Abstract
We densely genotyped, using 1000 Genomes Project pilot CEU and additional re-sequencing study variants, 183 reported immune-mediated disease non-HLA risk loci in 12,041 celiac disease cases and 12,228 controls. We identified 13 new celiac disease risk loci at genome wide significance, bringing the total number of known loci (including HLA) to 40. Multiple independent association signals are found at over a third of these loci, attributable to a combination of common, low frequency, and rare genetic variants. In comparison with previously available data such as HapMap3, our dense genotyping in a large sample size provided increased resolution of the pattern of linkage disequilibrium, and suggested localization of many signals to finer scale regions. In particular, 29 of 54 fine-mapped signals appeared localized to specific single genes - and in some instances to gene regulatory elements. We define a complex genetic architecture of risk regions, and refine risk signals, providing a next step towards elucidating causal disease mechanisms.
INTRODUCTION
Celiac disease is a common complex chronic immune-mediated disease with seroprevalence of ~1%1,2 in individuals of white European origin. A T-cell mediated small intestinal immune response is generated against gliadin fragments from wheat, rye and barley cereal proteins leading to villous atrophy. Its aetiology is poorly understood. Association with HLA variants was first shown in 1972, and predisposing HLA-DQ2 and -DQ8 sub-types are necessary but not sufficient to cause disease. Recent genome wide association studies (GWAS) have identified a further 26 non-HLA risk loci3-6. Many of these loci are also associated with other autoimmune or chronic immune-mediated diseases (albeit sometimes different markers and effect directions7), with particular overlap observed between celiac disease, type 1 diabetes8 and rheumatoid arthritis9.
Currently unanswered questions regarding the genetic predisposition to celiac disease, which are also relevant for other immune-mediated diseases, include explaining the remaining major fraction of heritability, including rare and additional common risk variants; and identification of causal variants and causal genes (or at least more finely localizing the risk signal). The Immunochip Consortium10 developed to explore these questions, taking advantage of emerging comprehensive common, low frequency, and rare variation datasets, and of a commercial offer of much lower per-sample custom genotyping costs for a very large project comprised of related diseases.
The Immunochip, a custom Illumina Infinium HD array, was designed to densely genotype, using 1000Genomes and any other available disease specific resequencing data, immune-mediated disease loci identified by common variant GWAS. The 1000 Genomes Project pilot CEU low-coverage whole genome sequencing dataset captures 95% of variants of MAF=0.05, and although underpowered to comprehensively detect variants of rarer allele frequency, still identifies 60% of variants of MAF=0.02, and 30% of variants of MAF=0.0111. The Consortium selected 186 distinct loci containing markers meeting genome wide significance criteria (P<5×10−8) from twelve such diseases (autoimmune thyroid disease, ankylosing spondylitis, Crohn’s disease, celiac disease, IgA deficiency, multiple sclerosis, primary biliary cirrhosis, psoriasis, rheumatoid arthritis, systemic lupus erythematosus, type 1 diabetes and ulcerative colitis). All 1000 Genomes Project low-coverage pilot CEU population sample variants11 (Sept 2009 release) within 0.1cM (HapMap3 CEU) recombination blocks around each GWAS region lead marker were submitted for array design. No filtering on correlated variants (linkage disequilibrium) was applied. Further case and control regional resequencing data were submitted by several groups (Online Methods, Supplementary Note), as well as a small proportion of investigator-specific undisclosed content including intermediate-significance GWAS results.
Most GWAS have been performed using common SNPs (typical minor allele frequency (MAF) >5%), further selected for low inter-marker correlation and/or even genomic spacing. In contrast to GWAS, the Immunochip presents a comprehensive in-depth opportunity to dissect the architecture of both rare and common genetic variation, at immuno-biologically relevant genomic regions, in human diseases. Due to the presence in our final Immunochip dataset of the majority of 1000 Genomes Project pilot CEU polymorphic genetic variants (and additional resequencing at some loci), the true causal variants from many risk loci may have been directly genotyped and analysed.
RESULTS
A total of 207,728 variants were submitted for Immunochip assay design and 196,524 passed manufacturing quality control at Illumina. After extensive and stringent data quality control (Online Methods), we analysed a near-complete dataset (overall 0.008% missing genotype calls) comprising 12,041 celiac disease cases and 12,228 controls (from 7 geographic regions, Table 1) and 139,553 polymorphic (defined here as ≥2 observed genotype groups) markers. 634 biallelic SNPs were assayed in duplicate, at these we observed 189 of 15,384,884 (0.0012%) genotype calls to be discordant. Considering the intended 207,728 variants submitted for design, and an observed ~9.1% non-polymorphic rate in our post-quality control data, we estimate we have high quality genotype data on ~74% of the complete 1000 Genomes Project pilot CEU true polymorphic variant set at the fine-mapped regions.
Table 1.
Sample Collections
| Population sample | Celiac cases | Controls |
|---|---|---|
| UK | 7728 | 8274b |
| The Netherlands | 1123 | 1147 |
| Poland | 505 | 533 |
| Spain - CEGECa | 545 | 308 |
| Spain - Madrida | 537 | 320 |
| Italy - Rome, Milan, Naples | 1374 | 1255 |
| India - Punjab | 229 | 391 |
| Total | 12041 | 12228 |
The two Spanish population samples were considered separately due to genotyping in different laboratories.
5430 UK 1958 Birth Cohort participants, and 2844 UK Blood Services-Common Controls.
Each of the collections from the UK, Netherlands, Poland, Spain (Madrid) and Italy contained essentially the same sample set as our 2010 celiac disease GWAS5, with now substantial additional samples from the UK and Netherlands and exclusion of amplified DNA samples from the Spanish collections. The Indian collection has not previously been studied. Our 2010 GWAS contained several collections not studied here.
