Abstract
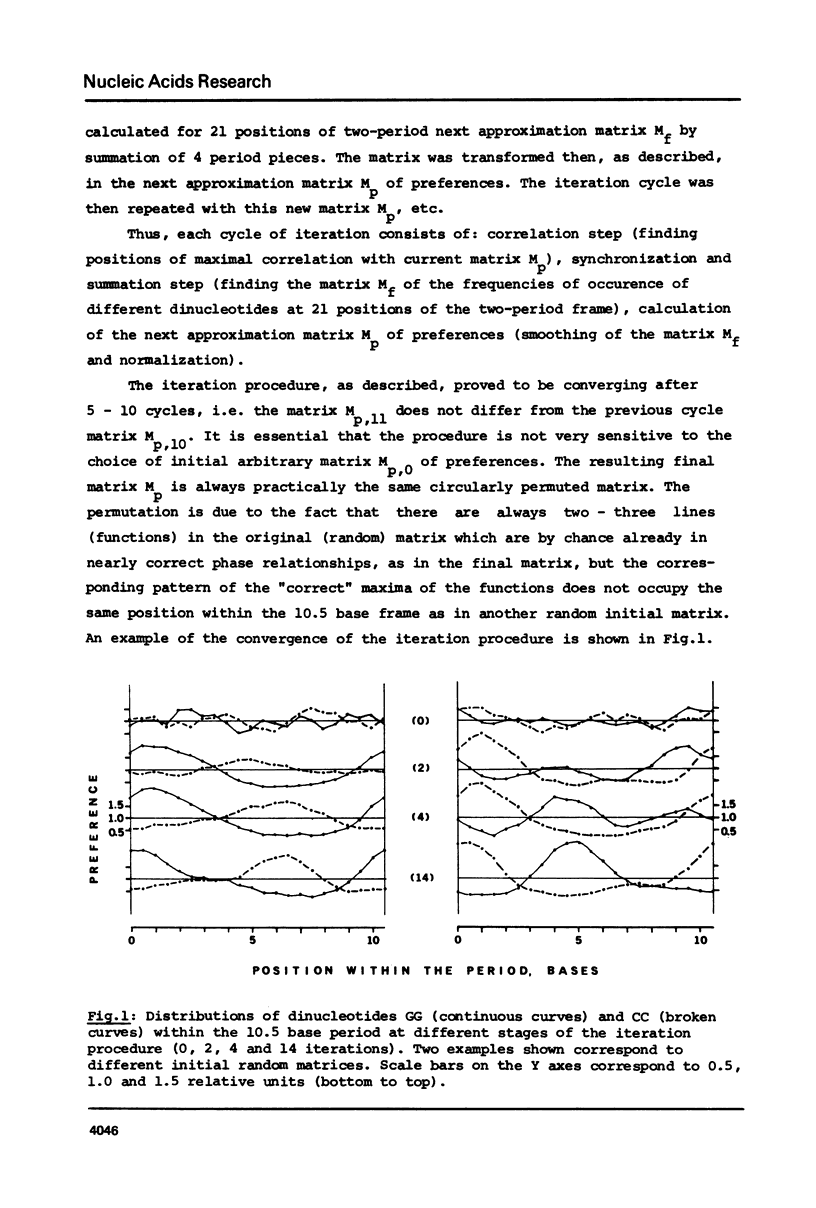
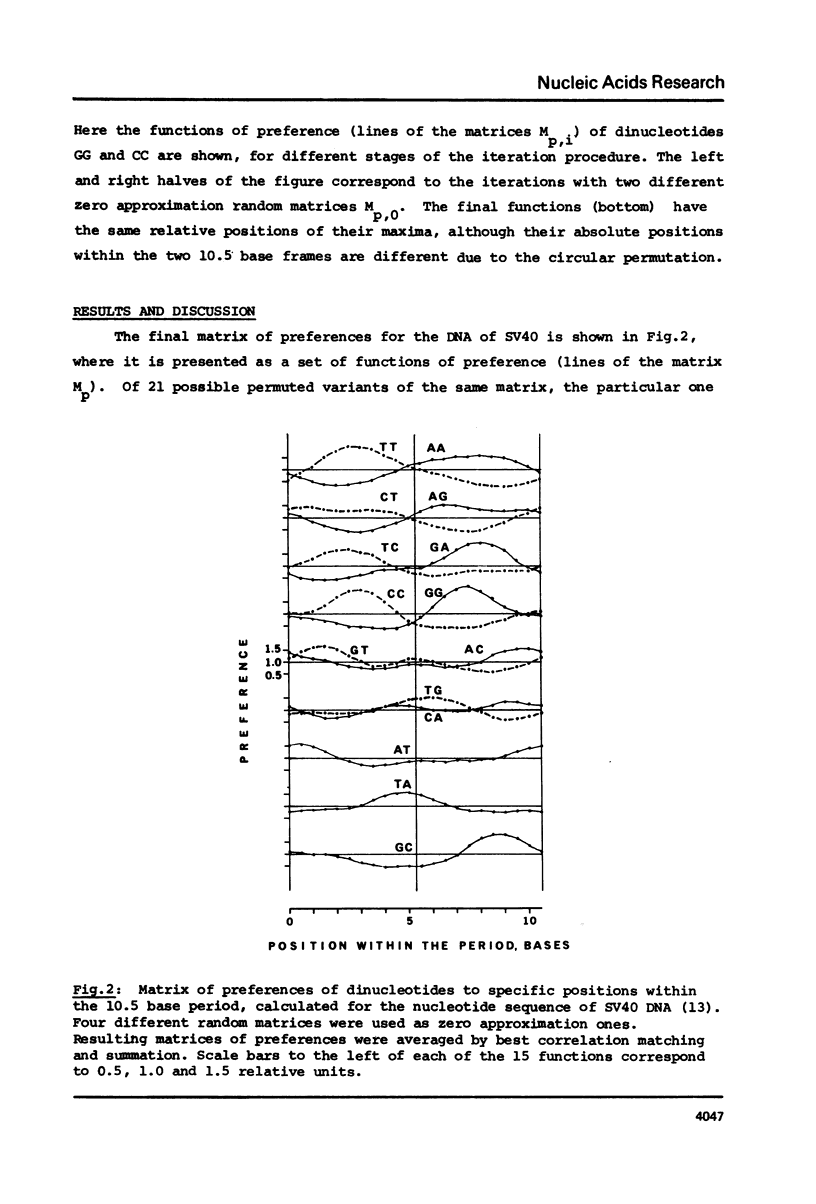
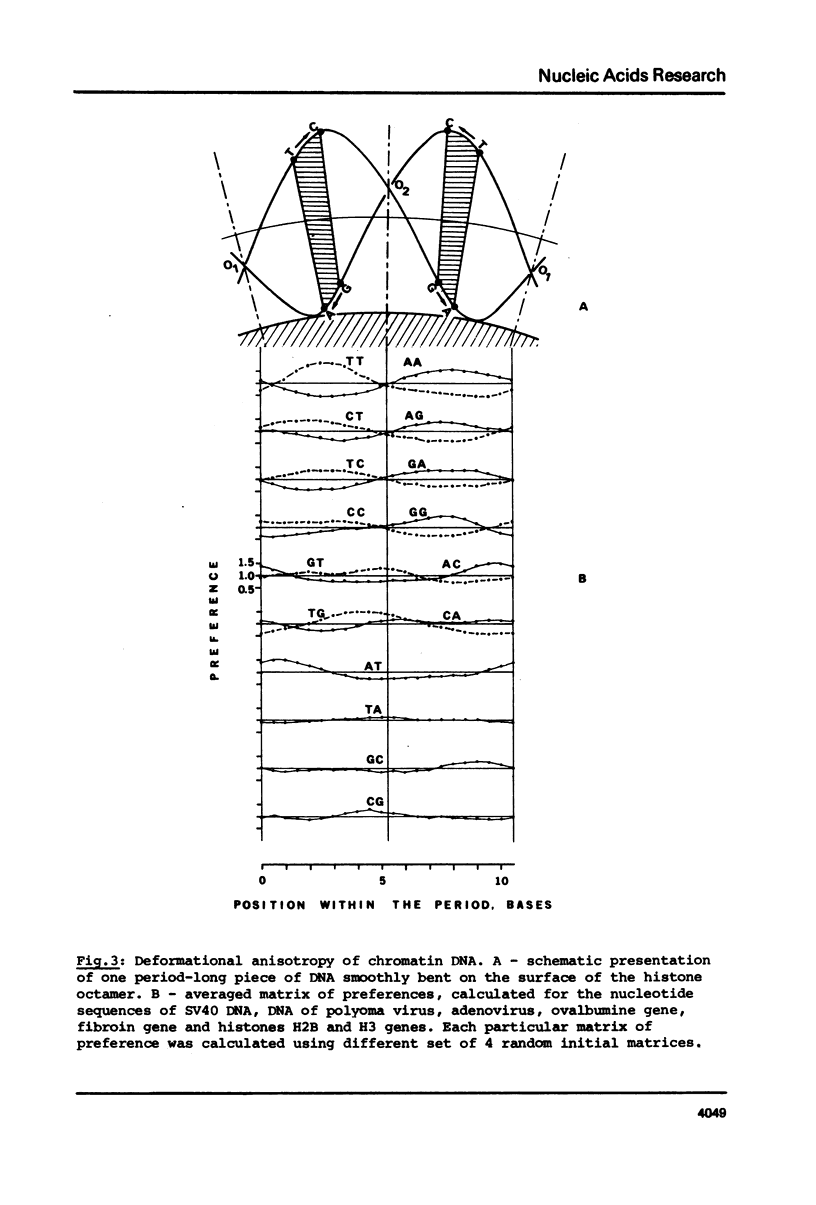
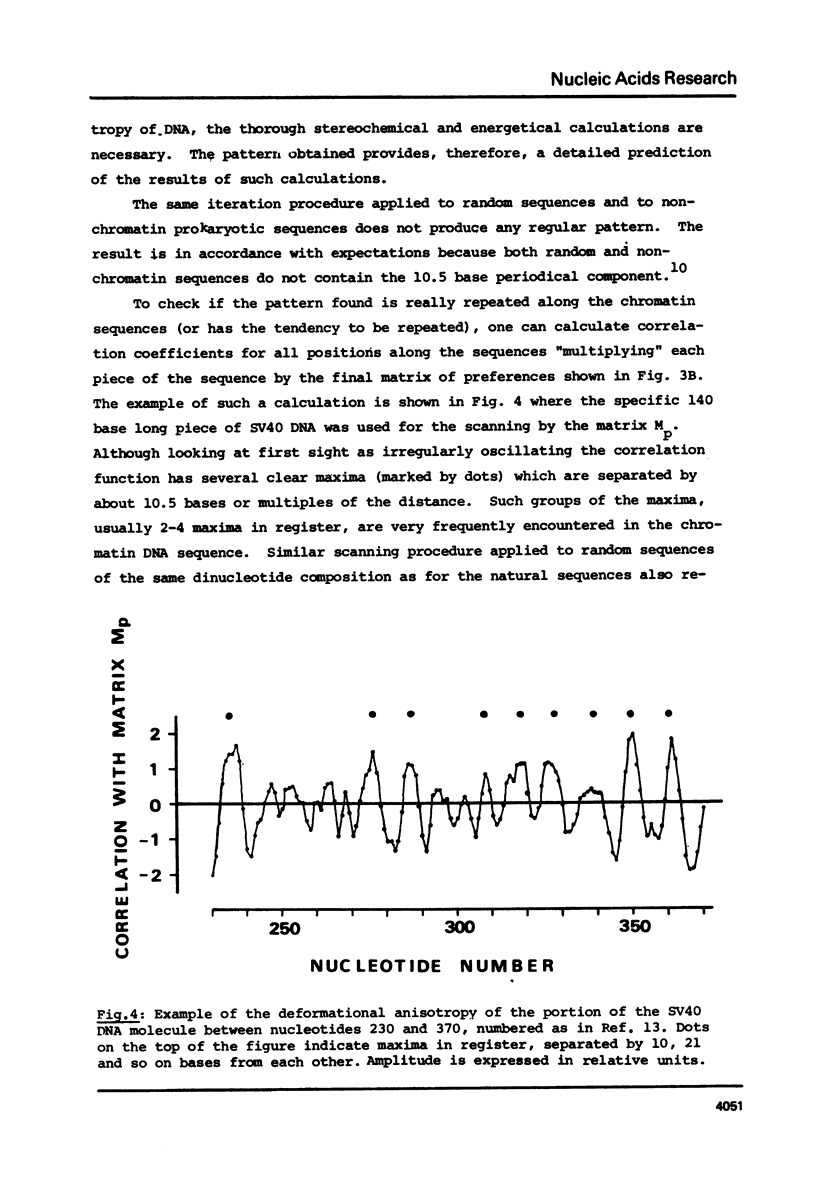
As found in previous work (E.N. Trifonov and J.L. Sussman, Proc. Natl. Acad. Sci. USA, in press) some dinucleotides of the chromatin DNA sequences have a clear tendency to be repeated along the sequences with a period of about 10.5 bases. A special iteration procedure is developed to find if there are phase relationships between different periodically repeating dinucleotides of chromatin DNA. A very specific symmetrical pattern of preferences of different dinucleotides to certain positions within a repeating 10.5 base frame is indeed found. This is interpreted as a manifestation of sequence-dependent deformational anisotropy of the chromatin DNA which facilitates its smooth folding in chromatin. The pattern found can be used for locating unidirectionally curved portions of the DNA molecules, possibly corresponding to nucleosomal DNA. This implies that the DNA is bound to the nucleosomes by one specific side which corresponds to the direction of the sequence-dependent curving of the DNA axis. The 10.5 base periodicity found can be considered as the second message present in chromatin DNA sequences together with 3 base frame coding message.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Hukins D. W. Refinement of the structure of B-DNA and implications for the analysis of x-ray diffraction data from fibers of biopolymers. J Mol Biol. 1973 Dec 5;81(2):93–105. doi: 10.1016/0022-2836(73)90182-4. [DOI] [PubMed] [Google Scholar]
