Abstract
Aberrant neocortical DNA methylation has been suggested to be a pathophysiological contributor to psychotic disorders. Recently, a growth arrest and DNA-damage-inducible, beta (GADD45b) protein-coordinated DNA demethylation pathway, utilizing cytidine deaminases and thymidine glycosylases, has been identified in the brain. We measured expression of several members of this pathway in parietal cortical samples from the Stanley Foundation Neuropathology Consortium (SFNC) cohort. We find an increase in GADD45b mRNA and protein in patients with psychosis. In immunohistochemistry experiments using samples from the Harvard Brain Tissue Resource Center, we report an increased number of GADD45b-stained cells in prefrontal cortical layers II, III, and V in psychotic patients. Brain-derived neurotrophic factor IX (BDNF IXabcd) was selected as a readout gene to determine the effects of GADD45b expression and promoter binding. We find that there is less GADD45b binding to the BDNF IXabcd promoter in psychotic subjects. Further, there is reduced BDNF IXabcd mRNA expression, and an increase in 5-methylcytosine and 5-hydroxymethylcytosine at its promoter. On the basis of these results, we conclude that GADD45b may be increased in psychosis compensatory to its inability to access gene promoter regions.
Keywords: epigenetic, histone, DNA methylation, CpG, schizophrenia, bipolar disorder
INTRODUCTION
The ability of 5′ cytosine methylation to endure an animal's lifespan, and even across generations, indicates its potential utility as a means for encoding information (Weaver et al, 2004; Rakyan et al, 2003). The informational content the methyl mark represents has been shown in animal studies to participate in learning and memory formation (Feng et al, 2010; Miller et al, 2008, 2010; Lubin et al, 2008; Miller and Sweatt, 2007; Gavin et al, 2011), while investigations using clinical samples reveal an important function in regulating gene expression in the human brain (Grayson et al, 2005; Abdolmaleky et al, 2005, 2006; Iwamoto et al, 2005). However, inconsistent results in relation to the pathophysiology of mental illnesses, including psychotic disorders, suggest an increasingly recognized higher level of complexity between the methyl mark, gene expression, and disease risk (Tochigi et al, 2008; Mill et al, 2008; Dempster et al, 2006).
This complexity can be in part accounted for by emerging data that the DNA methylation status of a given gene promoter is the consequence of a dynamic equilibrium between DNA methylation and putative demethylation activity (Szyf, 2010; Guidotti et al, 2011). Prior studies indicate global abnormalities may be present in psychosis. These include differences in expression of DNA methylating enzymes, the methyl donor S-adenosylmethionine (SAM), degradation of DNA methylation networks, and lack of coordination between restrictive histone modifications and DNA methylation (Mill et al, 2008; Guidotti et al, 2007; Veldic et al, 2005, 2007; Huang and Akbarian, 2007). It was our hypothesis that examining components of a DNA demethylation pathway may begin to help unravel the complexity of this system in psychosis.
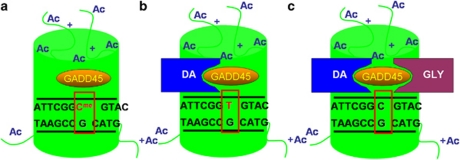
Increasing data support a means of active DNA demethylation that avoids the difficult task of breaking the covalent bonds between methyl groups and cytosines (Ooi and Bestor, 2008). This DNA demethylation pathway involves deaminating 5-methylcytosine (5MC) or 5-hydroxymethylcytosine (5HMC) to form thymine or 5-hydroxymethyluracil (5HMU), respectively (Rai et al, 2008; Cortellino et al, 2011). The resulting TpG:methyl-CpG or 5HMUpG:CpG mismatch is then excised using a DNA glycosylase (Figure 1). Brooks et al (1996) reported that glycosylases repairing T:G mismatches that result from deamination of 5MC counterbalance DNA methyltransferase activity in the brain. Later, the glycoslyase MBD4 was found to preferentially repair methyl-CpG:TpG mismatches (Hendrich et al, 1999), and mice deficient in this enzyme are incapable of demethylating DNA in reaction to parathyroid hormone treatment (Kim et al, 2009). Deaminases have also been shown to be necessary for active mammalian DNA demethylation (Bhutani et al, 2010; Popp et al, 2010). Finally, the GADD45 proteins (Figure 2) (Barreto et al, 2007; Schmitz et al, 2009) are thought to be master coordinators of this process by recruiting deaminases and glycosylases to promoter regions (Rai et al, 2008; Cortellino et al, 2011).
