Abstract
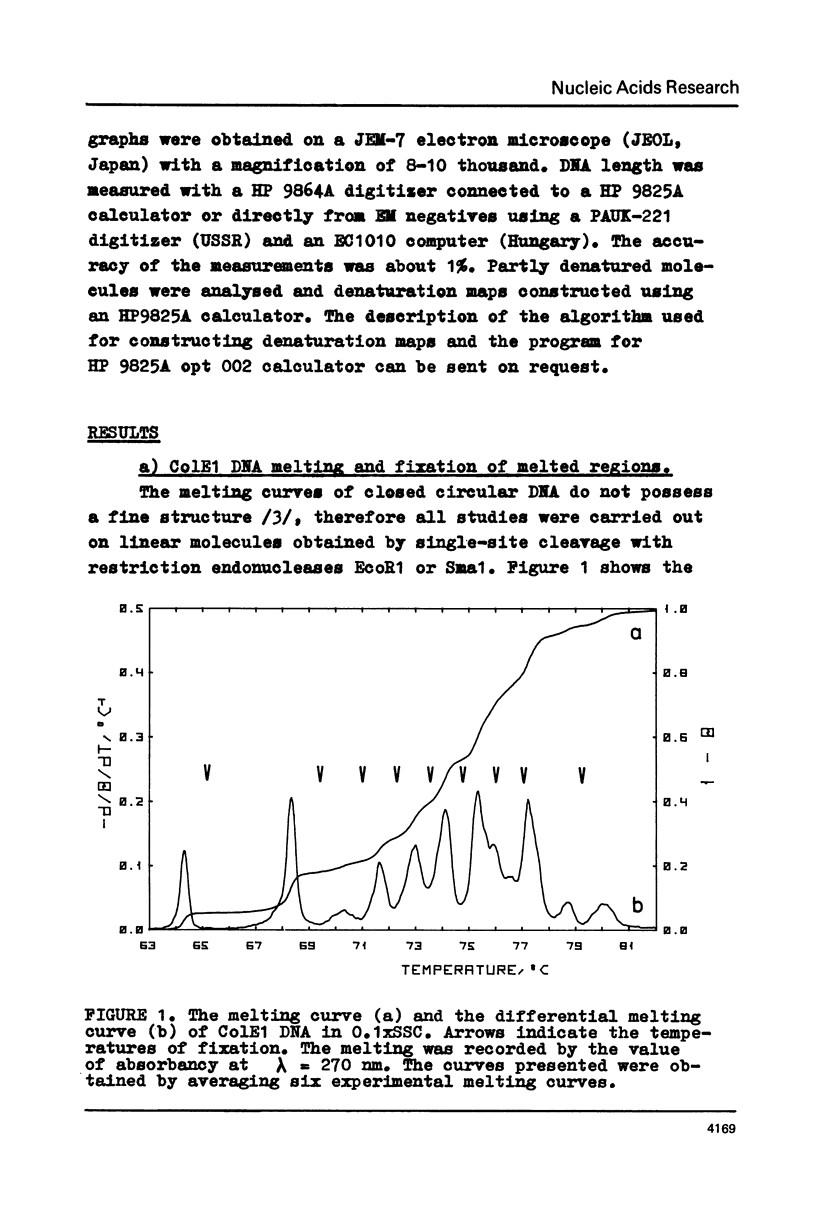
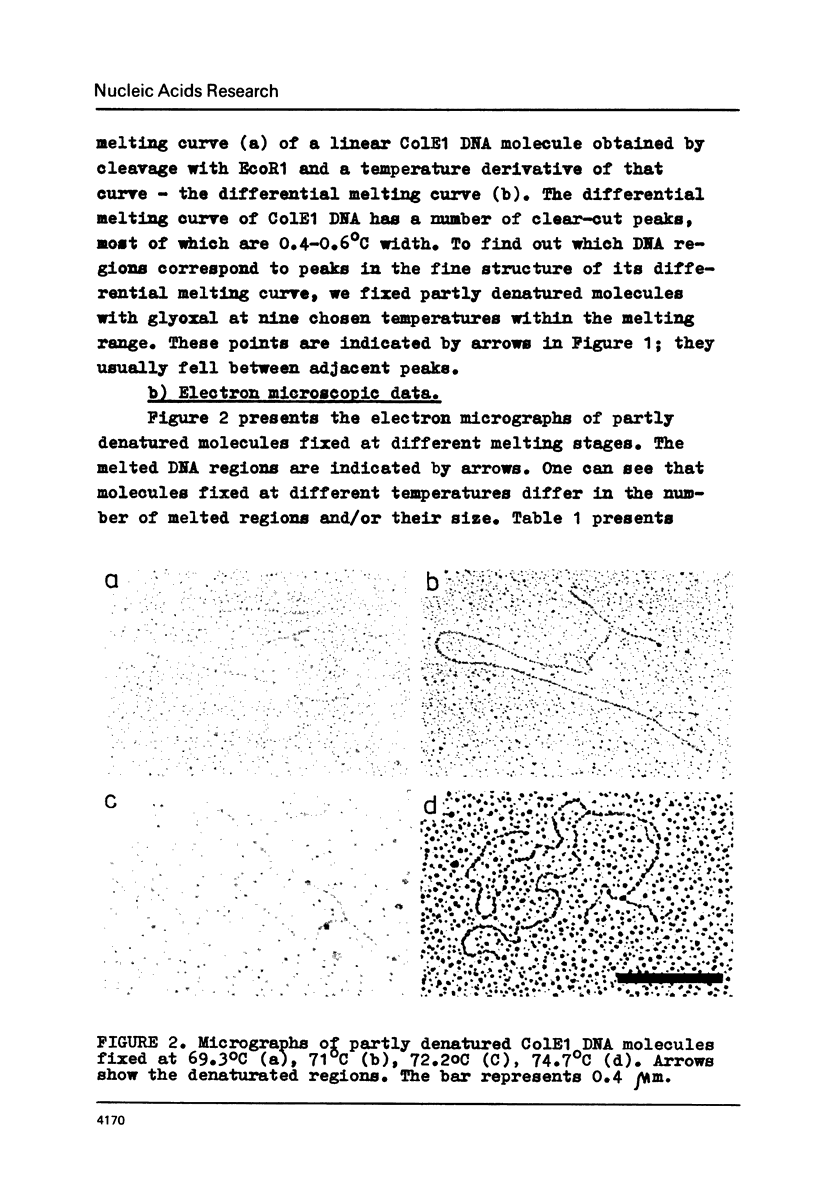
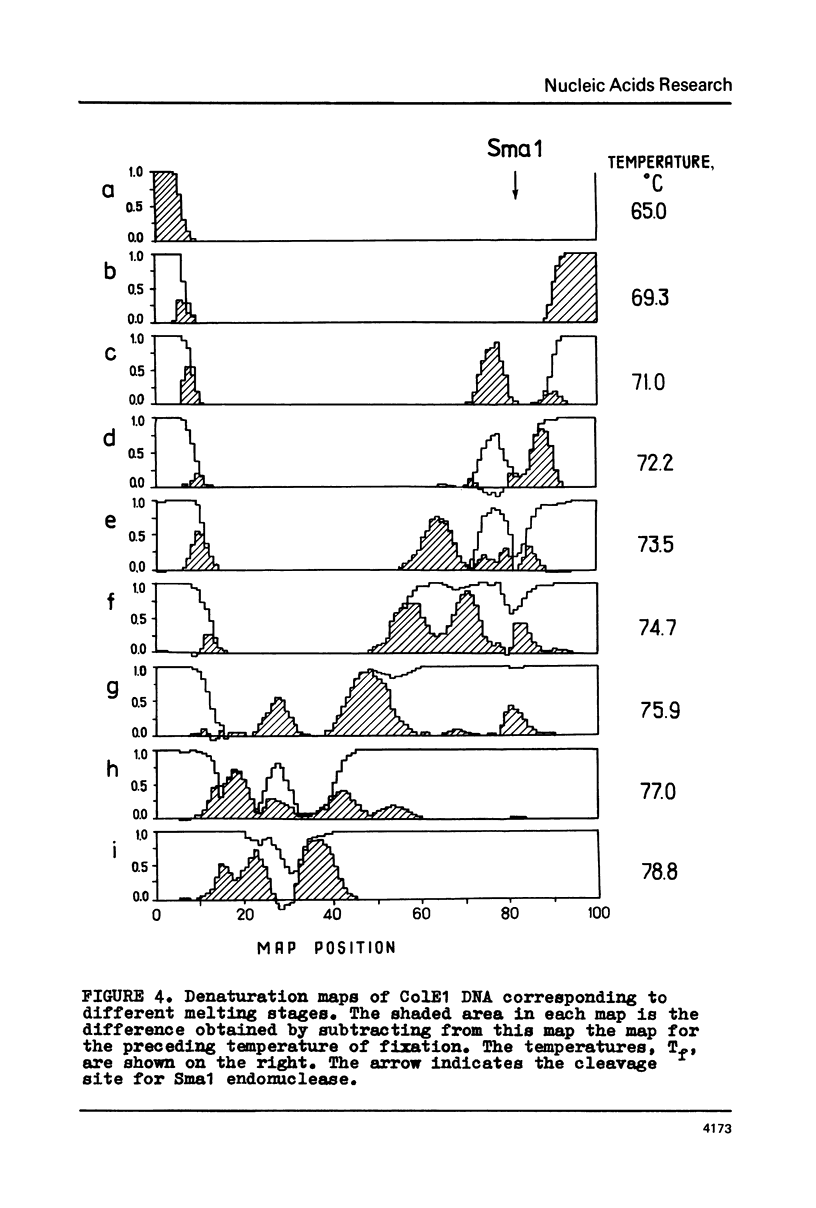
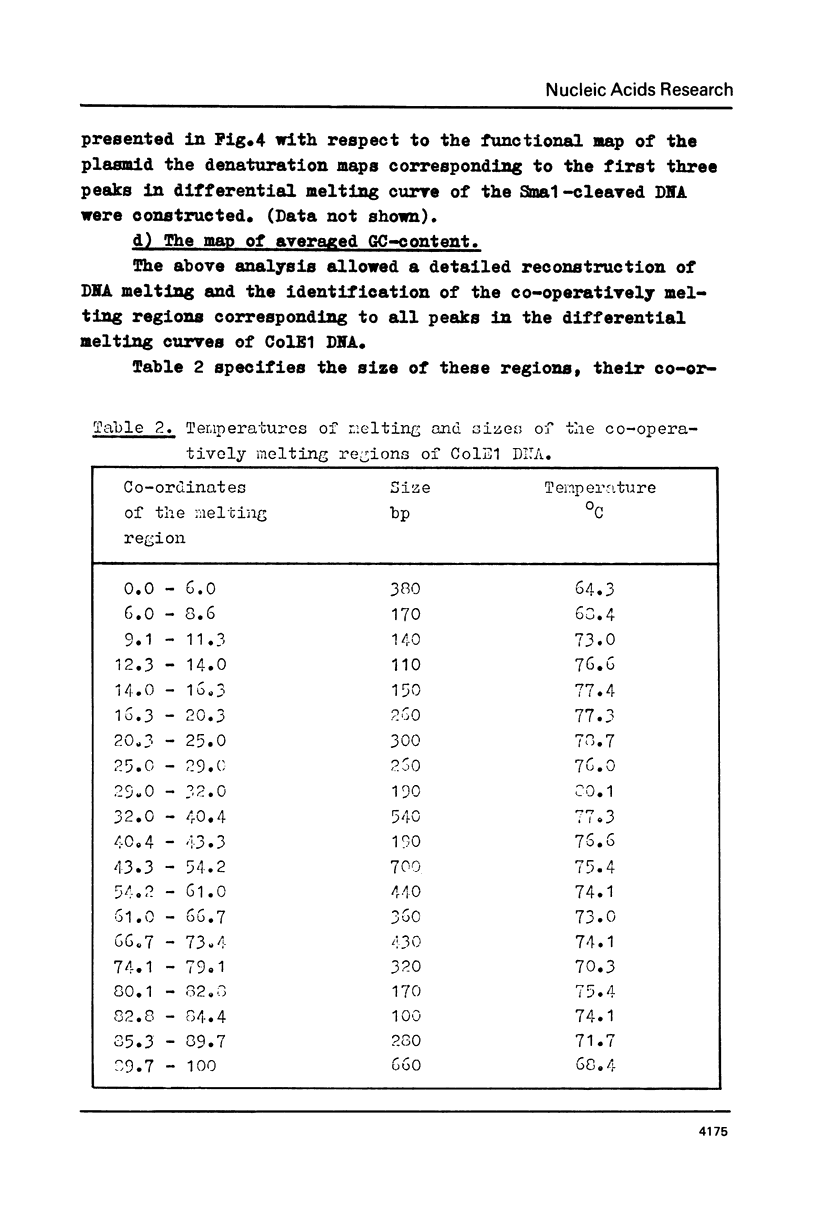
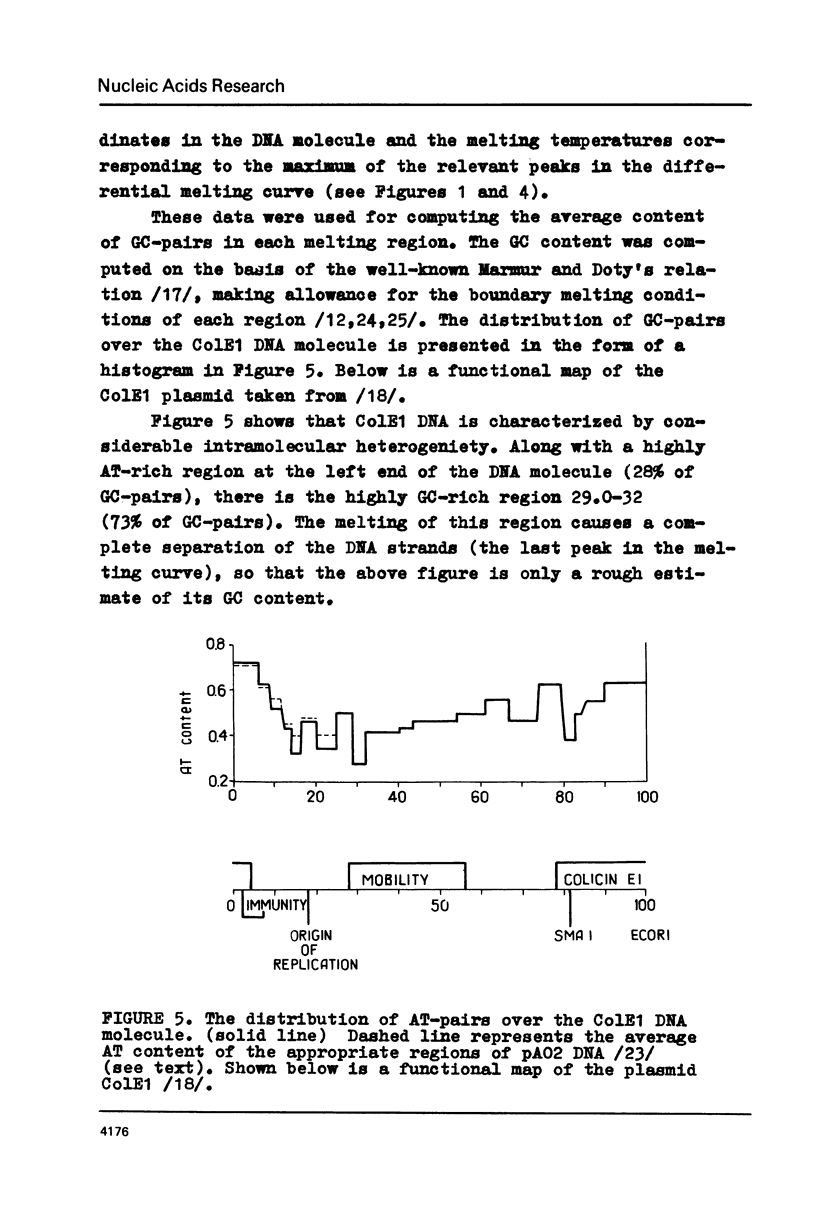
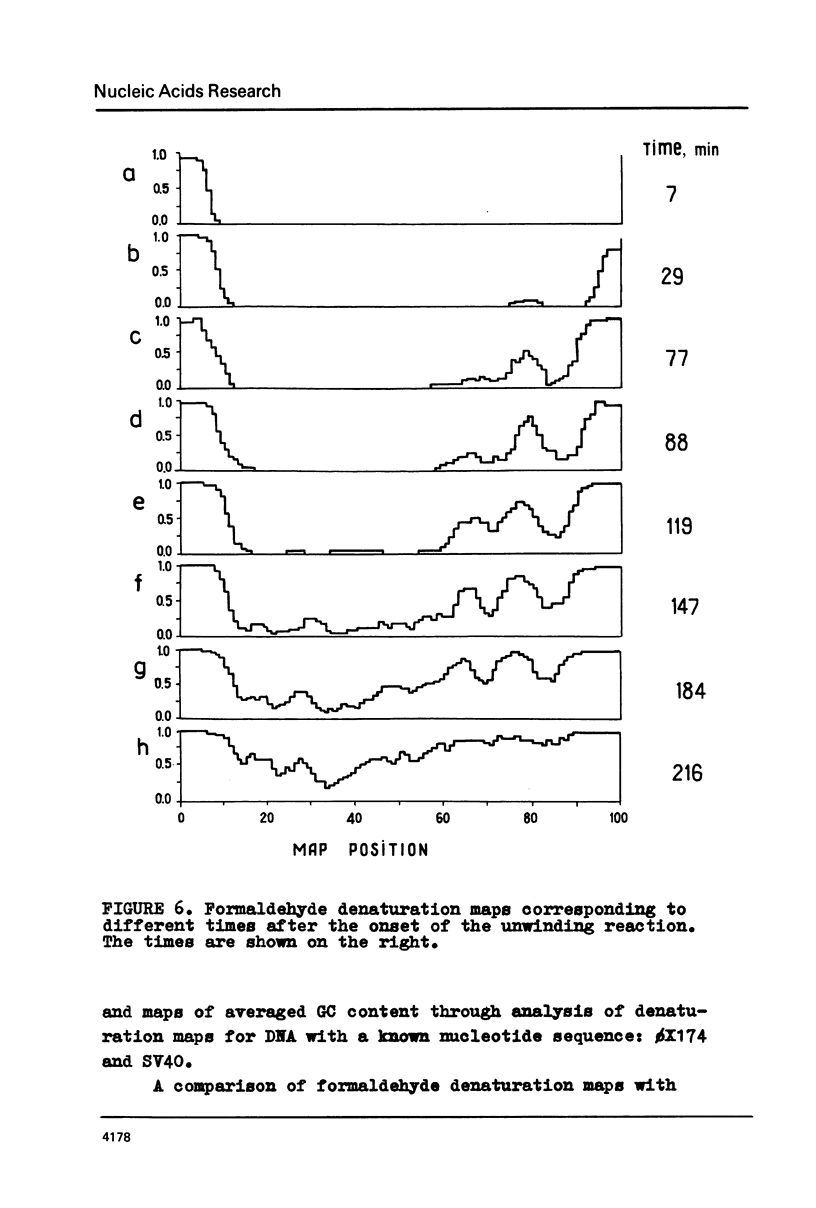
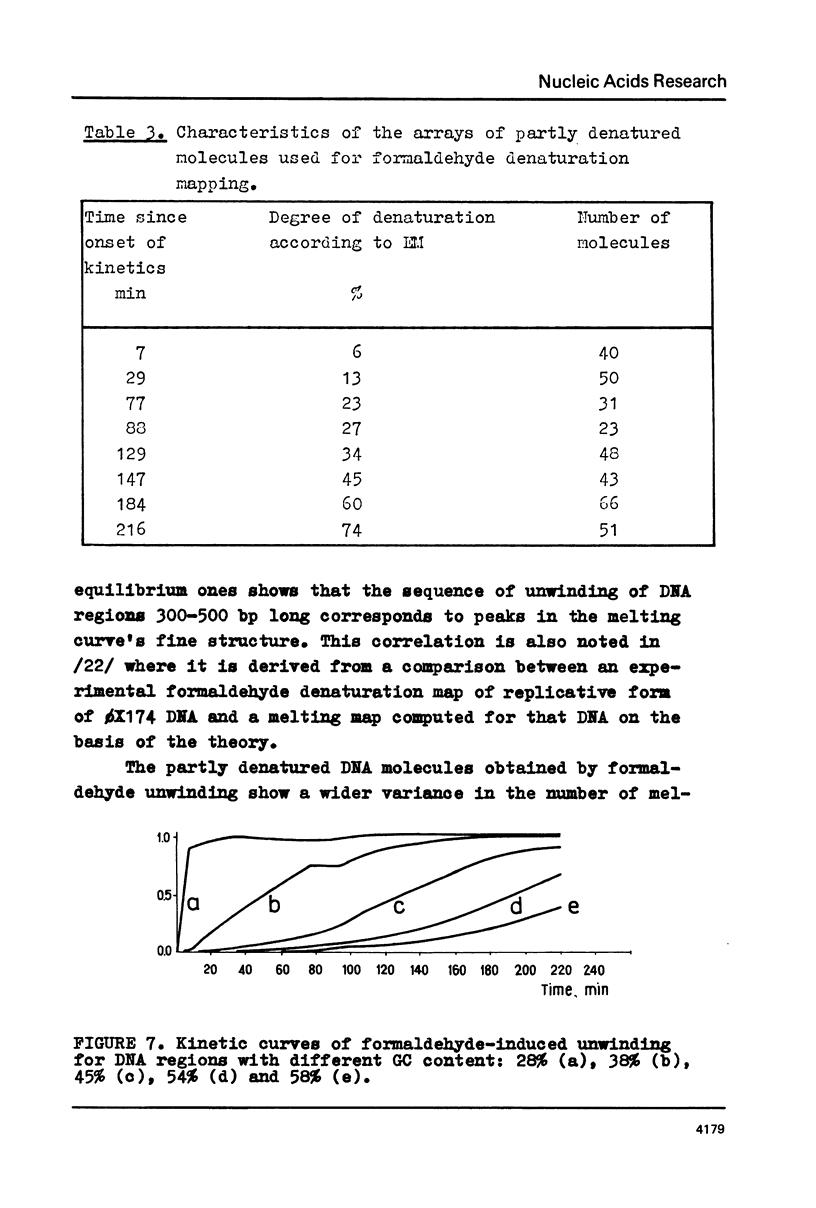
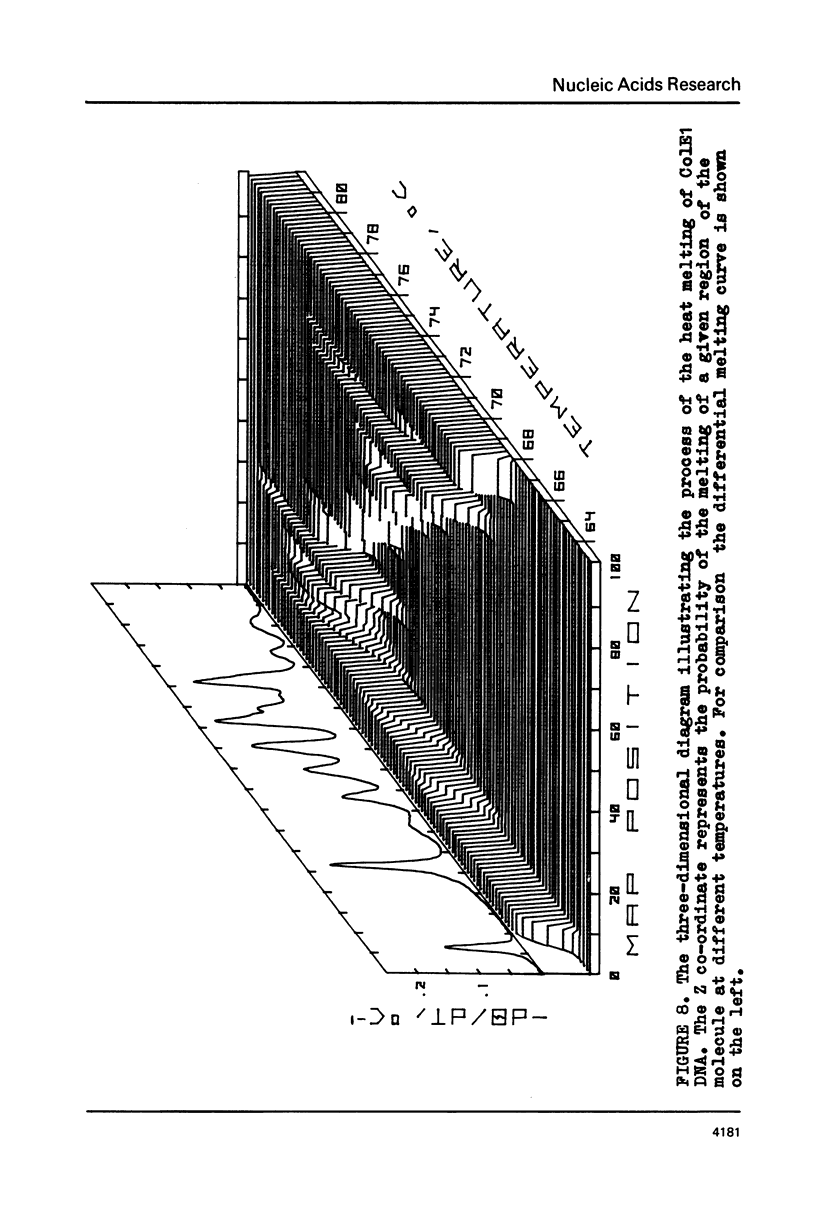
The fine structure of the melting curve for the linear colE1 DNA has been obtained. To find the ColE1 DNA regions corresponding to peaks in the melting curve's fine structure, we fixed the melted DNA regions with glyoxal /12/. Electron-microscopic denaturation