- Corden J., Engelking H. M., Pearson G. D. Chromatin-like organization of the adenovirus chromosome. Proc Natl Acad Sci U S A. 1976 Feb;73(2):401–404. doi: 10.1073/pnas.73.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H., Klug A. Kinky helix. Nature. 1975 Jun 12;255(5509):530–533. doi: 10.1038/255530a0. [DOI] [PubMed] [Google Scholar]
- Deininger P., Esty A., LaPorte P., Friedmann T. Nucleotide sequence and genetic organization of the polyoma late region: features common to the polyoma early region and SV40. Cell. 1979 Nov;18(3):771–779. doi: 10.1016/0092-8674(79)90130-2. [DOI] [PubMed] [Google Scholar]
- Feigon J., Kearns D. R. 1H NMR investigation of the conformational states of DNA in nucleosome core particles. Nucleic Acids Res. 1979;6(6):2327–2337. doi: 10.1093/nar/6.6.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Haegemann G., Rogiers R., Van de Voorde A., Van Heuverswyn H., Van Herreweghe J., Volckaert G., Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978 May 11;273(5658):113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- Friedmann T., Esty A., LaPorte P., Deininger P. The nucleotide sequence and genome organization of the polyoma early region: extensive nucleotide and amino acid homology with SV40. Cell. 1979 Jul;17(3):715–724. doi: 10.1016/0092-8674(79)90278-2. