Abstract
How the proto-oncogene c-Myc balances the processes of stem cell self-renewal, proliferation and differentiation in adult tissues is largely unknown. We explored c-Myc’s transcriptional roles at the Epidermal Differentiation Complex (EDC) a locus essential for skin maturation. Binding of c-Myc can simultaneously recruit (Klf4, Ovo1-1) and displace (C/EBPα, Mxi1 and Sin3A) specific sets of differentiation-specific transcriptional regulators to EDC genes. We found that Sin3A causes de-acetylation of c-Myc protein to directly repress c-Myc activity. In the absence of Sin3A, genomic recruitment of c-Myc to the EDC is enhanced, and re-activation of c-Myc-target genes drives aberrant epidermal proliferation and differentiation. Simultaneous deletion of c-Myc and Sin3A reverts the skin phenotype to normal. Our results identify how the balance of two transcriptional key regulators can maintain tissue homeostasis via a negative feedback loop.
Introduction
The wide-ranging biological roles of Myc oncogenes in normal and cancer cells has made c-Myc one of the most exhaustively studied genes, yet a unifying view on its functions has proven elusive 1. c-Myc and its often redundant paralogs modulate diverse and sometimes opposing cellular functions including cell growth and proliferation, metabolism, differentiation and apoptosis 2,3. Inhibition of all Myc proteins in somatic tissues with rapid turnover, including skin, bone, intestine and testis triggers a marked reduction in cellular proliferation 4. In some of these adult tissues, c-Myc is as a central regulator of self-renewal, proliferation and differentiation processes 2,5-9.
c-Myc may control multiple biological functions by direct regulation of two distinct sets of target genes: (i) those broadly involved in cell proliferation and growth, and (ii) those expressed tissue-specifically 1,2,10. For instance, c-Myc forces skin stem cells to exit their niche by reducing the adhesive interactions 11. Across the epidermis, c-Myc promotes proliferation and increases terminal differentiation into interfollicular epidermis and sebaceous gland lineages 12,13. How c-Myc induces epidermal differentiation remains unclear, but must depend in part on c-Myc’s transcriptional function.
Additional to its function as a transcription factor, c-Myc modulates the transcriptional machinery 14. c-Myc stimulates promoter-paused Polymerase II to enter into productive elongation at a large portion of active genes in embryonic stem cells 15,16. This observation, together with the finding that c-Myc preferentially binds to promoter regions carrying active chromatin marks 17,18, suggested that c-Myc contributes to expression of the majority of active genes in a given cell context. In skin, c-Myc is known to functionally regulate activated progenitor cells and might therefore contribute significantly to both proliferation and differentiation programs.
Skin differentiation is an excellent system to explore the mechanisms by which c-Myc regulates tissue homeostasis. Epidermal stem cells differentiate into hair follicles, sebaceous glands, and interfollicular epidermis (IFE) — and differentiation into IFE can be promoted by c-Myc 13. Proliferating IFE daughter cells execute terminal differentiation by migration through suprabasal layers with progressive cornification 19. IFE differentiation genes are clustered in mammalian genomes as the epidermal differentiation complex (EDC); the mouse EDC is a single 2.2 Mb locus on chromosome 3 20,21. The EDC concentrates tissue-specific genes, and thus facilitates the analysis of upstream regulatory networks.
We show that c-Myc determines the occurrence of transcriptional regulators and we characterize their collective effect on gene expression. Using a combination of mouse models and systems biology tools, we discover how a negative feedback loop between c-Myc and Sin3A is required to balance proliferation and differentiation.
Results
EDC genes are bound and directly regulated by c-Myc
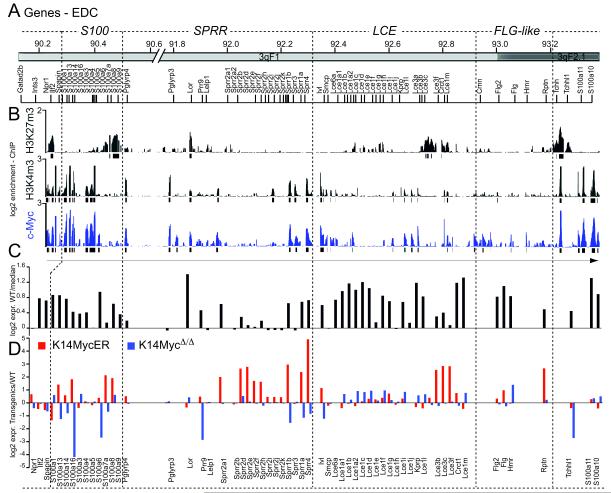
To better understand the paradox that activation of a proto-oncogene can lead to increased differentiation (Fig. S1A-D) 2, we asked whether c-Myc directly regulated genes involved in skin differentiation. We inducibly activated c-Myc in the basal, undifferentiated layer of the mouse epidermis (K14MycER) and performed chromatin immunoprecipitation combined with chromosome 3 tiling microarrays (ChIP on chip) capturing 61 genes in the EDC (http://www.ensembl.org/index.html) (Fig. 1A). We detected widespread binding of c-Myc to EDC genes, which over-lapped with H3K4me3, a histone mark that associates with active transcription start sites (Fig. 1B). With the exception of loricrin (Lor) and trichohyalin (Tchh), H3K4me3 and c-Myc binding excluded H3K27me3, an epigenetic mark that is closely linked to transcriptional repression (Fig. 1B). Genome-wide analysis using proximal promoter arrays confirmed that c-Myc and H3K4me3 occurrence overlapped by >80% and remained unchanged between wild-type and transgenic animals (Fig. S1E,F; Table S1).
Figure 1. Myc regulates genes involved in epidermal differentiation.
(A) Genomic organisation of the mouse EDC. (B) Schematic over-view of H3K27me3 and H3K4me3 occurrence and binding of c-Myc (blue) to EDC genes. (C) Expression of EDC RNA levels in wild-type skin. (D) Fold change of EDC gene expression in K14MycER (red) and K14MycΔ/Δ (red) mice normalized to wild-type animals.
Most EDC genes (47/55) were expressed above background in wild type skin (Fig. 1C), and 17/55 were up-regulated after c-Myc activation (Fig. 1D; red bars). Conversely, RNA levels of EDC genes were either unchanged or repressed in skin when c-Myc was conditionally deleted (K14MycΔ/Δ) (Fig. 1D; blue bars). Direct c-Myc regulation of epidermal differentiation was further evident when we analyzed RNA expression levels in c-Myc depleted skin (Fig. S2A-C). Almost half of all genes that were more than two-fold down-regulated when c-Myc was deleted were involved in epidermal maturation and differentiation (Fig. S2A-C; Table S2).
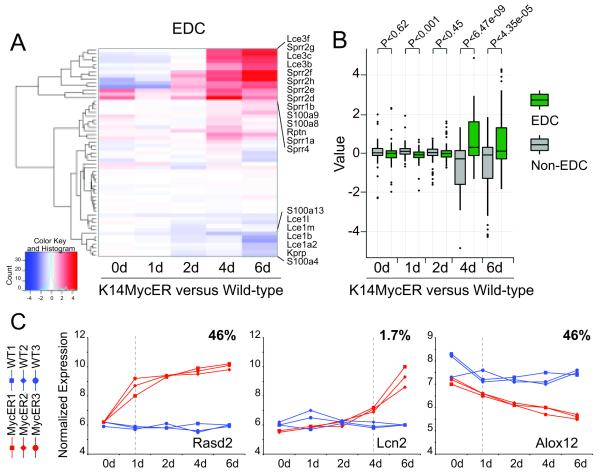
Cluster analysis confirmed that a set of EDC genes were up-regulated by 4-OHT driven c-Myc activation (Fig. 2A). We confirmed a specific regulatory function of c-Myc on EDC genes by comparing their change in RNA expression levels with randomly chosen non-EDC genes (Fig. 2B). During the six day time course, EDC genes were only up-regulated after 4 to 6 days of 4-OHT treatment in K14MycER mice (Fig. 2A). The delay in the EDC’s transcriptional response was unusual: virtually all transcriptional changes within the 5,000 genes most dependent on c-Myc activation occurred within one day of 4-OHT treatment (Fig. 2C, Rasd2, Alox12; Table S3). In contrast, only 1.7% of genes were up-regulated after 4 to 6 days, which included the EDC genes (Fig. 2C, Lcn2; Table S3).