We observed that 36 of the 183 non-HLA immune-mediated disease loci selected for Immunochip dense 1000Genomes-based genotyping achieved genome-wide significance (P<5×10−8) for celiac disease in either the current study or our previous GWAS5 (summary association statistics for all markers are available in T1DBase). All variants reaching genome wide significance were common (MAF >5%). We also observed marked enrichment for intermediate significance level celiac disease association signals (e.g. rs6691768, NFIA locus, P= 5.3×10−8) at a proportion of the remaining 147 dense-genotyped non-celiac autoimmune disease regions (Supplementary Figure 1). Variants from 3 dense-genotyped regions selected on Immunochip for a non-immune-mediated trait (bipolar disorder) showed no excess of association signals (Supplementary Figure 1).
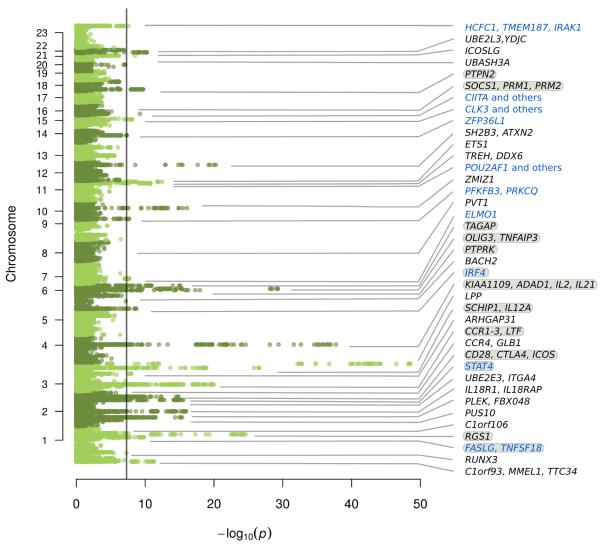
We identified 13 new celiac risk loci (P<5×10−8, Figure 1, Table 2, Supplementary Figure 2), 10 of which were from immune-mediated disease loci selected for Immunochip dense 1000Genomes-based genotyping. Several of these new loci were reported at lesser significance levels in our previous studies5,9, and almost all have been reported in at least one other immune-mediated disease. These, with HLA, bring to 40 the total number of reported (current and/or previous study5, which had an overlapping but slightly different sample set) genome wide significant celiac disease loci. Most contain candidate genes of immunological function, consistent with our previous findings at celiac disease loci3-5.
Figure 1. Manhattan plot of association statistics for known and novel celiac disease risk loci.
Novel loci indicated in blue, loci with multiple signals indicated with grey highlight. Significance threshold drawn at P=5×10−8.
Table 2. Risk variant signals at genome-wide significant celiac disease loci.
Non-HLA loci meeting genome-wide significance (P<5×10−8) in the current Immunochip dataset, or previous GWAS/replication dataset5, are shown. Loci reported for the first time for celiac disease at genome wide significance are shown in bold in the Top variant column.
| Top variant (dbSNP130 id) |
Chr | HapMap3 CEU LD blockb positions (hg18) (n markers, size) |
MAFc | Pd | OR | Highly correlated (r2>0.9) variants positions (hg18) (n markers, size) |
Localization: protein coding genes (RefSeq track UCSC/hg18) |
|---|---|---|---|---|---|---|---|
| rs4445406 | 1 | 2396747 - 2775531 (358, 379kb) |
0.344 | 5.4×10−12 | 0.87 | 2510162 - 2710035 (27, 200kb) |
C1orf93, MMEL1, TTC34 |
| rs72657048 | 1 | 25111876 - 25180863 (125, 69kb) |
0.498 | 3.8×10−6 | 0.92 | 25162321 - 25177139 (18, 15kb) |
0 - 10kb 5′ & 1st exon RUNX3 |
| rs12068671 | 1 | 170917308 - 171207073 (355, 290kb) |
0.185 | 1.4×10−10 | 0.86 | 170940206 - 170948695 (11, 8kb) |
35 - 43kb 5′ FASLG |
|
signal 2
rs12142280 |
1 | ” | 0.180 | 8.3×10−9d | 0.87 | 171129607 – 171131275 (2, 2kb) |
intergenic between FASLG and TNFSF18 |
| rs1359062 | 1 | 190728935 - 190814664 (181, 86kb) |
0.180 | 2.5×10−25 | 0.77 | 190786488 - 190811722 (17, 25kb) |
0 - 24kb 5′ & 1st exon RGS1 |
| signal 2 rs72734930 |
1 | ” | 0.022 | 3.7×10−4d | 1.23 | 190779182 (1) |
32kb 5′ RGS1 |
| rs10800746 | 1 | 199119734 - 199308949 (331, 189kb) |
0.305 | 2.6×10−8 | 0.89 | 199148015 (1) |
9th intron C1orf106 |
| rs13003464 | 2 | 60768233 - 61745913 (1047, 978kb) |
0.388 | 4.3×10−16 | 1.17 | 61040333 - 61058360 (3, 18kb) |
exons 5-11 PUS10 |
| rs10167650 | 2 | 68389757 - 68535760 (357, 146kb) |
0.266 | 1.3×10−4 | 0.92 | 68493221 - 68499064 (4, 6kb) |
intergenic between PLEK and FBX048 |
| rs990171 | 2 | 102221730 - 102573468 (894, 352kb) |
0.225 | 1.2×10−16 | 1.20 | 102338297 - 102459513 (45, 121kb) |
IL18R1, IL18RAP |
| rs1018326 | 2 | 181502502 - 181972196 (898, 470kb) |
0.418 | 3.1×10−16 | 1.16 | 181708291 - 181803246 (24, 95kb) |
intergenic between UBE2E3 and ITGA4 |
| rs6715106 | 2 | 191581798 - 191715979 (203, 134kb) |
0.058 | 8.4×10−9 | 0.79 | 191621279 - 191643278 (4, 22kb) |
exons 6-14 STAT4 |
|
signal 2
rs6752770 |
2 | ” | 0.296 | 1.3×10−6d | 1.10 | 191681808 (1) |
intron 3 STAT4 |
|
signal 3
rs12998748 |
2 | ” | 0.119 | 2.6×10−4d | 0.90 | 191656882 (1) |
intron 3 STAT4 |
| rs1980422 | 2 | 204154625 - 204524627 (642, 370kb) |
0.233 | 1.4×10−15 | 1.19 | 204318641 - 204320303 (2, 2kb) |
intergenic between CD28 and CTLA4 |
