demonstrated an association between the 939A>G polymorphism and HNMT enzyme activity in chronic urticaria patients.11
Histamine plays a key role in human allergic reactions. The increased local and systemic histamine levels consistently detected in AD patients suggest an important role for histamine in the pathogenesis of AD.22 HNMT catalyzes one of two major histamine metabolic pathways and is involved in the inactivation of intracellular histamine.6 Our data suggest that the 939G allele of the 939A>G polymorphism plays a protective role in AD by increasing HNMT enzyme activity, leading to reduced histamine levels. This is consistent with the action of histamine as a chemical mediator in the inflammation that contributes to AD disease pathogenesis. However, the association between the 939A>G polymorphism and AD phenotype became statistically insignificant after multiple correction. This may be partly explained by the contributions of additional inflammatory mediators to AD pathogenesis. Recent studies have demonstrated that activated mast cells can induce Th2 differentiation by modulating the function of maturing dendritic cells, and histamine functions in inducing Th2-promoting dendritic cells.23,24 IgE is an end product of the Th2 response, and thus histamine may influence IgE production through Th2 differentiation.23 The significant association of the 939G allele with lower IgE levels suggests that HNMT-mediated inactivation of histamine may affect serum IgE levels.
In conclusion, polymorphisms of the HNMT gene appear to confer susceptibility to AD in Korean children.
ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (2009-0075056).
Footnotes
There are no financial or other issues that might lead to conflict of interest.
References
- 1.Akdis CA, Akdis M, Bieber T, Bindslev-Jensen C, Boguniewicz M, Eigenmann P, Hamid Q, Kapp A, Leung DY, Lipozencic J, Luger TA, Muraro A, Novak N, Platts-Mills TA, Rosenwasser L, Scheynius A, Simons FE, Spergel J, Turjanmaa K, Wahn U, Weidinger S, Werfel T, Zuberbier T. Diagnosis and treatment of atopic dermatitis in children and adults: European Academy of Allergology and Clinical Immunology/American Academy of Allergy, Asthma and Immunology/PRACTALL Consensus Report. J Allergy Clin Immunol. 2006;118:152–169. doi: 10.1016/j.jaci.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 2.Williams H, Flohr C. How epidemiology has challenged 3 prevailing concepts about atopic dermatitis. J Allergy Clin Immunol. 2006;118:209–213. doi: 10.1016/j.jaci.2006.04.043. [DOI] [PubMed] [Google Scholar]
- 3.Hong SJ, Ahn KM, Lee SY, Kim KE. The prevalences of asthma and allergic diseases in Korean children. Korean J Pediatr. 2008;51:343–350. [Google Scholar]
- 4.Holloway JW, Yang IA, Holgate ST. Genetics of allergic disease. J Allergy Clin Immunol. 2010;125:S81–S94. doi: 10.1016/j.jaci.2009.10.071. [DOI] [PubMed] [Google Scholar]
- 5.Barnes KC. An update on the genetics of atopic dermatitis: scratching the surface in 2009. J Allergy Clin Immunol. 2010;125:16–29.e1-11. doi: 10.1016/j.jaci.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.García-Martín E, Ayuso P, Martínez C, Blanca M, Agúndez JA. Histamine pharmacogenomics. Pharmacogenomics. 2009;10:867–883. doi: 10.2217/pgs.09.26. [DOI] [PubMed] [Google Scholar]
- 7.Aksoy S, Raftogianis R, Weinshilboum R. Human histamine N-methyltransferase gene: structural characterization and chromosomal location. Biochem Biophys Res Commun. 1996;219:548–554. doi: 10.1006/bbrc.1996.0271. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Thomae B, Eckloff B, Wieben E, Weinshilboum R. Human histamine N-methyltransferase pharmacogenetics: gene resequencing, promoter characterization, and functional studies of a common 5'-flanking region single nucleotide polymorphism (SNP) Biochem Pharmacol. 2002;64:699–710. doi: 10.1016/s0006-2952(02)01223-6. [DOI] [PubMed] [Google Scholar]
- 9.Chen GL, Wang H, Wang W, Xu ZH, Zhou G, He F, Zhou HH. Histamine N-methyltransferase gene polymorphisms in Chinese and their relationship with enzyme activity in erythrocytes. Pharmacogenetics. 2003;13:389–397. doi: 10.1097/00008571-200307000-00004. [DOI] [PubMed] [Google Scholar]
