Abstract
Streptococcus sp. is gram-positive coccus that causes streptococcal infections in fish due to intensification of aquaculture and caused significant economic losses in fish farm industry. A streptococcal infection occurred from cultured diseased olive flounder (Paralichthys olivaceus) in May, 2005 at a fish farm in Jeju Island, Korea. The diseased flounder exhibited bilateral exophthalmic eyes and rotten gills; water temperature was 16~18℃ when samples were collected. Of the 22 fish samples collected, 3 samples were identified as Lactococcus garvieae and 18 samples were identified as Streptococcus parauberis by culture-based, biochemical test. Serological methods such as slide agglutination, hemolysis and antimicrobial susceptibility test were also used as well as multiplex PCR-based method to simultaneously detect and confirm the pathogens involved in the infection. S. parauberis and L. garvieae have a target region of 700 and 1100 bp., respectively. One fish sample was not identified because of the difference in the different biochemical and serological tests and was negative in PCR assay. In the present study, it showed that S. parauberis was the dominant species that caused streptococcosis in the cultured diseased flounder.
Keywords: Lactococcus garvieae, multiplex PCR assay, Streptococcus parauberis, Streptococcus sp.
Introduction
As the fish farming becomes a steadily growing industry, problems of controlling various fish infections are also increasing [1]. During the last decade, streptococcal infection has become a major problem in cultured fish populations [1,4,7,22,23,24] in many countries, including Israel [11,12], Italy [14], Japan [17,18], Spain [22] and the USA [4,12,25]. Through rapidly expanding aquaculture in Korean peninsula, the number of disease problem caused by bacterial pathogens has increased and fish farmers have experienced substantial economic losses due to its heavy stock mortality and impact on marine fish. In May 2005, populations of olive flounder (Paralichthys olivaceus) in a cage-culture facility, Jeju Island, Korea, experienced mortality, wherein the fish exhibited bilateral exophthalmic eyes and rotten gills. This study aims to isolate and identify streptococcal bacteria responsible for the infection of olive flounders.
Materials and Methods
Sample collections
In May 2005, a total of 22 samples of olive flounder (Paralichthys olivaceus) were collected in four different flounder farms; Nam-yang (North), Dun-ji (East), Jung-uo (South) and Yung-lime (West) in Jeju Island. The water temperature during sampling period ranged from 16~18℃.
Isolation and cultivation of bacterial pathogens from flounder
Sterile, swabs from liver, kidney and spleen of affected flounders were streaked on brain heart infusion agar plate (BHIA; Difco, USA) supplemented with 1.5% NaCl. The inoculated plates were incubated at 25℃ for 24 hr. Single colonies from plates with dense, virtually pure culture growth were re-streaked on the same media to obtain pure isolates.
Biochemical analysis
Biochemical tests (acidification of carbohydrates) and enzymatic tests were performed with API 20 STREP and API ZYM test (BioMerieux, France). Tests were inoculated with the pure isolates and read as described by the identification kit. The VITEK (BioMerieux, France) cards were also inoculated and allowed to incubate overnight for automated reading of the reactions.
Hemolysis test
A test for hemolysis was conducted in pure isolates using 5% sheep blood agar (Korea Media, Korea).
Slide agglutination test
The test was performed by mixing a small amount of bacterial colonies with several drops of mouse anti-Strep. iniae serum (Kyoritsu Seiyaku, Japan) diluted in phosphate buffered saline (PBS; Invitrogen, USA).
Antimicrobial susceptibility test
The susceptibility pattern of bacterial isolates to 19 antimicrobial drugs such as amikacin (30 µg), ampicillin (10 µg), carbenicillin (100 µg), cefixime (5 µg), cefoperazone (75 µg), centamicin (10 µg), ciprofloxacin (5 µg), colistin (10 µg), kanamycin (30 µg), nalidixic acid (30 µg), neomycin (30 µg), nitrofurantoin (300 µg), norfloxacin (10 µg), ofloxacin (5 µg), polymyxin b (300 IU/IE/UI), tetracycline (30 µg), tobramycin (10 µg), trimethoprim (5 µg), sulfamethoxazole (23.75 µg)/trimethoprim (1.25 µg) (BBL, USA) were tested and determined by using the standard method of Bauer and Kirby [3] on Muller Hinton agar (Difco, USA).