| signal 2 rs34037980 |
2 | ” | 0.217 | 1.6×10−5 d | 0.91 | 204470572 – 204478299 (2, 8kb) |
intergenic between CTLA4 and ICOS |
| signal 3 rs10207814 |
2 | ” | 0.039 | 1.3×10−4 d | 1.20 | 204158521 - 204168206 (5, 10kb) |
111 – 121 kb 5′ CD28 |
| rs4678523 | 3 | 32895606 - 33063377 (260, 168 kb) |
0.313 | 2.4×10−7 | 1.11 | 33012725 - 33012756 (2, 31bp) |
intergenic between CCR4 and GLB1 |
| rs2097282 | 3 | 45904804 - 46625997 (1343, 721kb) |
0.314 | 1.1×10−20 | 1.20 | 46321275 - 46377631 (27, 56kb) |
intergenic between CCR3 and CCR2 |
| signal 2 rs7616215 |
3 | ” | 0.361 | 8.6×10−9 d | 1.12 | 46162711 – 46180690 (2, 18kb) |
38 – 55 kb 3′ CCR1 |
| signal 3 rs60215663 |
3 | ” | 0.070 | 4.8×10−5 d | 1.16 | 46458634 – 46480319 (7, 22kb) |
exons 2-13 LTF (NM_002343.3) |
| rs61579022 | 3 | 120587671 - 120783345 (372, 196kb) |
0.390 | 9.9×10−9 | 1.11 | 120601187 - 120605968 (4, 5kb) |
intron 10 ARHGAP31 |
| [imm_3_161120372] | 3 | 161065075 - 161237201 (423, 168kb) |
0.111 | 2.6×10−27 | 1.36 | 161112778 - 161147744 (4, 35kb) |
intergenic between SCHIP1 and IL12A |
| signal 2 rs1353248 |
3 | ” | 0.288 | 9.8×10−9 d | 0.88 | 161106253 (1) | intergenic between SCHIP1 and IL12A |
| signal 3 rs2561288 |
3 | ” | 0.455 | 8.1×10−8 d | 1.12 | 161136316 – 161168494 (6, 32kb) |
intergenic between SCHIP1 and IL12A |
| rs2030519 | 3 | 189552054 - 189622323 (142, 70kb) |
0.486 | 3.0×10−49 | 0.76 | 189587750 - 189602595 (8, 15kb) |
intron 2 LPP |
| rs13132308 | 4 | 123192512 - 123784752 (1294, 592kb) |
0.166 | 1.9×10−38 | 0.71 | 123269042 - 123770564 (11, 502kb) |
multiple genes (KIAA1109, ADAD1, IL2, IL21) |
| signal 2 rs62323881 |
4 | ” | 0.073 | 8.6×10−5 d | 1.15 | 123257527 – 123722990 (87, 465kb) |
multiple genes (KIAA1109, ADAD1, IL2, IL21) |
| rs1050976 | 6 | 315547 - 402748 (199, 87kb) |
0.488 | 1.8×10−9 | 0.89 | 353079 - 355417 (3, 2kb) |
3′ UTR IRF4 (NM_002460.3) |
|
signal 2
rs12203592 |
6 | ” | 0.183 | 2.6×10−4 d | 0.91 | 341321 (1) |
intron 4 IRF4 (NM_002460.3) |
| rs7753008 | 6 | 90863556 - 91096529 (341, 233kb) |
0.380 | 2.7×10−7 | 1.10 | 90866360 - 90875874 (5, 10kb) |
intron 2 BACH2 (NM_001170794.1) |
| rs55743914 | 6 | 127993875 - 128382483 (572, 389kb) |
0.239 | 1.1×10−18 | 1.21 | 128332892 - 128335255 (2, 2kb) |
PTPRK last exon, 3′UTR (NM_002844.3) |
| signal 2 rs72975916 |
6 | ” | 0.150 | 1.2×10−5 d | 0.89 | 128307943 - 128339304 (15, 31kb) |
PTPRK exons 28-30, 3′UTR, to 24kb 3′ |
| rs17264332 | 6 | 137924568 - 138316778 (864, 392kb) |
0.211 | 5.0×10−30 | 1.29 | 138000928 - 138048197 (6, 47kb) |
intergenic between OLIG3 and TNFAIP3 |
|
signal 2 [imm_6_138043754] |
6 | ” | 0.190 | 2.1×10−7 d | 0.88 | 138015797 – 138043754 (4, 28kb) |
intergenic between OLIG3 and TNFAIP3 |
| rs182429 | 6 | 159242314 - 159461818 (514, 220kb) |
0.427 | 8.5×10−16 | 1.16 | 159385965 - 159390046 (4, 4kb) |
4kb 5′ and 5′ UTR TAGAP (NM_152133.1) |
|
signal 2 rs1107943 |
6 | ” | 0.071 | 2.8×10−6 d | 1.18 | 159418255 (1) |
32kb 5′ TAGAP (NM_152133.1) |
| [1kg_7_37384979] | 7 | 37330503 - 37406978 (213, 76kb) |
0.101 | 2.1×10−8 | 1.18 | 37366994 - 37404402 (31, 37kb) |
intron 1 ELMO1 |
| rs10808568 | 8 | 129211716 - 129368419 (400,157kb) |
0.256 | 2.2×10−5 | 0.91 | 129333242 - 129345888 (4, 13kb) |
151 - 163kb 3′ of PVT1 |
| rs2387397 | 10 | 6428077 - 6585110 (411, 157kb) |
0.229 | 1.9×10−8 | 0.88 | 6430198 (1) |
intergenic between PFKFB3 and PRKCQ |
| rs1250552 | 10 | 80690408 - 80774414 (223, 84kb) |
0.470 | 8.0×10−17 | 0.86 | 80728033 (1) |
intron 14 ZMIZ1 |
| rs7104791 | 11 | 110682429 - 110815769 (3, 133kb) |
0.209 | 1.9×10−11 | 1.16 | not high-density genotyped | [region: POU2AF1, C11orf93] |
| rs10892258 | 11 | 117847131 - 118270810 (466, 424kb) |
0.237 | 1.7×10−11 | 0.86 | 118080536 - 118085075 (5, 5kb) |
intergenic between TREH and DDX6 |
| rs61907765 | 11 | 127754640 - 127985723 (480, 231kb) |
0.213 | 3.4×10−13 | 1.18 | 127886184 - 127901948 (6, 16kb) |
5kb 5′ & 1st exon ETS1 (NM_001162422.1) |
| rs3184504 | 12 | 110183529 - 111514870 (938, 1331kb) |
0.488 | 5.4×10−21 | 1.19 | 110368991 - 110492139 (4, 123kb) |
5′ UTR & exons 1-3 SH2B3; exons 2-25 & 3′ UTR ATXN2 |
| rs11851414 | 14 | 68238574 - 68387815 (338, 149kb) |
0.221 | 4.7×10−8 | 1.13 | 68329159 - 68341722 (3, 13kb) |
1kb 5′ & 1st exon ZFP36L1 |
| rs1378938 | 15 | 72397784 - 73270664 (23, 873kb) |