- 10.García-Martín E, García-Menaya J, Sánchez B, Martínez C, Rosendo R, Agúndez JA. Polymorphisms of histamine-metabolizing enzymes and clinical manifestations of asthma and allergic rhinitis. Clin Exp Allergy. 2007;37:1175–1182. doi: 10.1111/j.1365-2222.2007.02769.x. [DOI] [PubMed] [Google Scholar]
- 11.Kim SH, Kang YM, Cho BY, Ye YM, Hur GY, Park HS. Histamine N-methyltransferase 939A>G polymorphism affects mRNA stability in patients with acetylsalicylic acid-intolerant chronic urticaria. Allergy. 2009;64:213–221. doi: 10.1111/j.1398-9995.2008.01795.x. [DOI] [PubMed] [Google Scholar]
- 12.Kennedy MJ, Loehle JA, Griffin AR, Doll MA, Kearns GL, Sullivan JE, Hein DW. Association of the histamine N-methyltransferase C314T (Thr105Ile) polymorphism with atopic dermatitis in Caucasian children. Pharmacotherapy. 2008;28:1495–1501. doi: 10.1592/phco.28.12.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanifin JM. Diagnostic criteria for atopic dermatitis: consider the context. Arch Dermatol. 1999;135:1551. [PubMed] [Google Scholar]
- 14.Johansson SG, Bieber T, Dahl R, Friedmann PS, Lanier BQ, Lockey RF, Motala C, Ortega Martell JA, Platts-Mills TA, Ring J, Thien F, Van Cauwenberge P, Williams HC. Revised nomenclature for allergy for global use: Report of the Nomenclature Review Committee of the World Allergy Organization, October 2003. J Allergy Clin Immunol. 2004;113:832–836. doi: 10.1016/j.jaci.2003.12.591. [DOI] [PubMed] [Google Scholar]
- 15.Preuss CV, Wood TC, Szumlanski CL, Raftogianis RB, Otterness DM, Girard B, Scott MC, Weinshilboum RM. Human histamine N-methyltransferase pharmacogenetics: common genetic polymorphisms that alter activity. Mol Pharmacol. 1998;53:708–717. doi: 10.1124/mol.53.4.708. [DOI] [PubMed] [Google Scholar]
- 16.Yan L, Galinsky RE, Bernstein JA, Liggett SB, Weinshilboum RM. Histamine N-methyltransferase pharmacogenetics: association of a common functional polymorphism with asthma. Pharmacogenetics. 2000;10:261–266. doi: 10.1097/00008571-200004000-00007. [DOI] [PubMed] [Google Scholar]
- 17.Deindl P, Peri-Jerkan S, Deichmann K, Niggemann B, Lau S, Sommerfeld C, Sengler C, Müller S, Wahn U, Nickel R, Heinzmann A. No association of histamine- N-methyltransferase polymorphism with asthma or bronchial hyperresponsiveness in two German pediatric populations. Pediatr Allergy Immunol. 2005;16:40–42. doi: 10.1111/j.1399-3038.2005.00218.x. [DOI] [PubMed] [Google Scholar]
- 18.Sharma S, Mann D, Singh TP, Ghosh B. Lack of association of histamine-N-methyltransferase (HNMT) polymorphisms with asthma in the Indian population. J Hum Genet. 2005;50:611–617. doi: 10.1007/s10038-005-0302-4. [DOI] [PubMed] [Google Scholar]
- 19.Darsow U, Pfab F, Valet M, Huss-Marp J, Behrendt H, Ring J, Ständer S. Pruritus and atopic dermatitis. Clin Rev Allergy Immunol. 2011;41:237–244. doi: 10.1007/s12016-010-8230-2. [DOI] [PubMed] [Google Scholar]
- 20.Petersen J, Drasche A, Raithel M, Schwelberger HG. Analysis of genetic polymorphisms of enzymes involved in histamine metabolism. Inflamm Res. 2003;52(Suppl 1):S69–S70. doi: 10.1007/s000110300059. [DOI] [PubMed] [Google Scholar]
- 21.Gould HJ, Sutton BJ, Beavil AJ, Beavil RL, McCloskey N, Coker HA, Fear D, Smurthwaite L. The biology of IGE and the basis of allergic disease. Annu Rev Immunol. 2003;21:579–628. doi: 10.1146/annurev.immunol.21.120601.141103. [DOI] [PubMed] [Google Scholar]
- 22.Ständer S, Steinhoff M. Pathophysiology of pruritus in atopic dermatitis: an overview. Exp Dermatol. 2002;11:12–24. doi: 10.1034/j.1600-0625.2002.110102.x. [DOI] [PubMed] [Google Scholar]
- 23.Kitawaki T, Kadowaki N, Sugimoto N, Kambe N, Hori T, Miyachi Y, Nakahata T, Uchiyama T. IgE-activated mast cells in combination with pro-inflammatory factors induce Th2-promoting dendritic cells. Int Immunol. 2006;18:1789–1799. doi: 10.1093/intimm/dxl113. [DOI] [PubMed] [Google Scholar]
- 24.Reuter S, Stassen M, Taube C. Mast cells in allergic asthma and beyond. Yonsei Med J. 2010;51:797–807. doi: 10.3349/ymj.2010.51.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]