Extraction of bacterial DNAs
The isolates were grown in BHIA supplemented with 1.5% NaCl. The colonies were picked and re-suspended in 500 µl of sterilized double distilled water; bacterial DNA was then extracted by boiling bacterial cells for 5 min and centrifuged at 6,000 g for 5min. Bacterial DNA was collected on the upper aqueous phase of the supernatant and then stored at -20℃ until used.
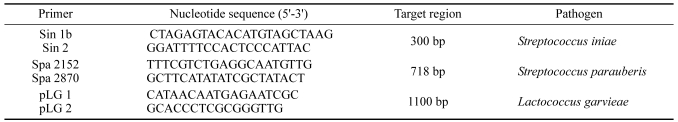
Primers and multiplex PCR amplification
A multiplex PCR assay was used for the simultaneous detection of Streptococcus iniae, Streptococcus parauberis and Lactococcus garviae from pure cultures. The target region and oligonucleotide primer set used for the detection of the three fish streptococcus pathogens in the multiplex PCR are indicated in Table 1. The multiplex PCR was performed in 20 µl reaction mixtures containing DNA template, a 0.05 µM concentration each primer (Bioneer, USA) and AccuPower PCR Premix (1 U Taq DNA polymerase, 250 µM dNTP, 10 mM Tris-HCl, 40 mM KCl, 1.5 mM MgCl2, stabilizer and tracking dye; Bioneer, USA). The amplifications were carried out in a thermocyclers (T-personal 48; Biometra, Germany) with the following parameters: an initial denaturation step of 94℃, 5 min; 30 serial cycles of a denaturation step of 94℃, 30 sec, annealing at 50℃, 30 sec, and extension at 72℃, 30 sec; and a final extension step of 72℃, 7 min. A negative control (no template DNA) and a positive control of S. iniae 0404M obtained from American Type Culture Collection were included in the PCR. The PCR products were analysed by 1.5% agarose gel electrophoresis in 1% Tris-borate-EDTA buffer. Gels were stained with ethidium bromide (0.5 µg/ml), visualized and photographed under UV illumination.
Table 1.
Oligonucleotide primers used in multiplex PCR assay
Results
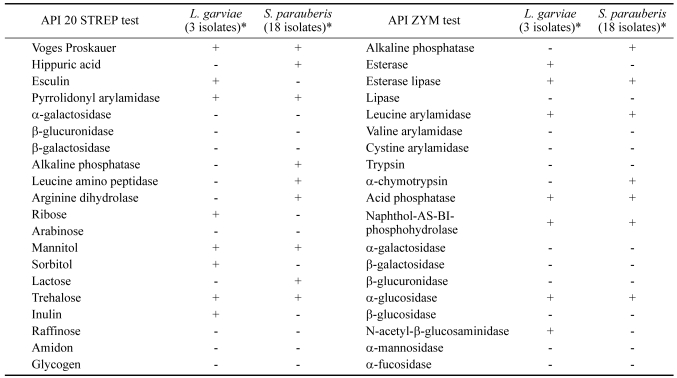
In the API 20 STREP test (Table 2), all 3 L. garviae isolates, were Voges-Proskauer (VP), esculin and pyrrolidonyl arylamidase (PYRA) positive. Ribose, mannitol, sorbitol, trehalose and inulin were acidified by fermentation. All other tests were negative. In all 18 S. parauberis isolates, were VP, hippuric acid, PYRA, alkaline phosphatase, leucine amino peptidase, arginine dihydrolas positive. Mannitol, lactose and trehalose were acidified by fermentation. All other tests were negative. In the API ZYM test (Table 2), all 21 isolates had positive reactions on the following enzymes: esterase lipase, leucine arylamidase, acid phosphatase, naphtholas-bi-phosphohydrolase and α-glucosidase. Three isolates of L. garviae had positive reactions for esterase and N-acetyl-β-glucosaminidase and 18 isolates of S. parauberis had positive reactions for alkaline phosphatase and α-chymotrypsin. All isolates had negative reactions for lipase, valine arylamidase, cystine arylamidase, trypsin, β-galactosidase, β-glucuronidase, β-glucosidase, α-mannosidase and α-fucosidase.