0.278 | 7.8×10−9 | 1.13 | not high-density genotyped | [region inc. CLK3, CSK and multiple genes] |
| rs6498114 | 16 | 10834038 - 10903351 (8, 69kb) |
0.246 | 5.8×10−10 | 1.14 | not high-density genotyped | [region: CIITA] |
| rs243323 | 16 | 11220552 - 11385420 (446, 165kb) |
0.300 | 2.5×10−5 | 0.92 | 11254549 - 11268703 (12, 14kb) |
11kb 5′, all of SOCS1, 1kb 3′ |
| signal 2 [imm_16_11281298] |
16 | ” | 0.004 | 1.3×10−4 d | 1.70 | 11281298 (1) |
intergenic between PRM1 and PRM2 |
| signal 3 rs9673543 |
16 | ” | 0.169 | 2.0×10−4 d | 1.10 | 11292457 (1) |
10kb 5′ PRM1 |
| rs11875687 | 18 | 12728413 - 12914117 (411, 186kb) |
0.150 | 1.9×10−10 | 1.17 | 12811903 - 12870206 (16, 58kb) |
exons 2-5 PTPN2 (NM_080422.1) |
| signal 2 rs62097857 |
18 | ” | 0.040 | 5.2×10−5 d | 1.20 | 12847758 (1) |
intron 2 PTPN2 (NM_080422.1) |
| rs1893592 | 21 | 42683153 - 42760214 (226, 77kb) |
0.282 | 3.0×10−9 | 0.88 | 42728136 (1) |
intron 9 UBASH3A (NM_018961) |
| rs58911644 | 21 | 44414408 - 44528088 (239, 114kb) |
0.193 | 6.2×10−7 | 0.89 | 44446245 - 44453549 (8, 7kb) |
18 - 25kb 3′ ICOSLG |
| rs4821124 | 22 | 20042414 - 20352005 (131, 310kb) |
0.186 | 5.7×10−11 | 1.16 | 20250903 - 20313260 (36, 62kb) |
UBE2L3, YDJC |
| rs13397 | X | 152825373 - 153043675 (88, 218kb) |
0.133 | 2.7×10−8 | 1.18 | 152872114 - 152937386 (4, 65kb) |
HCFC1, TMEM187, IRAK1 |
Only the most significantly associated risk variant from each region and independent signal is shown. Variant names shown are as in dbSNP130 where available. Otherwise, the Illumina Immunochip manifest name is shown in brackets (Supplementary Table 5 shows both names for variants).
Regions were first defined by linkage disequilibrium blocks, extending 0.1 cM to the left and right of the risk SNP as defined by the HapMap3 CEU recombination map. For loci with multiple different previously reported risk SNPs for different diseases, and overlapping blocks, the extended region is shown. Regions where additional case resequencing (as well as 1000Genomes) has been performed are shown, with boundaries of the resequencing effort(s). All chromosomal positions are based on NCBI build-36 (hg18) coordinates.
MAF shown for European controls. See Supplementary Table 4 for more detailed allele frequencies in cases and controls by collection. Low frequency and rare variants shown in bold.
Logistic regression association test. Tests for second (and third) independent signals are conditioned on the first (and second) reported variant(s). Per locus significance thresholds for second (and third) independent signals are shown in Supplementary Table 3.
Effect sizes (odds ratios, inverting protective effects) for the most significant marker per locus were median 1.155 (range 1.124 – 1.360) for the top signals from 26 non-HLA loci measured using Illumina Hap300/Hap550-chip linkage disequilibrium-pruned tag SNPs in our 2010 celiac disease GWAS5 and median 1.166 (range 1.087 – 1.408) for the corresponding most significant marker (for the same signal) per locus in the current high density fine-mapping Immunochip dataset (Wilcoxon test P=0.75, Supplementary Table 1). Although we observe no difference in effect sizes between GWAS lead SNPs and subsequent fine-mapped signals, we note that case resequencing in the current Immunochip dataset is limited (see also Discussion).
In all, we report 57 independent coeliac disease association signals (Table 2) from 39 separate loci, of which 18 (32%) were not efficiently (r2>0.9, Supplementary Table 2) tagged by our previous GWAS5 (Illumina Hap550, post quality control dataset) markers.
Multiple independent common and rare variant signals
In contrast to most GWAS chips, the Immunochip contains a substantial proportion of lower MAF polymorphic variants. Of 139,553 variants in our 11,837 European-origin controls, 24,661 variants are low frequency (defined11 as MAF 5% to 0.5%) and a further 22,941variants are rare (MAF<0.5%). We investigated the possibility of multiple independently associated variants (of all allele frequencies) at each locus, using stepwise logistic regression conditioning on the most significant variant at the locus (Online Methods, Supplementary Table 3). This analysis can be sensitive to genotype miscalling and missing data12, hence our use of extremely rigorous quality control measures for the dataset and manual inspection of genotype clusters for all reported markers.