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallenbach N. R., Appleby D. W., Bradley C. H. 31P magnetic resonance of DNA in nucleosome core particles of chromatin. Nature. 1978 Mar 9;272(5649):134–138. doi: 10.1038/272134a0. [DOI] [PubMed] [Google Scholar]
- Levitt M. How many base-pairs per turn does DNA have in solution and in chromatin? Some theoretical calculations. Proc Natl Acad Sci U S A. 1978 Feb;75(2):640–644. doi: 10.1073/pnas.75.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maat J., Van Ormondt H. The nucleotide sequence of the transforming HindIII-G fragment of adenovirus type 5 DNA. The region between map positions 4.5 (HpaI site) and 8.0 (HindIII site). Gene. 1979 May;6(1):75–90. doi: 10.1016/0378-1119(79)90086-6. [DOI] [PubMed] [Google Scholar]
- Namoradze N. Z., Goryunov A. N., Birshtein T. M. On conformations of the superhelix structure. Biophys Chem. 1977 Jun;7(1):59–70. doi: 10.1016/0301-4622(77)87015-4. [DOI] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Robertson M. A., Staden R., Tanaka Y., Catterall J. F., O'Malley B. W., Brownlee G. G. Sequence of three introns in the chick ovalbumin gene. Nature. 1979 Mar 22;278(5702):370–372. doi: 10.1038/278370a0. [DOI] [PubMed] [Google Scholar]