Table 2.
Biochemical characteristics of Streptococcus sp. isolated from diseased flounders
* +; positive, -; negative.
All 21 isolates reacted similarly in VITEK, and positive reactions were observed in peptone base, optochin, 10% bile, dextrose, mannitol, salicine, sorbitol, sucrose, trehalose, cellobiose, ribose. All isolates had negative reactions for bacitracin, 40% bile, arginine, urea, lactose, raffinose, arabinose, pyruvate, inulin, melibiose, melezitose, catalase and β-hemolysis. Three isolates of L. garviae also had positive reactions for hemicellulase, 6% sodium choloride, esculin, tetrazolium red, novobiocin, pullulan. A 18 isolates of S. parauberis had negative reactions for hemicellulase, 6% sodium choloride, esculin, tetrazolium red, novobiocin and pullulan.
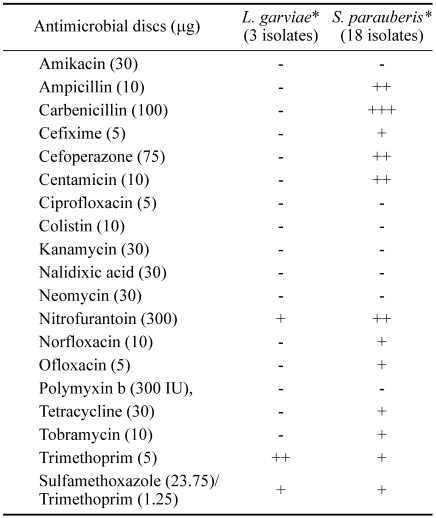
A test for hemolysis was conducted in blood agar plate incubated at 25℃ for 24 hr. All isolates were non β-hemolytic. By slide agglutination test, 3 isolates of L. garviae and 18 isolates of S. parauberis were all negative for agglutination against anti S. iniae. The susceptibility pattern of the bacterial isolates from 19 antimicrobial drugs is shown in Table 3. For L. garviae, three isolates were sensitive to trimethoprim, nitrofurantoin and sulfamethoxazole/trimethoprim. For S. parauberis, 18 isolates were sensitive to carbenicillin, ampicillin, cefoperazone, centamicin, nitrofurantoin, cefixime, norfloxacin, ofloxacin, tetracycline, tobramycin, trimethoprim and sulfamethoxazole/trimethoprim.
Table 3.
Antimicrobial susceptibility test of Streptococcus sp. isolated from diseased flounders
* -; resistant, +; susceptible, ++; more susceptible, +++; most susceptible.
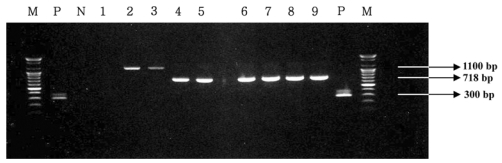
The multiplex PCR assay resulted in the amplification of bands of 3 samples at 1,100 bp for L. garviae and 18 samples at 718 bp for S. parauberis as shown in Fig. 1. One sample is negative for streptococcocal infection. Specific positive amplifications in all samples were consistently observed only for each corresponding pathogen, while no DNA amplifications were observed in other non-targeted bacteria.
Fig. 1.
Representative amplification products obtained using the multiplex PCR assay for detection of streptococcus species in flounder fish. Lanes M, 100-bp DNA ladder; lanes P, positive control (S. iniae 0404M, 300 bp); lane N, negative control; lane 1, negative for streptococcal infection; lanes 2 and 3, positive for L. garviae (1,100 bp); lanes 4~9, positive for S. parauberis (718 bp).