Figure 2. Distinct expression profile for EDC genes in response to activated c-Myc.
(A) Cluster analysis of RNA expression levels of EDC genes in K14MycER versus wild-type controls treated with 4-OHT for the indicated time points in days (d). (B) Comparison of EDC gene expression from (A) versus randomly selected non-EDC genes. (C) Common expression profiles of 5000 regulated genes in triplicates in epidermis from K14MycER (MycER1-3) and wild-type (WT1-3) mice. RNA expression of Rasd2 as one example for 46% of genes up-regulated after 1 day of 4-OHT treatment (left hand panel). Lcn2 as representative gene for a small group of genes (1.7%) up-regulated after 4 days of 4-OHT treatment (middle panel). Alox12 as an example for 46% of genes down-regulated after 1 day treatment with 4-OHT (right hand panel).
In contrast to the delay in transcriptional regulation, c-Myc bound to EDC genes within a single day of 4-OHT treatment (Fig. S2D-F). This disconnection between rapid binding followed by gradual up-regulation suggested that c-Myc control of EDC genes might require additional tissue-specific transcription factors.
Myc modulates binding of tissue-specific transcription factors to the EDC
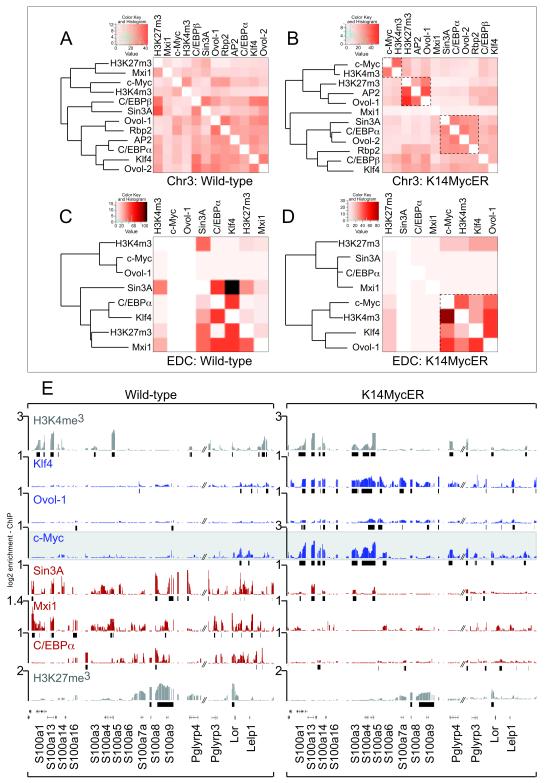
We characterized nine transcriptional regulators required for skin differentiation bind to the mouse genome; AP2γ, C/EBPα and β, Ovol-1 and 2, Klf4, Sin3A, and Mxi1 (Fig. 3A,B) 22-28. Rbp2 was chosen as an H3K4-specific de-methylase 29. In c-Myc over-expressing skin, specific clusters of transcription factors increased their shared occupancy of the mouse genome after c-Myc activation (Fig. 3A,B; inserts; Table S4), which was even more apparent within the 2.2 Mb containing the EDC (Fig. 3C,D). Here, we identified two distinct clusters of transcription factor binding only apparent in K14MycER mice: one contained Klf4, Ovol-1 and c-Myc, and the other Sin3A, Mxi1, and C/EBPα (Fig. 3C,D).
Figure 3. Myc determines specific regulatory networks at the EDC.
(A-D) Cluster analysis of binding events of the indicated tissue-specific transcription factors to chromosome 3 (Chr3) (A,B) and the EDC-only (C,D) in skin of wild-type (A,C) and K14MycER (B,D) mice. Heat maps show the number of shared binding events between two transcription factors (light orange: low similarity and dark orange: high similarity of binding sites occupied). Boxed inserts in (B) and (D) highlight specific clusters of transcriptional regulators sharing a high number of binding events only when c-Myc is over-expressed. The color codes are panel specific. (E) Occurrence of H3K4me3, H3K27me3 (grey) and occupancy of transcription factors indicated to a group of EDC genes in wild-type (left hand panel) and K14MyER mice (right hand panel). Over-expression of c-Myc leads to enhanced binding of Klf4 and Ovol-1 (blue) and loss of binding of Sin3A, Mxi1 and C/EBPα (red).
Occupancy of EDC genes by Klf4, Ovol-1, Sin3A, Mxi1 and C/EBPα depended directly on the levels of c-Myc (Fig. 3E). Whereas tri-methylation of H3K4 and H3K27 was comparable between transgenic and wild-type animals, binding of Ovol-1, Klf4, C/EBPα, Mxi1 and Sin3A changed considerably when c-Myc was over-expressed (Fig. 3E). In wild-type mice with low levels of c-Myc, EDC genes showed little Ovol-1 and Klf4 binding, but were highly bound by C/EBPα, Mxi1 and Sin3A (Fig. 3E, left hand panel). When c-Myc was over-expressed, binding of C/EBPα, Mxi1 and Sin3A was dramatically reduced; conversely, the binding of Ovol-1 and Klf4 were enhanced (Fig. 3E; right hand panel). The occurrence of transcription factors at EDC genes was not due to direct transcriptional up- or down-regulation of the transcription factors by c-Myc, and transcription factors profiled here were co-expressed within the same cellular sub-populations during epidermal differentiation (Fig S3A-O). Thus, c-Myc regulates EDC genes by controlling the identity of transcriptional partners present at EDC regulatory regions.
The dramatic binding changes of epidermal-specific transcription factors at the EDC induced by c-Myc suggest that these factors collectively mediate transcriptional activity (Fig. 3E). Among these regulators, only Sin3A principally acts as a transcriptional co-repressor; we therefore considered the possibility that Sin3A directly displaces c-Myc to antagonize its function.
Protein interaction with the Sin3A complex targets c-Myc for degradation
We first tested whether Sin3A and c-Myc interact on a protein level. Although we confirmed that Sin3A co-immunoprecipitated with c-Myc (Fig. 4A), Sin3A and c-Myc binding to the EDC was mutually exclusive in vivo (Fig. 3E), indicating that under physiological conditions any interaction of Sin3A with c-Myc was transient. Because c-Myc expression in wild-type epidermis is too low to perform co-immunoprecipitations, we asked how Sin3A might remove c-Myc from the promoters.
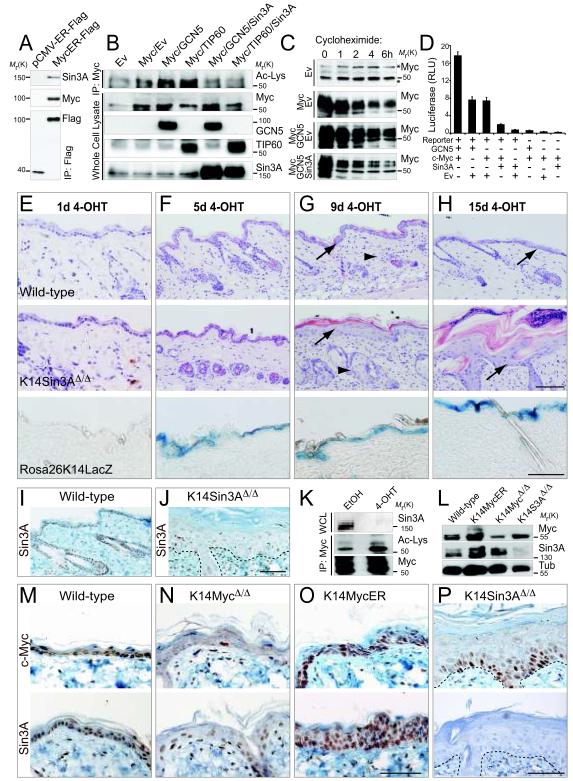
Figure 4. Sin3A causes de-acetylation of c-Myc and loss of Sin3A in skin causes increased thickness of IFE and sebaceous glands.