- Sobell H. M., Tsai C. C., Gilbert S. G., Jain S. C., Sakore T. D. Organization of DNA in chromatin. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3068–3072. doi: 10.1073/pnas.73.9.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sures I., Lowry J., Kedes L. H. The DNA sequence of sea urchin (S. purpuratus) H2A, H2B and H3 histone coding and spacer regions. Cell. 1978 Nov;15(3):1033–1044. doi: 10.1016/0092-8674(78)90287-8. [DOI] [PubMed] [Google Scholar]
- Sussman J. L., Trifonov E. N. Possibility of nonkinked packing of DNA in chromatin. Proc Natl Acad Sci U S A. 1978 Jan;75(1):103–107. doi: 10.1073/pnas.75.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifonov E. N., Bettecken T. Noninteger pitch and nuclease sensitivity of chromatin DNA. Biochemistry. 1979 Feb 6;18(3):454–456. doi: 10.1021/bi00570a011. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y., Suzuki Y. The DNA sequence of Bombyx mori fibroin gene including the 5' flanking, mRNA coding, entire intervening and fibroin protein coding regions. Cell. 1979 Oct;18(2):591–600. doi: 10.1016/0092-8674(79)90075-8. [DOI] [PubMed] [Google Scholar]
- Van Ormondt H., Maat J., De Waard A., Van der Eb A. J. The nucleotide sequence of the transforming HpaI-E fragment of adenovirus type 5 DNA. Gene. 1978 Dec;4(4):309–328. doi: 10.1016/0378-1119(78)90048-3. [DOI] [PubMed] [Google Scholar]
- Zhurkin V. B., Lysov Y. P., Ivanov V. I. Anisotropic flexibility of DNA and the nucleosomal structure. Nucleic Acids Res. 1979 Mar;6(3):1081–1096. doi: 10.1093/nar/6.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]