Discussion
Streptococcosis has been one of the infections associated with acute to chronic mortalities in aquaculture species such as olive flounders in Korea. High mortality rate usually occurs in many flounder farms especially during warm water season. Presumptive diagnosis of streptococcosis is based on clinical signs, including the isolation of gram-positive cocci in internal organs. In this study, the diseased flounders exhibited bilateral exopthalmic eyes, hemorrhages on the operculum and gills and distended abdomen. Bacterial isolates obtained from diseased flounders were gram positive cocci/ovoid cells in pairs or short chain, non-motile oxidase and catalase negative. Fish with warm-water streptococcocis exhibit very similar symptoms and clinical signs regardless of the etiological agent [5,10,11,12,13,19,21]. These species of gram positive cocci are warm-water streptococcocis-associated pathogens, because they occurred at water temperature between 16~18℃. Musquiz et al. [21] reported streptococcosis outbreaks that occur at water temperatures above 15℃ and warm-water streptococcosis are usually produced by L. garviae, S. parauberis, S. iniae and S. difficilis. Furthermore, characterization (based on their cultural, morphological and biochemical reactions using the API 20 STREP analytical profile index and VITEK microbiology reference manual) showed two types of streptococcal species that were presently infecting cultured flounders at Jeju Island. These isolates were phenotypically identified as L. garviae and S. parauberis.
The existence of different types of streptococcus species emphasizes the difficulties of definitive identification based on phenotypic traits alone. Therefore, final identification cannot be determined without the support of genetic data. In the present study, the multiplex PCR (m-PCR) assay confirmed and resulted in the amplification of bands of 1,100 bp for 3 isolates of L. garviae and 18 isolates of S. parauberis. The PCR amplification of species-specific isolates of L. garviae and S. parauberis in this study offers a rapid and sensitive method by which to identify both biochemically and serologically indistinguishable species. Moreover, individual PCR assays have been developed for detection and identification of the fish pathogens associated with warm-water streptoccococis [6,26,29]. A large number of individual PCR assays would be necessary if single primer sets are used on a large number of clinical samples, which can be relatively costly and time-consuming process. The simultaneous detection of several pathogens with an m-PCR that was developed by Mata et al. [20] is an effective tool for the rapid and specific detection of pathogens especially involved in warm-water streptococcosis.
Streptococcosis caused by L. garviae has already been reported in several species of cultured marine and freshwater fish such as olive flounder (Paralichthys olivaceus) and Korean rockfish (Sebastes schlegeli) [19]; yellowtail (Seriola quinqueradiata) [17,19]; grey mullet (Mugil cephalus) [8] and rainbow trout (Salmo gardneri) [14]. It was also reported that they cause infection in human and cattle [9,15]. The 3 isolates of L. garviae in this study showed almost similar result in the fermentation and hydrolysis reactions and corresponded to the investigations carried out for L. garviae in fish reported by Chen et al. [8], Eldar and Ghittino [13] and Eldar et al. [14]. Lactococcus garviae strains were mostly sensitive to trimethoprim antibiotics.
On the other hand, S. parauberis (formerly known as S. uberis genotype II) was a pathogen that causes bovine mastitis in cattle [27]. It was also reported that they cause infection in cultured juvenile and adult turbot fish Scopthalmus maximus [2,10]. As far as S. parauberis is concerned, Garvie and Bramley [16], Williams and Collins [27] and later Collins et al. [9] described that, after cultivation on sheep blood agar plate, S. parauberis appeared to be α-hemolytic or non-hemolytic. Domenech et al. [10] cultured α-hemolytic S. parauberis strains isolated from diseased turbots. Corresponding to these results, the isolates of S. parauberis strains of this study also exhibited an α-hemolysis. These results were comparable with the results of the study conducted by Domenech et al. [10], Garvie and Bramley [16], William and Collins [27] and the 18 S. parauberis strains of the this investigation exhibited almost identical fermentation and hydrolysis reactions. Furthermore, all 18 isolates of the this study were pyrrolidonyl arylamidase positive and β-glucuronidase enzyme negative. The present results also corresponded to the investigations carried out for S. parauberis reported by Domenech et al. [10] and Williams and Collins [28] S. parauberis strains were mostly sensitive to carbenicillin antibiotic.
Based on physiological, biochemical properties and molecular analysis documented by various authors and also the results of this study, it identifies and confirms that streptococcus species are major pathogens that cause outbreaks of disease in the cultured flounders in Jeju Island especially during warmwater season. The dominant strain causing streptococcosis is the S. parauberis. It is the first reported case associated with fish disease in Korea especially in Jeju Island. These findings must alarm the fish producers. Then should recognize that diseases due to streptococcosis are likely to become frequent in the future.