(A) Sin3A co-immunoprecipitates with Flag-tagged MycER (MycER-Flag) but not Flag-tagged ER only (ER-Flag). The Western Blots were labelled using antibodies for Sin3A, c-Myc and Flag. (B) Sin3A over-expression inhibits GCN5- and TIP60-mediated acetylation of c-Myc. Cells transfected with the constructs indicated were immunoprecipitated for c-Myc (IP; Myc). Acetylated c-Myc was detected by Western blotting using an antibody for acetylated lysines (Ac-Lys). The empty vector (Ev) served as negative control. Western Blot on whole cell lysates confirmed comparable protein levels of c-Myc, GCN5, TIP60 and Sin3A. (C) Western Blot for c-Myc of cells over-expressing the empty vector control (Ev) and c-Myc (Myc) or Myc and GCN5 or GCN5, c-Myc and Sin3A after treatment with cycloheximide for the indicated hours. All blots are exposed for 20 seconds. Star marks unspecific bands. (D) c-Myc luciferase reporter assay in cells expressing the empty vector control (Ev), c-Myc, GCN5 and Sin3A. (+) indicates transfected with the corresponding constructs; (-) means non-transfected. Error bars indicate s.d (n=4 experimental replicates). (E-H) Effect of 4-OHT treatment for 1 (E), 5 (F), 9 (G), and 15 (H) days on skin of wild-type (upper panels), K14Sin3AΔ/Δ (middle panels) and a Cre-reporter mouse (RosaK14LacZ). (I,J) Skin sections from wild-type (I) and K14Sin3AΔ/Δ (J) mice labelled with a Sin3A antibody. (K) Western Blot for Sin3A (upper panels), acetylated lysines of immuno-precipitated c-Myc (middle panel) and total c-Myc (lower panel) in K14Sin3AΔ/Δ primary mouse keratinocytes treated with 4-OHT or ethanol (EtOH) as negative control. (L) Protein expression of endogenous c-Myc and Sin3A in skin of the indicated transgenic or wild-type mice. Tubulin (Tub) served as a loading control. (M-P) Skin sections from wild-type and the indicated transgenic mice were stained for c-Myc (upper panels) and Sin3A (lower panels). Dotted lines in (J,P) mark the basement membrane. Scale bars: 100 μm (E-H, M-P). Uncropped images of blots are shown in Supplementary Figure S9.
c-Myc activates transcription by forming a heterodimer with Max and recruitment of multi-protein complexes including the histone acetyl transferases GCN5 and TIP60 30-32. Unlike c-Myc, Max also heterodimerizes with Myc-antagonists of the Mxd (Mad) family 27,33,34. Mxd-Max heterodimers repress transcription by recruiting Sin3A/histone deacetylase complexes (HDACs) 35-37. Since the enzymatic activity of the Sin3A co-repressor complex is mediated via HDACs, we speculated that the complex may inhibit acetylation of c-Myc. Acetylation of c-Myc by GCN5 and TIP60 increases its protein stability and transactivation 38.
Over-expression of GCN5 and TIP60 led to increased acetylation of c-Myc compared to controls (Fig. 4B; Ac-Lys) 38. In contrast, acetylation of c-Myc was decreased in cells over-expressing both the acetyltransferases and Sin3A (Fig. 4B; Ac-Lys). Western Blotting for c-Myc, GCN5, TIP60 and Sin3A confirmed that the proteins were expressed at similar levels (Fig. 4B). To test whether de-acetylation of c-Myc caused its degradation, we inhibited protein translation using cycloheximide. The protein half-life of c-Myc was reduced when Sin3A was co-expressed (Fig. 4C; Fig. S4A). c-Myc RNA levels were the same when GCN5 or Sin3A were over-expressed and GCN5 and Sin3A proteins were expressed equally (Fig. S4B,C). Reporter assays demonstrated that transactivation of c-Myc was induced by GCN5 and reduced by Sin3A (Fig. 4D).
Depletion of Sin3A in skin induces proliferation and differentiation
To test whether Sin3A negatively regulates c-Myc target genes in vivo, we generated an inducible, conditional knockout mouse model for Sin3A in the epidermis (Methods) (Fig. S4D). We removed Sin3A by topical application of 4-OHT to the mouse back skin and confirmed down-regulation of Sin3A on both RNA and protein level (Fig. S4E-G,L; Fig. 4I,J,L).
Application of 4-OHT in a time course led to an increase of thickness in the IFE (arrows) and sebaceous glands (arrowheads) only in Sin3A-depleted skin (Fig. 4E-H; upper and middle panels). Activation of Cre-recombinase in RosaLacZ reporter mice confirmed homogenous recombination in the IFE after 5 days of 4-OHT treatment (Fig. 4E-H; lower panels). Neither control mice nor mice with deleted Sin3B (K14Sin3BΔ/Δ) showed an epidermal phenotype in response to 4-OHT treatment for 14 or 28 days, respectively (Fig. S4H-J), excluding the possibility that the phenotype was due to nonspecific effects of Cre-activity, and demonstrating that Sin3B is dispensable for skin homeostasis.
In wild-type skin, Sin3A was expressed throughout the IFE, the sebaceous glands and the hair follicles (Fig. 4I). In tail skin IFE, Sin3A-positive nuclei were enriched towards the basal, undifferentiated layers (Fig. S4K). Higher expression of Sin3A in undifferentiated cells of the IFE was confirmed on RNA levels (Fig. S3I; left hand panel).
To confirm a transcriptional interaction between Sin3A and c-Myc in vivo, we isolated epidermal cells from K14Sin3AΔ/Δ mice and tested whether acetylation levels of c-Myc changed with deletion of Sin3A (Fig. 4K). Using ImageJ, we measured a 1.4-fold increase in overall acetylation of c-Myc (Ac-Lys) relative to total c-Myc when Sin3A was absent (Fig. 4K). Accordingly, nuclear c-Myc was enriched in the IFE when Sin3A was deleted (Fig. 4L-P; upper panels). Sin3A was highly expressed in both c-Myc depleted and over-expressing skin (Fig. 4L; middle panel and M-P; lower panels).
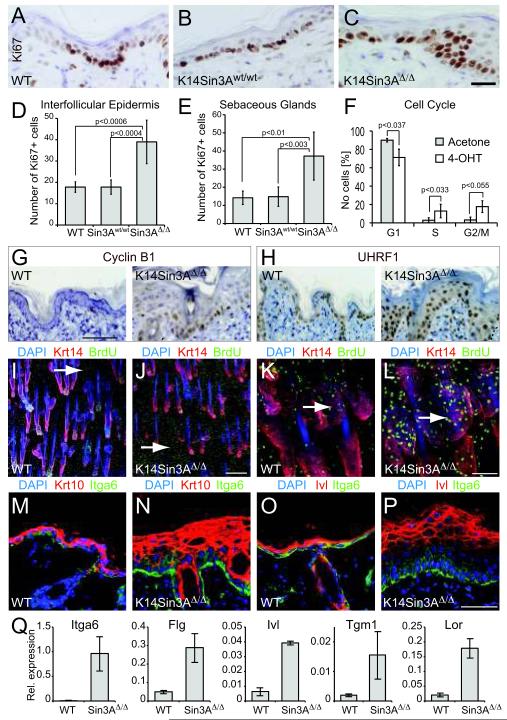
Up-regulation of c-Myc in the absence of Sin3A led to enhanced proliferation in the IFE (Fig. 5A-C). The number of Ki67-positive cells increased around 2-fold in both the IFE and sebaceous glands (Fig. 5D,E). Analysis of the cell cycle revealed a significant increase in number of cells in S- and G2/M-phase upon treatment with 4-OHT (Fig. 5F), leading to accumulation of Cyclin B1, UHRF1 and Mcm2 positive nuclei (Fig. 5G,H; Fig. S4M).
Figure 5. Sin3A-deletion causes epidermal proliferation and differentiation.