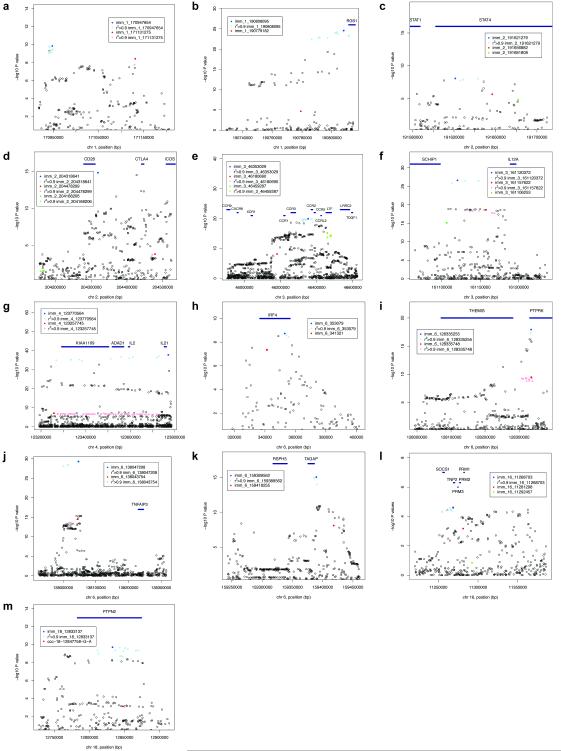
We observed two or more independent signals at 13 of 36 high-density genotyped non-HLA loci (Figure 2). Four of these loci each had three independent signals (STAT4, the chromosome 3 CCR region, IL12A, SOCS1/PRM1/PRM2, Table 2). Low frequency and/or rare variant signals were seen at four separate loci (RGS1, CD28/CTLA4/ICOS, SOCS1/PRM1/PRM2, PTPN2). Notably, the strongest effect (OR 1.70) was seen at the rare variant imm_16_11281298 (SOCS1/PRM1/PRM2 locus) with genotype counts (AA/AG/GG) of 1/136/11904 (MAF 0.57%) in all celiac cases and 0/91/12136 (MAF 0.37%) in all controls (detailed genotype count and allele frequency data for top signals by collection are shown in Supplementary Table 4).
Figure 2. Loci with multiple independent signals.
Non-conditioned P values shown for loci with multiple independent signals (from Table 2). The most associated variant for a signal shown in bold colour, further variants in r2>0.90 (calculated from the 24,249 sample Immunochip dataset) shown in normal colour. First signal coloured blue, second coloured red, third coloured green. Squares indicate markers present in our previous celiac disease GWAS post quality control dataset (Illumina Hap550)5.
We next performed haplotype analysis on all loci with multiple independent signals, to investigate whether the multiple signals were due to multiple causal effects or a single effect best tagged by several variants. For all but one locus (PTPN2) the haplotype association tests (not shown) were of similar significance to the single SNP association tests, suggesting that for each signal we have genotyped either the causal variant, or markers very strongly correlated with it. These findings contrast with those from a recent resequencing study13, probably because of the much greater variant density of our study. However, at the PTPN2 locus, the imm_18_12833137(T) + ccc-18-12847758-G-A(G) haplotype was considerably more associated (P=4.8×10−14, OR 0.84) than either SNP alone (imm_18_12833137 P=1.9×10−10; ccc-18-12847758-G-A P= 0.0008).
Interestingly at the SOCS1 locus, the third independent signal imm_16_11292457 shows association only after conditioning on the two other signals (P=2.0×10−4) but not in the single SNP non-conditioned association analysis (P=0.15). Further inspection revealed the protective imm_16_11292457(A) allele to be correlated (in linkage disequilibrium) with the risk (A) allele of the first signal imm_16_11268703, thus although there are indeed three independent signals, the effect of the third signal is only revealed after conditioning on the first. A similar statistical effect (Simpson’s paradox) was recently shown at a Parkinson’s disease locus14.
Fine-mapping to localize causal signals
GWAS signals are typically reported within relatively large linkage disequilibrium blocks. We tested whether our much denser genotyping strategy would allow finer-scale localization, and the pinpointing of association signals. We found that markers strongly correlated (r2>0.9) with the most significant independent variant clustered together, and defined regions that are a median 12.5x smaller than the relevant HapMap3 CEU 0.1cM linkage disequilibrium blocks (Table 2, Figure 2, Supplementary Figure 2). Localization was highly successful for some regions (e.g. PTPRK, TAGAP), but not possible at others (e.g. IL2-IL21). At many loci, the localized regions comprised only a handful of markers in close physical proximity.
Considering the 36 high density genotyped loci, we have localized to a single gene 29 of the total 54 independent non-HLA signals (Table 2, Supplementary Figure 2). We identified all markers strongly correlated (r2>0.9) with the independent non-HLA variants reported in our analyses (from Table 2), and on functional annotation (Supplementary Table 2) identified only a handful of markers in exonic regions and of these only three are protein altering variants (nsSNPs: imm_1_2516606 (MMEL1), imm_12_110368991 (SH2B3), 1kg_X_152937386 (IRAK1). In contrast, a number of signals appeared to be more finely localized around the transcription start site of specific genes (which we defined as the first exon, and 10kb 5′ of the first exon), including signals at RUNX3, RGS1, ETS1, TAGAP, ZFP36L1; and around the 3′ UTR region (and 10kb 3′) including signals at IRF4, PTPRK and ICOSLG.
Overlap between multiple independent signal regions was seen at some loci (Figure 2), suggesting that causal variants might be functioning through a shared mechanism e.g. within a 2kb region of the PTPRK 3′ UTR; within a 11kb region 5′ of IL12A; or within a 28kb region of TNFAIP3. In contrast, multiple independent signals were observed that spread between the three immune genes of the CD28/CTLA4/ICOS region.