Acknowledgments
This study was supported by the Korea Research Foundation Grant (KRF-005-E00076), the Seoul National University (550-20040025) and the Research Institute for Veterinary Science.
References
- 1.Al-Harbi, Ahmed H. First Isolation of Streptococcus sp. from hybrid tilapia (Orcheochromis niloticus x O. aureus) in Saudi Arabia. Aquaculture. 1994;128:195–201. [Google Scholar]
- 2.Austin B, Austin DA, editors. Diseases of Farmed and Wild Fish. Berlin: Springer-Verlag; 1990. pp. 13–15. [Google Scholar]
- 3.Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- 4.Baya AM, Lupiani B, Hetrick FM, Robertson BS, Lukacovic R, May E, Poukish C. Association of Streptococcus sp. with mortalities in the Chesapeake Bay and its tributaries. J Fish Dis. 1990;13:251–253. [Google Scholar]
- 5.Bercovier H, Ghittino C, Eldar A. Immunization with bacterial antigens: infectious with streptococci and related organism. Dev Biol Stand. 1997;90:153–160. [PubMed] [Google Scholar]
- 6.Berridge BR, Bercovier H, Frelier PF. Streptococcus agalactiae and Streptococcus difficile 16S-23S intergenic rDNA: genetic homogeneity and species-specific PCR. Vet Microbiol. 2001;78:165–173. doi: 10.1016/s0378-1135(00)00285-6. [DOI] [PubMed] [Google Scholar]
- 7.Chang PH, Plumb JA. Histopathology of experimental Streptococcus sp. infection in Tilapia, Oreochromis niloticus (L.), and channel catfish, Ictalurua punctatus (Rafinesque) J Fish Dis. 1996;19:235–241. [Google Scholar]
- 8.Chen SC, Liaw LL, Su HY, Ko SC, Wu CY, Chang HC, Tsai YH, Yang KL, Chen YC, Chen TH, Lin GR, Cheng SY, Lin YD, Lee JL, Lai CC, Weng YJ, Chu SY. Lactococcus garvieae, a cause of disease in grey mullet, Mugil cephalus L., in Taiwan. J Fish Dis. 2002;25:727–732. [Google Scholar]
- 9.Collins MD, Farrow JAE, Phillips BA, Kandler O. Streptococcus garvieae sp. nov. and Streptococcus plantarum sp. nov. J Gen Microbiol. 1984;129:3427–3431. doi: 10.1099/00221287-129-11-3427. [DOI] [PubMed] [Google Scholar]
- 10.Domenech a, Fernandez-Garayzabal JF, Pascual C, Garcia JA, Cutuli MT, Moreno MA, Collins MD, Dominguez L. Streptococcosis in cultured turbot, Scopthalmus maximus (L.), associated with Streptococcus parauberis. J Fish Dis. 1996;19:33–38. [Google Scholar]
- 11.Eldar A, Bejerano Y, Bercovier H. Streptococcus shiloi and Streptococcus difficile: two new streptococcal species causing a meningoencephalitis in fish. Curr Microbiol. 1994;28:139–143. [Google Scholar]
- 12.Eldar A, Frelier PF, Assenta L, Varner PW, Lawhon S, Bercovier H. Streptococcus shiloi, the name for an agent causing septicemic infection in fish, is a junior synonym of Streptococcus iniae. Int J Syst Bacteriol. 1995;45:840–842. doi: 10.1099/00207713-45-4-840. [DOI] [PubMed] [Google Scholar]
- 13.Eldar A, Ghittino C. Lactococcus garvieae and Streptococcus iniae infections in rainbow trout, Oncorhynchus mykiss: similar but different diseases. Dis Aquat Org. 1999;36:227–231. doi: 10.3354/dao036227. [DOI] [PubMed] [Google Scholar]
- 14.Eldar A, Ghittino C, Asanta L, Bozzetta E, Goria M, Prearo M, Bercovier H. Enterococcus seriolicida is a junior synonym of Lactococcus garviae, a causative agent of septicemia and meningoencephalitis in fish. Curr Microbiol. 1996;32:85–88. doi: 10.1007/s002849900015. [DOI] [PubMed] [Google Scholar]