maps were obtained for nine temperature points within the melting range. Thereby the whole process of colE1 DNA melting was reconstructed in detail. Spectrophotometric and electron microscopic data were used for mapping the distribution of Gc-pairs over the DNA molecule. The most AT-rich DNA regions (28 and 37% of GC-pairs), 380 and 660 bp long resp., are located on both sides of the site of ColE1 DNA's cleavage by EcoR1 endonuclease. The equilibrium denaturation maps are compared with maps obtained by the method of Inman /20/ for eight points of the kinetic curve of ColE1 DNA unwinding by formaldehyde.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Broude N. E., Budowsky E. I. The reaction of glyoxal with nucleic acid components. V. Denaturation of DNA under the action of glyoxal. Biochim Biophys Acta. 1973 Feb 4;294(1):378–384. doi: 10.1016/0005-2787(73)90092-0. [DOI] [PubMed] [Google Scholar]
- Funnell B. E., Inman R. B. Comparison of partial denaturation maps with the known sequence of simian virus 40 and phi X174 replicative form DNA. J Mol Biol. 1979 Jun 25;131(2):331–340. doi: 10.1016/0022-2836(79)90079-2. [DOI] [PubMed] [Google Scholar]
- Gómez B., Lang D. Denaturation map of bacteriophage T7 DNA and direction of DNA transcription. J Mol Biol. 1972 Sep 28;70(2):239–251. doi: 10.1016/0022-2836(72)90536-0. [DOI] [PubMed] [Google Scholar]
- Karataev G. I., Permogorov V. I., Vologodskii A. V., Frank-Kamenetskii M. D. Denaturation maps of DNA: experimental and theoretical maps of phiX174 DNA. Nucleic Acids Res. 1978 Jul;5(7):2493–2500. doi: 10.1093/nar/5.7.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubchenko Y. L., Frank-Kamenetskii M. D., Vologodskii A. V., Lazurkin Y. S., Gause G. G., Jr Fine structure of DNA melting curves. Biopolymers. 