DISCUSSION
We show that fine mapping of GWAS regions using dense resequencing data, e.g. (as here) from the 1000Genomes project, is feasible and generates substantial additional information at many loci. We identify a complex architecture of multiple common and rare genetic risk variants at around a third of the now 40 proven celiac disease loci. The design of our study has allowed us to find many more such complex regions than the ~10% with multiple signals seen in our previous study5 and a recent large GWAS for human height15. It seems probable that if larger sample sizes than in the current study were to be tested, additional loci might be shown to have a similarly rich multiple risk variant architecture. Multiple independent risk signals for celiac disease have also long been known in the HLA region16. Our success in celiac disease might be partly due to the extensive selective pressures for haplotypic diversity that have taken place at immune gene loci17. Previous studies have reported independently associated common and rare variants at individual loci for a handful of phenotypes e.g. fetal haemoglobin13, sick sinus syndrome18, Crohn’s disease19, hypertriglyceridemia20. To the best of our knowledge, ours is the first study to have comprehensively surveyed the genetic architecture of all known risk loci for a trait.
In part, our identification of rare variants at risk regions relies on the prior discovery of a genome-wide significant common variant association signal at each locus. This then permits a per-locus rather than genome-wide multiple testing correction when searching for additional independent association signals. Only particularly strong rare variant signals would, on their own, generate significance levels reaching the genome-wide threshold typically used in GWAS studies (P<5×10−8). Alternative methods, such as collapsing rare variant signals across a gene or functional categories of genes have therefore been suggested as approaches to the same problem21. Although a rare variant may have occurred on a recent haplotypic background, and thus show linkage disequilibrium at substantially longer range than common variants, we deliberately restricted our search to around the common variant linkage disequilibrium blocks as to do otherwise would have incurred a considerably greater penalty from multiple testing. Therefore, although our study provides considerable encouragement for exome and whole genome sequencing efforts aimed at identifying rare risk variants (not necessarily restricted to GWAS loci) in common complex diseases, it further highlights the statistical challenges of establishing rare variant associations.
We used a dense genotyping strategy and stepwise conditional association analysis, but did not identify any rare highly penetrant variants that might explain the genome-wide significant common SNP signals at any of the 39 loci. Our study does have limitations in this regard, particularly i) analysis restricted to 0.1cM linkage disequilibrium blocks; ii) the limited control resequencing sample size of the 1000 Genomes Project pilot CEU dataset; iii) the limited case resequencing sample size; and iv) case resequencing limited to three loci for celiac disease, and selected loci for other immune diseases. We observed a weak trend towards lower MAF (P=0.042, Wilcoxon test, Supplementary Table 1) for the best fine-mapping SNP (Immunochip experiment) versus the lead SNP from our 2010 tag SNP GWAS (measuring MAF in a subset of samples genotyped in both datasets). One signal showed substantially higher MAF (>25% change) on fine-mapping, four signals showed substantially lower MAF on fine mapping (Supplementary Table 1), yet all fine-mapping variants corresponding to lead GWAS SNPs remained common (MAF>0.10). We suggest that these changes in MAF upon fine-mapping of lead GWAS SNPs simply reflect more precise measurement of common frequency risk haplotypes. Although we cannot exclude the possibility that a single high-penetrance lower-frequency variant explains most of the association signal at a locus, especially without more comprehensive case resequencing, we find no evidence in support this possibility in the current fine-mapping experiment. Nor can our stepwise selection procedure robustly refute the “synthetic association” hypothesis - in particular that a combination of multiple rare variants jointly explains the association signal22 - although similarly we have not observed so far evidence supporting this possibility.
We established at genome wide significance 13 new loci for celiac disease, most of which have been reported previously at lesser significance or for another immune-mediated disease. The Illumina Hap550 chip (used in our 2010 GWAS) should have detected 10 of the 13 new loci, and in total 39 of the 57 independent non-HLA signals that we report. A current genotyping platform, the Illumina Omni2.5 chip would have detected 12 of the 13 new loci, and in total 50 of the 57 independent non-HLA signals that we report. Neither chip would have provided the finer scale localization of the Immunochip. The thirteen new loci contain many candidate genes of immunological function (P=0.0002 for enrichment of the Gene Ontology term “immune system process”23), in line with expectations from our previous studies. We also show evidence suggesting substantial additional signals at other immune-mediated disease loci, which lie beneath the genome wide significance reporting threshold applied to the current dataset. It is a point of debate whether such strict (P<5×10−8) criteria should apply - a Bayesian analyst might apply a higher prior at a locus already reported in another immune-mediated disease. Alternatively, an Immunochip-wide P value with a Bonferroni correction for independent SNPs, as used recently for the Cardiochip custom genotyping project24, of P<1.9 × 10−6 (Online Methods) would yield 16 additional celiac disease loci. These 16 loci also mostly contain immune system genes. An analysis of these currently intermediate significance signals would gain substantial additional power by a meta-analysis across the several hundred thousand samples from multiple immune-mediated disease collections presently being run on Immunochip,
We found that our previous GWAS using tag SNPs gave very similar estimates of effect size to our current fine-mapping experiment (Supplementary Table 1), in contrast to a simulation study which suggested that GWAS markers often underestimate risk14. We have, however, found substantial evidence for multiple additional signals at known loci and report many new loci. In Europeans, the current 39 non-HLA loci now explain 13.7% of coeliac disease genetic variance (HLA accounts for a further ~40%). We also show a long tail of likely effects of weaker significance, which will explain substantial additional heritability.
Only one of the variants reported here was discovered by a disease-specific resequencing study: ccc-18-12847758-G-A (rs62097857), a marker identified by the WTCCC group’s resequencing of Crohn’s disease cases and controls (Supplementary Note) and also present in the Watson genome. We submitted for Immunochip ~4,000 variants from high throughput resequencing of pools of 80 celiac disease cases for extended genomic regions at three loci (RGS1, IL12A, IL2- IL21, Supplementary Note). These did not contribute additional signals over and above those obtained from the 1000 Genomes Project pilot CEU variants, although did contribute to increase the numbers of variants correlated with each signal (i.e the set of markers that likely contains the causal variant(s)) and more precisely define the bounds of the signal localization. We note that larger scale case resequencing (e.g. many hundreds of samples) would identify a rarer spectrum of variants than the current study, and has previously been used with success at selected genes and phenotypes.