- 15.Elliot JA, Collins MD, Piggott NE, Facklam RR. Differentiation of Lactococcus lactis and Lactococcus garvieae from humans by comparison of whole cell protein profile patterns. J Clin Microbiol. 1991;29:2731–2734. doi: 10.1128/jcm.29.12.2731-2734.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garvie EI, Bramley A. Streptococcus uberis: an approach to its classification. J Appl Bacteriol. 1979;46:295–304. doi: 10.1111/j.1365-2672.1979.tb00855.x. [DOI] [PubMed] [Google Scholar]
- 17.Kitao T. Streptococcal infection. In: Inglis V, Roberts RJ, Bromage NR, editors. Bacterial Diseases of Fish. Oxford: Blackwell; 1993. pp. 196–210. [Google Scholar]
- 18.Kusuda R, Kawai K, Toyoshima T, Komatsu I. A new pathogenic bacterium belonging to genus Streptococcus isolated from an epizootic of cultured yellowtail. Bull Jpn Soc Sci Fish. 1976;42:1345–1352. [Google Scholar]
- 19.Lee DC, Lee JI, Park CI, Park SI. The study on the causal agent of streptococcosis (Lactococcus garviae), isolated from cultured marine fish. J Fish Pathol. 2001;14:71–80. [Google Scholar]
- 20.Mata AI, Gibello A, Casamayor A, Blanco MM, Dominguez L. Multiplex PCR assay for detection of Bacterial Pathogens associated with warm-water streptococcosis in fish. Appl Environ Microbiol. 2002;68:5177–5180. doi: 10.1128/AEM.70.5.3183-3187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muzquiz JL, Royo FM, Ortega C, De Blas I, Ruiz I, Alonso JL. Pathogenicity of streptococcosis in rainbow trout (Orcorhynchus mykiss): dependence on age of diseased fish. Bull Eur Assoc Fish Pathol. 1999;19:114–119. [Google Scholar]
- 22.Nieto JM, devesa S, Quiroga A, Toranzo AE. Pathology of Enterococcus sp. in farmed turbot, Scopthalmus maximus L. J Fish Dis. 1995;18:21–30. [Google Scholar]
- 23.Pier GB, Madin SH. Streptococcus iniae sp. nov., a beta-hemolytic streptococcus isolated from an Amazon freshwater dolphin, Inia geoffrensis. Int J Syst Bacteriol. 1976;26:545–553. [Google Scholar]
- 24.Plumb JA. Major diseases of striped bass and redfish. Vet Hum Toxicol. 1991;33:34–39. [PubMed] [Google Scholar]
- 25.Rasheed V, Plumb J. Pathogenicity of a non-hemolytic group B Streptococcus sp. in gulf killifish, Fundulus grandis (Baird and Girard) Aquaculture. 1984;37:97–105. [Google Scholar]
- 26.Riffon R, Sayasith K, Khalil H, Dubreuil P, Drolet M, Lagace J. Development of a rapid and sensitive test for identification of major pathogens in bovine mastitis by PCR. J Clin Microbiol. 2001;39:2584–2589. doi: 10.1128/JCM.39.7.2584-2589.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams AM, Collins MD. Molecular taxonomic studies on Streptococcus uberis types I and II. Description of Streptococcus parauberis sp. nov. J Appl Bacteriol. 1990;68:485–490. doi: 10.1111/j.1365-2672.1990.tb02900.x. [DOI] [PubMed] [Google Scholar]
- 28.Williams AM, Collins MD. DNA fingerprinting of Streptococcus uberis based on polymorphism of DNA encoding rRNA. Lett Appl Microbiol. 1991;12:23–28. doi: 10.1111/j.1472-765x.1991.tb00493.x. [DOI] [PubMed] [Google Scholar]
- 29.Zlotkin A, Eldar A, Ghittino C, Bercovier H. Identification of Lactococcus garveiae by PCR. J Clin Microbiol. 1998;36:983–985. doi: 10.1128/jcm.36.4.983-985.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]