(A-C) Labelling of cycling cells in IFE of wild-type mice (WT) (A), mice expressing K14CreER only (K14Sin3Awt/wt) (B) and mice with deleted Sin3A (K14Sin3AΔ/Δ) (C) using an antibody for Ki67 (brown). (D) Quantification of (A-C). (E) Quantification of Ki67-positive cells in sebaceous glands. (F) Cell cycle profile of K14Sin3AΔ/Δ epidermal cells treated with 4-OHT or acetone as a control. (G,H) Immunohistochmistry for Cyclin B1 (G) and UHRF1 (H) in wild-type (WT) (left hand panels) and K14Sin3AΔ/Δ (right hand panels) skin. (I-L) Whole mount labelling of wild-type (WT) (I,K) and K14Sin3AΔ/Δ (J,L) tail skin for cycling cells using an antibody for BrdU (green), keratin 14 (Krt14) (red). Arrows indicate BrdU-positive nuclei in IFE (I,J) and sebaceous glands (K,L). (M-P) Skin sections of wild-type (WT) (M,O) and K14Sin3AΔ/Δ (N,P) labelled for Itgα6 (green), keratin 10 (Krt10) (red) (M,N) and involucrin (Ivl) (red) (O,P). Nuclei are counter-stained with DAPI (blue) (M-P). (Q) QPCR for RNA expression for the indicated genes in skin of wild-type (WT) and K14Sin3AΔ/Δ (Sin3AΔ/Δ) animals. Scale bars: 25 μm (A-C); 100 μm (G,H); 250 μm (I,J); 75 μm (K,L); 50 μm (M-P). Error bars in indicate s.d (D-F: n=5 animals of each mouse line; Q: n= 3 biological replicates averaged over 3 technical replicates each).
Whole mount labelling of tail skin demonstrated that BrdU incorporation in the IFE and sebaceous glands was higher in K14Sin3AΔ/Δ mice than wild-type animals (Fig. 5I-L; arrows). Thus, in contrast to recent findings in mouse embryonic fibroblasts in which deletion of Sin3A caused proliferation arrest and apoptosis 39,40, loss of Sin3A in the epidermis promoted cellular proliferation. This effect was not skin-specific and was also found in testis and salivary glands (Fig. S5A-J).
To analyse whether the increased proliferation led to induced differentiation, we labelled sections of back skin for the terminal differentiation markers keratin 10 (Krt10) and involucrin (Ivl). Both markers increased in skin lacking Sin3A (Fig. 5M-P). Itgα6-positive, undifferentiated layers of the epidermis were also increased upon Sin3A deletion (Fig. 5M-P). Induced expression of markers for both undifferentiated and differentiated keratinocytes in skin of K14Sin3AΔ/Δ mice was confirmed by QPCR (Fig. 5Q).
In summary, deletion of Sin3A in the epidermis increased cellularity in the IFE, which was due to increased proliferation. This increase in cell division was accompanied by enhanced differentiation, demonstrating that Sin3A is dispensable for cells to complete the epidermal terminal differentiation program.
Loss of Sin3A leads to increased promoter occupancy of c-Myc at EDC genes
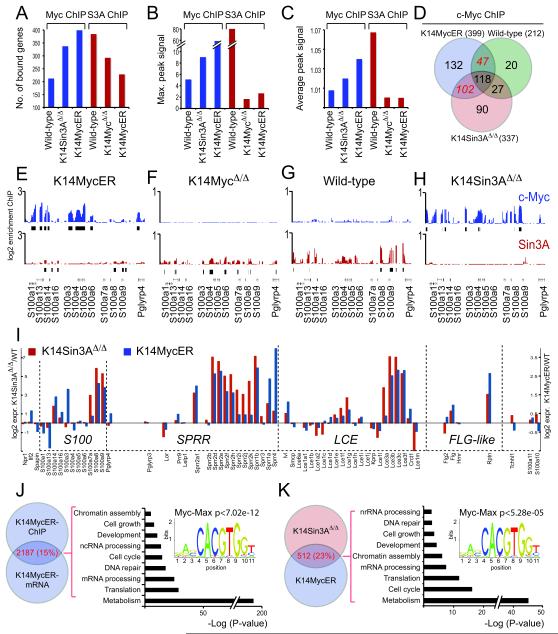
To test whether loss of Sin3A led to up-regulation of c-Myc-target genes, we performed ChIP on chip experiments on chromosome 3 for c-Myc in skin of K14Sin3AΔ/Δ mice as well as wild-type and K14MycER animals (Fig. 6A-C, blue bars; Table S5). Both, the number of c-Myc bound genes and the peak signal, measured as maximal and average peak signal of the combined data, increased when Sin3A was deleted compared to wild-type skin (Fig. 6A-C, blue bars). The number of c-Myc bound genes commonly found in K14Sin3AΔ/Δ and K14MycER but not wild-type epidermis increased more than two-fold compared to the number of genes with c-Myc binding only found in wild-type and K14MycER skin (Fig. 6D; Table S5).
Figure 6. Re-activation of Myc-target genes in response to Sin3A-deletion.
(A-C) ChIP on chip analysis on chromosome 3 using c-Myc (blue) and Sin3A (red) antibodies using skin of wild-type and indicated transgenic animals. Shown are the total number of bound genes (A), the maximal peak signal (B), and the average peak signal (C) of the combined replicates. (D) Venn diagram (http://www.pangloss.com/seidel/Protocols/venn.cgi) shows increase of c-Myc-binding to genes on chromosome 3 in the absence of Sin3A (K14Sin3AΔ/Δ) compared to control (wild-type). (E-H) Binding of c-Myc (blue) and Sin3A (red) to EDC genes in skin of K14MycER (E), K14MycΔ/Δ (F), wild-type (G) and K14Sin3AΔ/Δ (H) mice. (I) Change of RNA expression levels of EDC genes in K14Sin3AΔ/Δ (red) and K14MycER (blue) mice relative to wild-type mice. (J) Gene ontology categorization of c-Myc-regulated genes by comparing ChIP on chip data with gene expression arrays obtained from K14MycER and control skin after treatment with 4-OHT for 4 days. P-value indicates the statistical significance for over-representation of c-Myc-Max binding sites in the 2187 c-Myc-regulated genes. (K) Gene ontology categorization of c-Myc and Sin3A regulated genes by comparing c-Myc-regulated genes from (I) with differentially expressed genes in K14Sin3AΔ/Δ mice compared to controls after treatment with 4-OHT for 14 days. P-value for over-representation of c-Myc-Max binding sites in the 512 c-Myc/Sin3A-regulated genes.
We also compared Sin3A genomic occupancy in skin of wild-type mice or when c-Myc was either deleted (K14MycΔ/Δ) or over-expressed (K14MycER) (Fig. 6A-C, red bars; Table S5). The number of Sin3A peaks as well as the intensity of the signal in wild-type skin was higher than in epidermis that either over-expressed or lacked c-Myc (Fig. 6A-C, red bars; Fig. S6), indicating that a substantial number of Sin3A target genes required the presence of c-Myc.
If our hypothesis that Sin3A detects and removes c-Myc from promoter regions replacing it with antagonists of the Mxd-family was correct, binding of Mxi1 should similarly to Sin3A depend on the presence and absence of c-Myc. Indeed, the overall number of binding events of Mxi1 remained unchanged in wild-type versus K14Sin3AΔ/Δ epidermis (Fig. S7A, D-G; Table S6). As expected, when c-Myc was over-expressed, the number of Mxi1-binding events was reduced (Fig. S7A; Table S6). Furthermore, deletion of c-Myc led to an increase of both maximum and average peak signal of Mxi1 across chromosome 3 (Fig. S7B,C). Thus, similarly to Sin3A, enrichment of Mxi1 depended on the presence and absence of c-Myc.
As described earlier, at EDC genes occurrence of Sin3A was highest in wild-type, lower in epidermis that lacked c-Myc and absent in c-Myc over-expressing skin (Fig. 6E-G). In contrast, binding of endogenous c-Myc was enriched on EDC genes when Sin3A was deleted (Fig. 6H). Thus, increased differentiation in K14Sin3AΔ/Δ transgenic mice might be due to enhanced c-Myc-binding to EDC genes. Indeed, we found a highly similar pattern in RNA expression levels of EDC genes in epidermis that either lacked Sin3A or over-expressed c-Myc (Fig. 6I). Remarkably, c-Myc bound genes in K14Sin3AΔ/Δ epidermis over-lapped up to 71% with Sin3A binding sites in wild-type skin (Fig. S8A; Table S5) and only 18% of all c-Myc bound genes in K14MycER skin were never occupied by Sin3A in wild-type or c-Myc-transgenic skin (Fig. S8B; Table S5).
Thus, absence of the Sin3A co-repressor complex in the epidermis can lead to an enhanced genomic occupancy of c-Myc.