The possibility of performing fine-scale mapping of GWAS regions using e.g. 1000 Genomes Project data has been discussed as a natural follow-on strategy for such studies25,26 and has been recently used to identify risk variants in APOL1 in African-Americans with renal disease27. Our current report is the first to test such a strategy on a large scale in a complex disease. At multiple regions, we were able to refine the signal to a handful of variants over a few kilobases or tens of kilobases, although some regions (e.g. IL2-IL21) were resistant to this approach presumably due to particularly strong linkage disequilibrium. Most GWAS publications report signals mapping to a “LD block” based on HapMap recombination rates (sample size, 60 CEU families). In our data, where we have both i) much denser genotyping than GWAS chips (mean 13.6x at celiac loci versus the Illumina Hap550 chip) and ii) nearly 25,000 genotyped samples for the linkage disequilibrium calculations, we are able to observe much finer scale recombination and more precisely estimate of the bounds of no/minimal recombination intervals. Our findings are similar in terms of genotyping density and the resulting fine-mapped region size and lack of haplotype-specific effects to an earlier study of the IL2RA locus in type 1 diabetes26. At the majority of regions a tight block of highly correlated variants was seen, rather than a gradual decay of correlation (e.g. Figure 2 plots for IL12A, PTPRK). At many loci, we have now defined a handful of likely candidates to be the causal variant(s) to be taken forward into functional studies, although we may have missed candidate variants at some regions due to the sample size of the 1000 Genomes Project pilot CEU dataset (60 individuals), their status as controls, and our estimate that ~25% of these variants were excluded from our final dataset. These might be assessed by imputation methods28, but our approach – particularly with regards to the more sensitive conditional regression analysis - has been to prefer the more accurate direct genotyping of all assayable variants. As and when much larger whole genome resequencing based reference datasets become available (e.g. the main 1000 Genomes Project), these might be used to impute into our Immunochip dataset, including substantially lower frequency variants29. We also investigated whether our use of multiple ethnic subgroups within Europe (e.g. southern European Spanish versus northern European UK) or the relatively small Indian collection contributed to fine mapping, and found that in most cases, the same degree of localization was possible with just the UK collection alone (data not shown).
Our data suggest that most common risk variants might function by influencing regulatory regions, consistent with those previously reported in other immune-mediated diseases, and complex traits in general11. The exception is the SH2B3 nsSNP imm_12_110368991 (rs3184504), reported in our 2008 celiac GWAS4, which even with the fine-mapping of 938 polymorphic variants from the SH2B3 region remains the strongest signal at this locus thus suggesting it may be the causal variant. The same variant has been associated with other immune diseases, and a functional immune phenotype5. Interestingly, we observed a common ~980bp intergenic deletion between IL2 and IL21 (DGV40686, accurately genotyped by Infinium assay with control MAF 7.3%) correlated with the second independent signal at this region, although we have no evidence to suggest causality.
Our fine-scale localization approach has identified likely causal genes at many loci, and at eight genes signals localized around the 5′ or 3′ regulatory regions. For example, at the THEMIS/PTPRK locus, two independently associated sets of variants cluster in the 3′ UTR of the PTPRK gene (one, imm_6_128332892/rs3190930 in a predicted binding site for miRNA hsa-miR-1910). PTPRK, a TGF-beta target gene, is involved in CD4+ T cell development and a deletion mutation causes T helper deficiency in the LEC rat strain30. The signal at TAGAP lies within a 4kb region immediately 5′ of the transcription start site, presumably containing promoter elements. At ETS1, the signal comprises 6 variants overlapping the promoter and 1st exon of the T cell expressed isoform NM_001162422.1, and one of the variants (imm_11_127897147/rs61907765) has predicted regulatory potential and overlaps multiple transcription factor binding sites (UCSC GenomeBrowser ChipSeq and ESPERR tracks, Supplementary Table 2). Similarly interesting variants are observed in regulatory regions of RUNX3 (imm_1_25165788/rs11249212), and RGS1 (imm_1_190807644/rs1313292, imm_1_190811418/rs2984920) (Supplementary Table 2). Such an approach to identify the functional potential of risk variants was recently successful used to define a causal systemic lupus erythematosus TNFAIP3 variant31. Although we have localized signals at many loci, and recent research suggests the likely causal gene is often located near the most strongly associated variant15, only more detailed functional studies (e.g. transcription factor binding assays31 and transcriptional activity assays of constructs with individual single nucleotide alterations at risk SNPs32), will prove precisely which gene variants might be causal.
We conclude that dense fine mapping of regions identified through GWAS studies can uncover a complex genetic architecture of independent common and rare variants, and often successfully localize risk variant signals to a small set of SNPs to be taken forward into functional assays. Denser fine mapping studies, utilising larger resequencing sample sizes from both cases and controls over broader regions, might provide further resolution of GWAS signals.
Supplementary Material
ACKNOWLEDGMENTS
We thank Coeliac UK for assistance with direct recruitment of celiac disease individuals, and UK clinicians (L.C. Dinesen, G.K.T. Holmes, P.D. Howdle, J.R.F. Walters, D.S. Sanders, J. Swift, R. Crimmins, P. Kumar, D.P. Jewell, S.P.L. Travis, K. Moriarty) who recruited celiac disease blood samples described in our previous studies. We thank the Dutch clinicians for recruiting celiac disease blood samples described in our previous studies (C.J. Mulder, G.J. Tack, W.H.M. Verbeek, R.H.J. Houwen, J.J. Schweizer). We thank the genotyping facility of the UMCG (Pieter van der Vlies) for helping in generating part of immunochip data and S. Jankipersadsing, A. Maatman, at UMCG for preparation of samples. We thank R. Scott for preparing samples for genotyping and the University of Pittsburgh Genomics and Proteomics Core Laboratories for performing genotyping. We thank C. Wallace for assistance with Immunochip SNP selection, and J. Stone for co-ordinating Immunochip design and production at Illumina. We thank the members of each disease consortium who initiated and sustained the cross-disease Immunochip project. We especially thank all individuals with celiac disease and control individuals for participating in this study.