1976 Jun;15(6):1019–1036. doi: 10.1002/bip.1976.360150602. [DOI] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Michel F. Hysteresis and partial irreversibility of denaturation of DNA as a means of investigating the topology of base distribution constraints: application to a yeast rho- (petite) mitochondrial DNA. J Mol Biol. 1974 Oct 25;89(2):305–326. doi: 10.1016/0022-2836(74)90521-x. [DOI] [PubMed] [Google Scholar]
- Oka A., Nomura N., Morita M., Sugisaki H., Sugimoto K., Takanami M. Nucleotide sequence of small ColE1 derivatives: structure of the regions essential for autonomous replication and colicin E1 immunity. Mol Gen Genet. 1979 May 4;172(2):151–159. doi: 10.1007/BF00268276. [DOI] [PubMed] [Google Scholar]
- Pavlov V. M., Lyubchenko Y. L., Borovik A. S., Lazurkin Y. S. Specific fragmentation of T7 phage DNA at low-melting sites. Nucleic Acids Res. 1977 Nov;4(11):4053–4062. doi: 10.1093/nar/4.11.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert M., Van Assel S. Base compisition heterogeneity in kinetoplast DNA FROM FOUR SPECIES OF HEMOFLAGELLATES. Biochem Biophys Res Commun. 1974 Dec 23;61(4):1249–1255. doi: 10.1016/s0006-291x(74)80418-3. [DOI] [PubMed] [Google Scholar]