Sin3A and c-Myc co-operate in regulating genes involved in differentiation and proliferation
We tested whether Sin3A also inhibited Myc-activity at target genes involved in cell growth and proliferation. To identify genes that were commonly regulated by Sin3A and c-Myc, we compared mRNA transcription in skin of both transgenic mouse models (Fig. 6J,K). First, we identified 2187 genes that were potentially directly regulated by c-Myc by comparing c-Myc binding with genes transcriptionally regulated in skin of K14MycER mice (Fig. 6J; left hand panel; Table S7). Then, we identified 512 genes within this set that were also differentially expressed in K14Sin3AΔ/Δ mice compared to control mice (Fig. 6K; left hand panel; Table S7). Gene ontology analysis revealed highly similar gene categories for c-Myc-only and c-Myc-Sin3A regulated genes (Fig. 6J,K; right hand panels) (http://genetrail.bioinf.uni-sb.de/) 41. Promoter motif searches using the 2187 c-Myc-regulated and the 512 c-Myc-Sin3A-regulated genes revealed a statistical significant over-representation of c-Myc-Max binding sites (Fig. 6J,K; right hand panels) (http://159.149.109.9/pscan/) 42. In line with our previous data Klf4 binding sites were also enriched in those genes (Table S8).
We concluded that in addition to EDC genes many other genes involved in cell growth and proliferation were regulated by c-Myc and Sin3A.
Deletion of c-Myc in K14Sin3AΔ/Δ mice restores tissue homeostasis
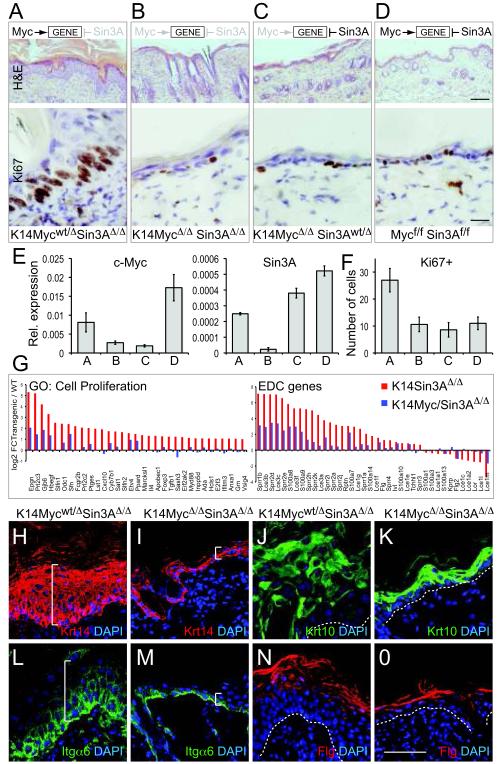
To test whether the phenotype in K14Sin3AΔ/Δ mice was solely dependent on c-Myc, we generated a conditional double knockout mouse for Sin3A and c-Myc in the epidermis (Methods) (Fig. 7). Deletion of c-Myc in K14Sin3AΔ/Δ transgenic mice (K14MycΔ/ΔSin3AΔ/Δ) rescued the skin phenotype: the epidermis resembled that of control mice (Fig. 7A-D). Down-regulation of c-Myc and Sin3A in this transgenic mouse was confirmed by QPCR (Fig. 7E).
Figure 7. Deletion of c-Myc reverts proliferation and differentiation to normal in skin lacking Sin3A.
(A-D) Hematoxylin and Eosin staining (upper panels) and Ki67 labelling (lower panels) of skin sections when Sin3A is deleted (A) (K14Mycwt/Δ Sin3A Δ/Δ), when both c-Myc and Sin3A are deleted (B) (K14MycΔ/Δ Sin3AΔ/Δ), when only c-Myc is deleted (C) (K14MycΔ/ΔSin3Awt/Δ), and when no Cre-recombinase is expressed (D) (Mycf/f Sin3Af/f). Effect of c-Myc and/or Sin3A-deletion on gene-expression is indicated in upper row. (E) QPCR for c-Myc (left hand panel) and Sin3A (right hand panel) RNA in skin of mice indicated in (A-D). (F) Quantification of Ki67 staining from (A-D). Error bars indicate s.d (E: n= 3 biological replicates averaged over 3 technical replicates each; F: n=5 animals of each mouse line). (G) Fold change of expression of genes categorised as cell proliferation (left hand panel) and EDC genes (right hand panel) in skin when only Sin3A is deleted (K14Sin3AΔ/Δ, red) or both c-Myc and Sin3A are deleted (K14Myc/Sin3AΔ/Δ, blue) normalized to the corresponding wild-type (WT) controls. (H-O) Protein expression of keratin 14 (Krt14) (red) (H,I) and Itgα6 (green) (L,M) as markers for undifferentiated epidermal cells and Krt 10 (green) (J,K) and filaggrin (Flg) (red) (N,O) as markers for terminal differentiated cells are reverted to normal when both c-Myc and Sin3A are deleted (K14MycΔ/ΔSin3AΔ/Δ) (I,M,K,O) compared to single deletion of Sin3A (K14Mycwt/Δ Sin3AΔ/Δ) (H,L,J,N). White line in (H,I,L,M) indicates thickness of the IFE. Dotted white line in (J,K,N,O) indicates the basement membrane. Nuclei are counter-stained with DAPI (blue) (H-O). Scale bars: 100 μm (A-D; upper hand panels); 25 μm (A-D; lower hand panels); 50 μm (H-O).
We confirmed that epidermal proliferation in the absence of c-Myc and Sin3A was similar to controls by labelling cycling cells for Ki67 (Fig. 7A-D; lower panels and Fig. 7F). Indeed after deletion of both Sin3A and c-Myc, a number of phenotypes reverted to near normal levels: (i) expression of proliferation and cell cycle genes two-fold up-regulated by deletion of Sin3A alone were now either wild-type levels or decreased (Fig 7G, left hand panel; Fig. S8C); (ii) EDC gene expression perturbed by removal of Sin3A were closer to wild-type levels (Fig. 7G; right hand panel); and, (iii) the balance of both undifferentiated and differentiated epidermal layers was reverted to normal when both genes were deleted (Fig. 7H-O).
Together, the opposing effects of c-Myc and Sin3A on gene transcription are essential to maintain normal skin homeostasis.
Discussion
The underlying mechanisms for c-Myc-induced cellular differentiation are generally thought to be indirect 2. Our data in mammalian epidermis indicates instead that c-Myc directly controls genes involved in regulating differentiation. Myc directly binds to EDC genes at regions enriched for trimethylation of H3K4 (H3K4me3), a chromatin mark found at active genes, as well as a strong determinant for Myc-binding 17,18. The chromatin state does not change after activation of c-Myc, supporting a model whereby the deposition of H3K4me3 is independent of c-Myc 1,17.
The majority of transcriptional changes driven by c-Myc in the genome are immediate. In contrast, the EDC genes directly bound by c-Myc demonstrate a substantial temporal lag in their activation. One explanation for the delay in activation is that the regulatory landscape within the EDC requires remodelling. We demonstrate that specific sets of transcription factors must be removed, and others newly placed, after c-Myc activation. Thus, it appears that c-Myc controls EDC transcription at two levels: it first directly binds EDC genes, but to regulate gene expression, it then requires the formation of a differentiation-specific regulatory transcription factor network.
The transcription factors recruited to the EDC included Klf4 and Ovol-1, both of which are essential for proper barrier function in skin and both negatively regulate Myc-expression 24,26,43. Thus, in later stages of differentiation, Klf4 and Ovol-1 may replace c-Myc at EDC genes to complete the terminal differentiation program.
The transcription factors displaced from the EDC after c-Myc activation include C/EBPα, Mxi1 and its co-repressor Sin3A. We further identify Sin3A as the major opposing factor to c-Myc in controlling cellular proliferation and differentiation processes in skin. Sin3A is an important regulator for the maintenance of the adult muscle and testis 44,45; and Sin3 complexes have been shown to spread immediately down-stream of transcription start sites of both transcribed and repressed genes during myotube differentiation 46.