Funding was provided by the Wellcome Trust (084743 to D.A.vH.); by grants from the Coeliac Disease Consortium, an Innovative Cluster approved by the Netherlands Genomics Initiative, and partially funded by the Dutch Government (BSIK03009 to C.W.) and the Netherlands Organisation for Scientific Research (NWO, VICI grant 918.66.620 to C.W.); by NIH grant 1R01CA141743 (to R.H.D); Fondo de Investigación Sanitaria FIS08/1676 and FIS07/0353 (to E.U.). This research utilizes resources provided by the Type 1 Diabetes Genetics Consortium, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Allergy and Infectious Diseases, the National Human Genome Research Institute, the National Institute of Child Health and Human Development, and the Juvenile Diabetes Research Foundation International and is supported by NIH grant U01-DK062418. We acknowledge use of DNA from The UK Blood Services collection of Common Controls (UKBS-CC collection), funded by the Wellcome Trust grant 076113/C/04/Z and by NIHR programme grant to NHSBT (RP-PG-0310-1002). The collection was established as part of the Wellcome Trust Case Control Consortium (WTCCC)33. We acknowledge use of DNA from the British 1958 Birth Cohort collection, funded by the UK Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02.
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
URLs
Database of Genomic Variants, http://projects.tcag.ca/variation/?source=hg18
T1Dbase: www.t1dbase.org
SIFT: sift.jcvi.org
BioGPS: biogps.gnf.org
REFERENCES
- 1.Bingley PJ, et al. Undiagnosed coeliac disease at age seven: population based prospective birth cohort study. BMJ. 2004;328:322–3. doi: 10.1136/bmj.328.7435.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.West J, et al. Seroprevalence, correlates, and characteristics of undetected coeliac disease in England. Gut. 2003;52:960–5. doi: 10.1136/gut.52.7.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Heel DA, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39:827–9. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunt KA, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubois PC, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trynka G, et al. Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-kappaB signalling. Gut. 2009;58:1078–83. doi: 10.1136/gut.2008.169052. [DOI] [PubMed] [Google Scholar]
- 7.Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nature reviews. Genetics. 2009;10:43–55. doi: 10.1038/nrg2489. [DOI] [PubMed] [Google Scholar]
- 8.Smyth DJ, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med. 2008;359:2767–77. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhernakova A, et al. Meta-Analysis of Genome-Wide Association Studies in Celiac Disease and Rheumatoid Arthritis Identifies Fourteen Non-HLA Shared Loci. PLoS genetics. 2011;7:e1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis research & therapy. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clayton DG, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nature genetics. 2005;37:1243–6. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 13.Galarneau G, et al. Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nature genetics. 2010;42:1049–51. doi: 10.1038/ng.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spencer C, Hechter E, Vukcevic D, Donnelly P. Quantifying the underestimation of relative risks from genome-wide association studies. PLoS genetics. 2011;7:e1001337. doi: 10.1371/journal.pgen.1001337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–8. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Heel DA, Hunt K, Greco L, Wijmenga C. Genetics in coeliac disease. Best Pract Res Clin Gastroenterol. 2005;19:323–39. doi: 10.1016/j.bpg.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Zhernakova A, et al. Evolutionary and functional analysis of celiac risk loci reveals SH2B3 as a protective factor against bacterial infection. Am J Hum Genet. 2010;86:970–7. doi: 10.1016/j.ajhg.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holm H, et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nature genetics. 2011 doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lesage S, et al. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. American journal of human genetics. 2002;70:845–57. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansen CT, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nature genetics. 2010;42:684–7. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asimit J, Zeggini E. Rare variant association analysis methods for complex traits. Annual review of genetics. 2010;44:293–308. doi: 10.1146/annurev-genet-102209-163421. [DOI] [PubMed] [Google Scholar]
- 22.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS biology. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng Q, Wang XJ. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic acids research. 2008;36:W358–63. doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanktree MB, et al. Meta-analysis of Dense Genecentric Association Studies Reveals Common and Uncommon Variants Associated with Height. American journal of human genetics. 2011;88:6–18. doi: 10.1016/j.ajhg.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–31. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- 26.Lowe CE, et al. Large-scale genetic fine mapping and genotype-phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nature genetics. 2007;39:1074–82. doi: 10.1038/ng2102. [DOI] [PubMed] [Google Scholar]
- 27.Genovese G, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–5. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shea J, et al. Comparing strategies to fine-map the association of common SNPs at chromosome 9p21 with type 2 diabetes and myocardial infarction. Nature genetics. 2011;43:801–5. doi: 10.1038/ng.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jostins L, Morley KI, Barrett JC. Imputation of low-frequency variants using the HapMap3 benefits from large, diverse reference sets. European journal of human genetics : EJHG. 2011;19:662–6. doi: 10.1038/ejhg.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asano A, Tsubomatsu K, Jung CG, Sasaki N, Agui T. A deletion mutation of the protein tyrosine phosphatase kappa (Ptprk) gene is responsible for T-helper immunodeficiency (thid) in the LEC rat. Mammalian genome : official journal of the International Mammalian Genome Society. 2007;18:779–86. doi: 10.1007/s00335-007-9062-0. [DOI] [PubMed] [Google Scholar]
- 31.Adrianto I, et al. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nature genetics. 2011;43:253–8. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Musunuru K, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–9. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Revised criteria for diagnosis of coeliac disease. Report of Working Group of European Society of Paediatric Gastroenterology and Nutrition. Archives of disease in childhood. 1990;65:909–11. doi: 10.1136/adc.65.8.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romanos J, et al. Six new coeliac disease loci replicated in an Italian population confirm association with coeliac disease. Journal of medical genetics. 2009;46:60–3. doi: 10.1136/jmg.2008.061457. [DOI] [PubMed] [Google Scholar]
- 36.Plaza-Izurieta L, et al. Revisiting genome wide association studies (GWAS) in coeliac disease: replication study in Spanish population and expression analysis of candidate genes. Journal of medical genetics. 2011;48:493–6. doi: 10.1136/jmg.2011.089714. [DOI] [PubMed] [Google Scholar]
- 37.Megiorni F, et al. HLA-DQ and risk gradient for celiac disease. Human immunology. 2009;70:55–9. doi: 10.1016/j.humimm.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 38.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;405:847–56. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.