- Sümegi J., Breedveld D., Hossenlopp P., Chambon P. A rapid procedure for purification of EcoRI endonuclease. Biochem Biophys Res Commun. 1977 May 9;76(1):78–85. doi: 10.1016/0006-291x(77)91670-9. [DOI] [PubMed] [Google Scholar]
- Tachibana H., Wada A., Gotoh O., Takanami M. Location of the cooperative melting regions in bacteriophage fd DNA. Biochim Biophys Acta. 1978 Feb 16;517(2):319–328. doi: 10.1016/0005-2787(78)90198-3. [DOI] [PubMed] [Google Scholar]
- Vizard D. L., Ansevin A. T. High resolution thermal denaturation of DNA: thermalites of bacteriophage DNA. Biochemistry. 1976 Feb 24;15(4):741–750. doi: 10.1021/bi00649a004. [DOI] [PubMed] [Google Scholar]
- Vologodskii A. V., Frank-Kamenetskii M. D. Theoretical melting profiles and denaturation maps of DNA with known sequence: fdDNA. Nucleic Acids Res. 1978 Jul;5(7):2547–2556. doi: 10.1093/nar/5.7.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki S., Gotoh O., Wada A. Fine structures in denaturation curves of bacteriophage lambda DNA. Their relation to the intramolecular heterogeneity in base compositon. Biochim Biophys Acta. 1975 Jul 7;395(3):258–273. doi: 10.1016/0005-2787(75)90196-3. [DOI] [PubMed] [Google Scholar]