The exact biological role Sin3A plays in tissue-specific regulatory networks is not entirely understood. Sin3A was originally identified as a co-repressor associated with the Mxd family of c-Myc antagonists, and was shown to suppress epidermal proliferation 27,35-37. We confirm this hypothesis by showing that tissue-specific deletion of Sin3A in the epidermis induces cellular proliferation via a Myc-dependent pathway. Although, we cannot exclude the possibility that loss of Sin3A in skin leads to a destabilization of the Myc-antagonist repressor complex at chromatin, and thereby allows c-Myc to bind to promoter sequences, we find that at least Mxi1’s binding to DNA is unaffected by the absence of Sin3A.
Sin3A is absent from promoter regions when c-Myc is over-expressed and is therefore unable to form a stable repressor complex on chromatin when high levels of c-Myc are bound. Thus, the interaction of Sin3A and c-Myc is most likely indirect via core proteins mediating the de-acetylase enzymatic activity. Although we show that over-expression of Sin3A causes de-acetylation of c-Myc, leading to decreased protein stability and reduced transactivation, further studies will have to reveal the exact molecular pathway of how de-acetylated c-Myc is degraded. Acetylation of c-Myc decreases its ubiquitination, and thus de-acetylation may be linked to the ubiquitin proteosome pathway 47,48.
Although Sin3A limits cell proliferation, Sin3A is entirely dispensable for completion of the terminal differentiation program. The inhibition of proliferation by Mxd-Sin3A complex has been suggested as a mechanism to induce differentiation of primary human keratinocytes 28. However, many non-proliferating cells in vivo are not terminally differentiated cells; for example, in skin a large number of non-cycling, undifferentiated cells are tightly attached to the basement membrane 49. Thus, our data indicates that Sin3A is a proliferation inhibitor, but this function is not coupled to terminal differentiation in skin.
Maintaining homeostasis in mammalian tissues is a balance to avoid over-proliferation or under-proliferation, leading to cancer or tissue failure. How the mechanistic interplay between transcriptional activators and repressors results in stability within tissues has been poorly understood. We identify c-Myc and Sin3A as two master regulators in a mammalian tissue, which can maintain cellular proliferation and differentiation processes by controlling transcriptional balance between activation and repression.
Supplementary Material
Acknowledgements
We thank the CRI Genomics and Bioinformatics Core Facilities. We are most grateful to Patrick McDonel and everybody else who provided us with reagents. In particular we thank Alan Clarke for providing us with the Mycf/f mouse line and Austin G. Smith for advice and helpful comments on the manuscript. We further thank Peter Humphreys, Margaret McLeish and Nigel Miller for their technical support. We gratefully acknowledge the support of the Cambridge Stem Cell Initiative, Stephen Evans-Freke, the ERC (DTO), and the EMBO Young Investigator Programme (DTO). This work was funded by Cancer Research UK (CR-UK) and the Medical Research Council (MRC).
Abbreviation for mouse lines
- f/f
floxed alleles
- Δ/Δ
deleted alleles
Appendix
Methods
Mouse lines and treatment with 4-OHT
All mouse husbandry and experiments were carried out according to the local ethics committee under the terms of a UK Home Office license. All mouse lines were bred to a mixed genetic background of CBA x C57BL/6J. The mouse line K14MycER was kindly provided by F.M. Watt 13. KRT14-cre/Esr1 (The Jackson Laboratory) and Rosa26RLacZ-Cre 50 lines were crossed with mice carrying floxed alleles for Sin3A, Sin3B, and c-Myc and genotyped according to published protocols: Sin3Af/f 39, Sin3Bf/f 51 and Mycf/f 52. Mouse back or tail skin was treated with 4-OHT as described previously 11,53. If not stated otherwise K14MycER mice were treated with 4-OHT for four days, K14MycΔ/Δ mice for 21 days and K14Sin3AΔ/Δ mice for 15 days. Mice were either compared to 4-OHT treated wild-type or vehicle treated (acetone) transgenic littermates.
Illumina gene expression microarrays
250 ng of total RNA was converted to complementary RNA (cRNA) target using the Illumina TotalPrep-96 Kit (Ambion 4397949). Total RNA was reverse transcribed and converted to double-stranded cDNA using a T7 promoter-Oligo(dT) primer and purified with magnetic oligo(dT) beads. This formed the template for an in vitro transcription (IVT) reaction which included a biotinylated nucleotide / ribonucleotide mix for both cRNA amplification and biotin labeling. After purification, quality control and quantity normalization the cRNA was hybridized to arrays (3 Sentrix BeadChip Array MouseWG-6 v2 Part #11278593). Hybridization, washing, staining and scanning were performed according to standard Illlumina protocols (Illumina WGGX DirectHyb Assay Guide 11286331 RevA).
ChIP on chip analysis
Back skin of six mice was pooled and used for one ChIP experiment. Isolation of primary mouse keratinocytes and ChIP was performed as described previously 54. Sonicated fractions were incubated with 10 to 15 μg of the following antibodies: c-Myc (sc-764, Santa Cruz), H3K27me3 (07-449, Millipore), H4K3me3 (ab71998, Abcam or 04-745, Millipore), C/EBPα (sc-9314, Santa Cruz), C/EBPβ (sc-150, Santa Cruz), Sin3A (sc-994, Santa Cruz), Klf4 (sc-20691, Santa Cruz), Ovol-1 and Ovol-2 (kindly provided by X. Dai), AP2γ (05-909, Millipore), Mxi1 (sc1042, Santa Cruz), and Rbp-2 (kindly provided by E.V. Benevolenskaya).
Immunoprecipitated samples were washed at least four times with RIPA buffer [50 mM HEPES pH 7.6, 1 mM EDTA, 0.7% (v/v) Na-deoxycholate, 1% (v/v) NP-40 and 0.5 M LiCl] and once with TE buffer [50 mM Tris pH 8.0, 1 mM EDTA and 1% (w/v) SDS] at 4°C. Samples were eluted in 200 μl of SDS based elution buffer, diluted 1 volume in TE buffer and treated with RNAse and Proteinase K. DNA was extracted with phenol:chloroform and ethanol precipitated. ChIP samples were amplified by ligation mediated PCR (LM-PCR) and Cy3-Cy5 labelled as described previously 55.
Microarray hybridization, washing and scanning was performed at the Genomics core facility of CR-UK (CRI, Cambridge, UK) according to the standard protocols (http://www.genomics.agilent.com/files/Manual/G4481-90010_MammalianProtocol_10.11.pdf). ChIP on chip experiments were performed on mouse whole genome chromosome 3 (Amadid 15317) or proximal promoter Agilent arrays using a minimum of two independent biological replicates.
Microarray and Data processing
Analysis of gene expression data involved the use of scripts written in R (R Development Core Team 2008; www.r-project.org) and Bioconductor (www.bioconductor.org) 56. Differential gene expression analysis was performed using limma 57 and time course analysis using the time course package 58.
ChIP on chip data analysis was also carried out in R using bioconductor, specifically the Ringo package 59. The R script used to analyse the ChIP on Chip data is provided as supplementary material. To generate the scatterplots for the ChIP on chip raw data, the mouse binding events for c-Myc were used as targets to center each window. A half size window corresponding to 3000 bp was defined and we set a minimum number of 3 probes to consider a binding event. Targets were predicted using a 0.90 quantile and genomic regions between 1.0 kb upstream and 0.3 kb downstream of the transcription start site (TSS) of annotated genes from the mouse genome (mm9) were used.
Clustering, annotation, and visualization
Microarray data were clustered using Euclidian distance and the “complete” agglomeration method. These were visualized in R, using the gplots package (Warnes et al. 2010; http://CRAN.R-project.org/package=gplots). Visualization of the time course of expression for individual genes was also done in R.
Histology, BrdU labelling, tissue staining and cell cycle analysis
Tissues were fixed in 4% (w/v) paraformaldehyde and paraffin embedded or for cryosections directly embedded in OCT (Raymond A Lamb). The tissue was sectioned at 5 to 10 μm for hematoxylin and eosin (H&E) staining or immunostaining as described elsewhere 11,12.
Immunohistochemistry was performed using Ventana Discovery (Ventana Medical Systems, Inc) on paraffin embedded tissues fixed in 4% (w/v) paraformaldehyde, following manufacturer’s guidelines. Antigen retrieval was performed using Ventana Cell Conditioning 1 solution (Roche) for 40 minutes at 99°C. Primary antibody incubation was performed for 54 minutes at 37°C. Secondary antibody incubation (Donkey anti-rabbit Ig biotinylated, Jackson or Horse anti-goat Ig biotinylated Vector Labs) (1:1000) was performed for 30 minutes at 37°C. Antibody detection was performed using the DAB Map kit and sections were counterstained using hematoxylin and bluing reagent (Roche).
The primary antibodies used were Sin3A (1:50; sc-767, Santa Cruz), c-Myc (1:25; sc-769, Santa Cruz), Ki67 (1:100; SP6, Vector Labs), Krt14 (1:250; PRB-155P Covance), BrDU (1:100; Abcam), Krt10 (1:250; PRB-159P, Covance), Itga6 (1:250; GoH3 clone, Serotec), Ivl (1:250; ERLI-3)53, Flg (1:250; PRB-417-P, Covance). Cyclin B1 (1:100; LS-C95967, Lifespan Biosciences), UHRF1 (1:100; ab100810, Abcam), Mcm2 (1:100; LS B391, Lifespan Biosciences), C/EBPα (1:100; sc-9314, Santa Cruz).
To determine the pattern of recombination at the Rosa26R reporter locus, β-galactosidase analysis was performed in sectioned skin as described previously 9. For BrdU labeling, mice were injected with 50 mg of BrdU per kg body weight and killed after 2 hours. Whole mounts of mouse tail epidermis were prepared and labeled for immunofluorescence analysis as described previously 53.
White field images were acquired using an Olympus IX80 microscope and a DP50 camera. Confocal images were acquired on a Leica TCS SP5 confocal microscope. Z-stacks were acquired at 100 Hz with an optimal stack distance and 1024x1024 dpi resolutions. Z-stack projections were generated using the LAS AF software package (Leica Microsystems). All the images were processed with Photoshop CS4 (Adobe) software.
For cell cycle analysis, cells were fixed with cold 70% (v/v) ethanol and washed with PBS before being stained with propidium iodide (PI). After incubating the cells for 1 hour in RNase, analysis was carried out on a CyAN ADP analyzer (Beckman Coulter). Cell cycle profile was analysed using FlowJo software.
RNA extraction and semi-quantitative RT-PCR (QPCR)
RNA was isolated from total skin (epidermis and dermis) or cultured cells using trizol reagent (Invitrogen) according to manufacturer’s instructions. Double stranded cDNA was generated from 1 μg RNA using superscript III reverse transcriptase (Invitrogen) following the manufacturer’s instructions. Real-time PCR amplification and analysis was conducted using the 7900HT Real-Time PCR System (Applied Biosystems). The standard amplification protocol was used with pre-designed probe sets and TaqMan Fast Universal PCR Master Mix (2×) (Applied Biosystems). RNA levels were normalized to GAPDH expression using the ΔCt method.
Cell culture, transient transfection, co-immunoprecipitation
The following constructs were cloned into eukaryotic expression vectors: estrogen receptor domain fused to a Flag-tag (ER-Flag), human c-Myc fused to ER and Flag-tag (MycER-Flag) 60, and full-length cDNA for Sin3A. The cytomegalovirus-driven expression vector containing Tip60 was a kind gift from S. Khochbin, hGCN5 was kindly provided by S. Dent and c-Myc was a kind gift of S. McMahon 38.
For luciferase assays, c-Myc, GCN5, Sin3A and the empty vector control were transfected into HEK293 cells and the Myc reporter assays were performed according to the manufacturer’s instructions (Cignal Myc Reporter; Qiagen).
For c-Myc acetylation analysis, COS7 cells were grown in 150 mm2 dishes and transfected at 50% confluence with the empty vector, or c-Myc, Tip60, hGCN5 and Sin3A constructs with Lipofectamine LTX and Plus Reagent (Invitrogen) and harvested after 24 hours. To detect endogenous acetylation levels of c-Myc, epidermal cells were isolated from back skin of K14Sin3Af/f mice and cultured as described previously 11. Cells were treated with 4-OHT or ethanol as a negative control and harvested after 48 hours.
All cells were harvested following trypsinisation and lysed in lysis buffer consisting of PBS without Ca2+ and Mg2+, 0.5% (v/v) NP-40, 0.1% (w/v) sodium deoxycholate, 0.05% (w/v) SDS and protease inhibitors (Roche). Lysates were then centrifuged at 16,000 x g for 10 minutes at 4°C and the supernatant was then added to Protein G Dynabeads (Invitrogen) which had been pre-incubated with 4 μg of rabbit polyclonal anti c-Myc antibody (sc-764; Santa Cruz) for 1 hour at 4°C. Following 2 hour incubation at 4°C, beads were washed 5 times with lysis buffer and immunoprecipitated protein was then eluted with SDS sample buffer at 70°C. SDS-PAGE electrophoresis was then performed on 10% (w/v) gels and proteins were then transferred onto nitrocellulose membranes. Western blotting was followed by incubation with mouse monoclonal anti acetyl-lysine antibody (1:1000; Cell Signalling Technology) to detect the amount of acetylated c-Myc. As controls, Western blotting was also performed on whole cell lysates to detect the amount of total c-Myc (1:250; sc-764, Santa Cruz), Tip60 (1:500: sc-5725, Santa Cruz), hGCN5 (1:500; sc-6303, Santa Cruz), Sin3A (1:250; sc-994, Santa Cruz) and Tubulin (1:500; ab15246, Abcam). To detect the Flag-tag we used ANTI-FLAG (1:500; M2, Sigma). Relative intensity of the bands after Western blotting was performed using ImageJ.
Statistical Analysis
The significance of quantitative data was tested using the unpaired, two-tailed Student’s T test.
Footnotes
COMPETING FINANCIAL INTEREST
The authors declare no competing financial interest.
References
- 1.Eilers M, Eisenman RN. Myc’s broad reach. Genes & development. 2008;22:2755–2766. doi: 10.1101/gad.1712408. doi:22/20/2755 [pii] 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watt FM, Frye M, Benitah SA. MYC in mammalian epidermis: how can an oncogene stimulate differentiation? Nature reviews. 2008;8:234–242. doi: 10.1038/nrc2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sodir NM, Evan GI. Nursing some sense out of Myc. J Biol. 2009;8:77. doi: 10.1186/jbiol181. doi:jbiol181 [pii] 10.1186/jbiol181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soucek L, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson A, Laurenti E, Trumpp A. Balancing dormant and self-renewing hematopoietic stem cells. Current opinion in genetics & development. 2009;19:461–468. doi: 10.1016/j.gde.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Habib T, et al. Myc stimulates B lymphocyte differentiation and amplifies calcium signaling. The Journal of cell biology. 2007;179:717–731. doi: 10.1083/jcb.200704173. doi:jcb.200704173 [pii] 10.1083/jcb.200704173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conacci-Sorrell M, Ngouenet C, Eisenman RN. Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell. 2010;142:480–493. doi: 10.1016/j.cell.2010.06.037. doi:S0092-8674(10)00727-0 [pii] 10.1016/j.cell.2010.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoelzle T, Schwarb P, Trumpp A, Hynes NE. c-Myc affects mRNA translation, cell proliferation and progenitor cell function in the mammary gland. BMC Biol. 2009;7:63. doi: 10.1186/1741-7007-7-63. doi:1741-7007-7-63 [pii] 10.1186/1741-7007-7-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muncan V, et al. Rapid loss of intestinal crypts upon conditional deletion of the Wnt/Tcf-4 target gene c-Myc. Mol Cell Biol. 2006;26:8418–8426. doi: 10.1128/MCB.00821-06. doi:MCB.00821-06 [pii] 10.1128/MCB.00821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawlor ER, et al. Reversible kinetic analysis of Myc targets in vivo provides novel insights into Myc-mediated tumorigenesis. Cancer research. 2006;66:4591–4601. doi: 10.1158/0008-5472.CAN-05-3826. doi:66/9/4591 [pii] 10.1158/0008-5472.CAN-05-3826. [DOI] [PubMed] [Google Scholar]
- 11.Frye M, Gardner C, Li ER, Arnold I, Watt FM. Evidence that Myc activation depletes the epidermal stem cell compartment by modulating adhesive interactions with the local microenvironment. Development (Cambridge, England) 2003;130:2793–2808. doi: 10.1242/dev.00462. [DOI] [PubMed] [Google Scholar]
- 12.Frye M, Watt FM. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr Biol. 2006;16:971–981. doi: 10.1016/j.cub.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 13.Arnold I, Watt FM. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr Biol. 2001;11:558–568. doi: 10.1016/s0960-9822(01)00154-3. [DOI] [PubMed] [Google Scholar]
- 14.Cole MD, Cowling VH. Transcription-independent functions of MYC: regulation of translation and DNA replication. Nat Rev Mol Cell Biol. 2008;9:810–815. doi: 10.1038/nrm2467. doi:nrm2467 [pii] 10.1038/nrm2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cowling VH, Cole MD. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol Cell Biol. 2007;27:2059–2073. doi: 10.1128/MCB.01828-06. doi:MCB.01828-06 [pii] 10.1128/MCB.01828-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. doi:S0092-8674(10)00318-1 [pii] 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guccione E, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nature cell biology. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 18.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. doi:S0092-8674(08)00328-0 [pii] 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs E. Finding one’s niche in the skin. Cell stem cell. 2009;4:499–502. doi: 10.1016/j.stem.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marenholz I, et al. Genetic analysis of the epidermal differentiation complex (EDC) on human chromosome 1q21: chromosomal orientation, new markers, and a 6-Mb YAC contig. Genomics. 1996;37:295–302. doi: 10.1006/geno.1996.0563. [DOI] [PubMed] [Google Scholar]
- 21.Volz A, et al. Physical mapping of a functional cluster of epidermal differentiation genes on chromosome 1q21. Genomics. 1993;18:92–99. doi: 10.1006/geno.1993.1430. doi:S0888-7543(83)71430-8 [pii] 10.1006/geno.1993.1430. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Pasolli HA, Williams T, Fuchs E. AP-2 factors act in concert with Notch to orchestrate terminal differentiation in skin epidermis. The Journal of cell biology. 2008;183:37–48. doi: 10.1083/jcb.200804030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez RG, et al. C/EBPalpha and beta couple interfollicular keratinocyte proliferation arrest to commitment and terminal differentiation. Nature cell biology. 2009;11:1181–1190. doi: 10.1038/ncb1960. [DOI] [PubMed] [Google Scholar]
- 24.Nair M, et al. Ovol1 regulates the growth arrest of embryonic epidermal progenitor cells and represses c-myc transcription. The Journal of cell biology. 2006;173:253–264. doi: 10.1083/jcb.200508196. doi:jcb.200508196 [pii] 10.1083/jcb.200508196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wells J, et al. Ovol2 suppresses cell cycling and terminal differentiation of keratinocytes by directly repressing c-Myc and Notch1. The Journal of biological chemistry. 2009;284:29125–29135. doi: 10.1074/jbc.M109.008847. doi:M109.008847 [pii] 10.1074/jbc.M109.008847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nature genetics. 1999;22:356–360. doi: 10.1038/11926. doi:10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 27.Hurlin PJ, et al. Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. The EMBO journal. 1995;14:5646–5659. doi: 10.1002/j.1460-2075.1995.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurlin PJ, et al. Regulation of Myc and Mad during epidermal differentiation and HPV-associated tumorigenesis. Oncogene. 1995;11:2487–2501. [PubMed] [Google Scholar]
- 29.Klose RJ, et al. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. doi:S0092-8674(07)00194-8 [pii] 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 30.Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science (New York, N.Y. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 31.Prendergast GC, Lawe D, Ziff EB. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- 32.McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayer DE, Kretzner L, Eisenman RN. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. doi:0092-8674(93)90661-9 [pii] [DOI] [PubMed] [Google Scholar]
- 34.Zervos AS, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. doi:0092-8674(93)90662-A [pii] [DOI] [PubMed] [Google Scholar]
- 35.Rao G, et al. Mouse Sin3A interacts with and can functionally substitute for the amino-terminal repression of the Myc antagonist Mxi1. Oncogene. 1996;12:1165–1172. [PubMed] [Google Scholar]
- 36.Laherty CD, et al. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 37.Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. doi:S0092-8674(00)80214-7 [pii] [DOI] [PubMed] [Google Scholar]
- 38.Patel JH, et al. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol. 2004;24:10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. doi:24/24/10826 [pii] 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dannenberg JH, et al. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes & development. 2005;19:1581–1595. doi: 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cowley SM, et al. The mSin3A chromatin-modifying complex is essential for embryogenesis and T-cell development. Mol Cell Biol. 2005;25:6990–7004. doi: 10.1128/MCB.25.16.6990-7004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keller A, et al. GeneTrailExpress: a web-based pipeline for the statistical evaluation of microarray experiments. BMC Bioinformatics. 2008;9:552. doi: 10.1186/1471-2105-9-552. doi:1471-2105-9-552 [pii] 10.1186/1471-2105-9-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zambelli F, Pesole G, Pavesi G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic acids research. 2009;37:W247–252. doi: 10.1093/nar/gkp464. doi:gkp464 [pii] 10.1093/nar/gkp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiol Rev. 2010;90:1337–1381. doi: 10.1152/physrev.00058.2009. doi:90/4/1337 [pii] 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Payne CJ, et al. Sin3a is required by sertoli cells to establish a niche for undifferentiated spermatogonia, germ cell tumors, and spermatid elongation. Stem cells (Dayton, Ohio) 2010;28:1424–1434. doi: 10.1002/stem.464. doi:10.1002/stem.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Oevelen C, et al. The mammalian Sin3 proteins are required for muscle development and sarcomere specification. Mol Cell Biol. 2010 doi: 10.1128/MCB.00975-10. doi:MCB.00975-10 [pii] 10.1128/MCB.00975-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Oevelen C, et al. A role for mammalian Sin3 in permanent gene silencing. Molecular cell. 2008;32:359–370. doi: 10.1016/j.molcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vervoorts J, et al. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 2003;4:484–490. doi: 10.1038/sj.embor.embor821. doi:10.1038/sj.embor.embor821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Popov N, Schulein C, Jaenicke LA, Eilers M. Ubiquitylation of the amino terminus of Myc by SCF(beta-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nature cell biology. 2010;12:973–981. doi: 10.1038/ncb2104. doi:ncb2104 [pii] 10.1038/ncb2104. [DOI] [PubMed] [Google Scholar]
- 49.Blanpain C, Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol. 2009;10:207–217. doi: 10.1038/nrm2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature genetics. 1999;21:70–71. doi: 10.1038/5007. doi:10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 51.David G, et al. Specific requirement of the chromatin modifier mSin3B in cell cycle exit and cellular differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4168–4172. doi: 10.1073/pnas.0710285105. doi:0710285105 [pii] 10.1073/pnas.0710285105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Alboran IM, et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. doi:S1074-7613(01)00088-7 [pii] [DOI] [PubMed] [Google Scholar]
- 53.Braun KM, et al. Manipulation of stem cell proliferation and lineage commitment: visualisation of label-retaining cells in wholemounts of mouse epidermis. Development (Cambridge, England) 2003;130:5241–5255. doi: 10.1242/dev.00703. [DOI] [PubMed] [Google Scholar]
- 54.Hussain S, et al. The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. The Journal of cell biology. 2009;186:27–40. doi: 10.1083/jcb.200810180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ren B, et al. Genome-wide location and function of DNA binding proteins. Science (New York, N.Y. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. doi:10.1126/science.290.5500.2306 290/5500/2306 [pii] [DOI] [PubMed] [Google Scholar]
- 56.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. doi:gb-2004-5-10-r80 [pii] 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smyth G. In: Bioinformatics and Computational Biology Solutions using R and Bioconductor. Carey V, Gentleman R, Dudoit S, Huber W, Irizarry R, editors. Springer; 2005. pp. 397–420. [Google Scholar]
- 58.Tai YC, Speed TP. On gene ranking using replicated microarray time course data. Biometrics. 2009;65:40–51. doi: 10.1111/j.1541-0420.2008.01057.x. doi:BIOM1057 [pii] 10.1111/j.1541-0420.2008.01057.x. [DOI] [PubMed] [Google Scholar]
- 59.Toedling J, et al. Ringo--an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics. 2007;8:221. doi: 10.1186/1471-2105-8-221. doi:1471-2105-8-221 [pii] 10.1186/1471-2105-8-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic acids research. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. doi:5w0032 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.