Figure 1.
Proposed mechanism of activity-dependent DNA demethylation. Following depolarization GADD45 protein binds to a methylated promoter region proximal to an acetylated histone (a). GADD45 recruits a deaminase (DA), which converts 5-methylcytosine (5MC) to thymine leading to a T:G mismatch (b). GADD45 recruits a DNA glycosylase (GLY), which removes thymine from the T:G mismatch. Thymine is later replaced with an unmethylated cytosine (c) (Guidotti et al, 2011).
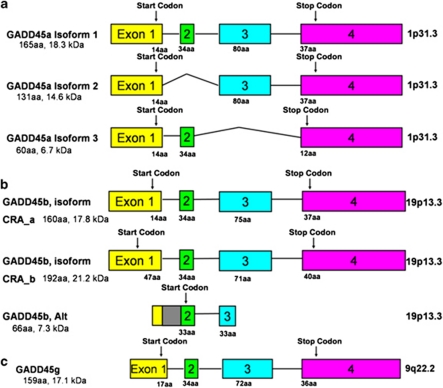
Figure 2.
Exon–intron structure and alternative transcripts of human GADD45 genes. Exons are shown as boxes and introns are shown as lines. Despite the three GADD45 genes being located on different chromosomes there are substantial similarities in terms of gene structure and protein composition. All three proteins are 17–18 kDa in mass and highly acidic. They share 56% protein sequence identity. In (a), are the three known splice variants of GADD45a. Isoform 1 contains four exons, while the other two splice variants are missing the second or third exon respectively. In (b), are the three splice variants of GADD45b. Isoform CRA_a and CRA_b differ based on their start sites. Another splice variant, referred to here as ‘GADD45b, Alt' includes the first intron (gray box), and translation begins at the second exon. This isoform is noted on the NCBI database (AAT38867.1) and in (Ying et al, 2005). In (c), is the gene structure of GADD45g.
We investigated the expression and promoter binding of components of a DNA demethylation pathway, and their association with the DNA methylation status at a brain-derived neurotrophic factor (BDNF) gene promoter in pathogenetically related disorders; bipolar disorder with psychotic features (BP) and schizophrenia (SZ), referred to together as ‘major psychosis' (Mill et al, 2008; Guidotti et al, 2007; Veldic et al, 2005; Craddock et al, 2006; Connor and Akbarian, 2008; Pope and Yurgelun-Todd, 1990; Tsuang, 1991; Reichenberg et al, 2009). We performed our initial investigations on inferior parietal lobule (IPL) samples from the Stanley Medical Research Institute. We selected this cortical area for several reasons. First, early neuropathological investigations noted that the IPL is among the most seriously damaged cortical regions in SZ (von Angyal, 1934). Second, imaging studies have reported decreased volume and differences in asymmetry as compared with controls (Goldstein et al, 1999; Niznikiewicz et al, 2000; Frederikse et al, 2000). In addition, many of the IPL functions, such as sensory integration, body image, concept of self, visuospatial abilities, and executive functions have been found to be disturbed in SZ (Jimenez et al, 2010; Torrey, 2007; Naghavi and Nyberg, 2005). Also, GADD45 proteins have been shown to be highly regulated in this region of the cortex (Chen et al, 1998; Zhu et al, 1997). Subsequently, we sought to confirm that gene expression abnormalities are not limited to this region using prefrontal cortical sections from the Harvard Brain Tissue Resource Center.
MATERIALS AND METHODS
Demographic Details
We obtained fresh-frozen parietal cortex (BA39-40) tissue from the neuropathology consortium of the Stanley Foundation Neuropathology Consortium (SFNC) (Bethesda, MD). The demographics associated with each patient population are presented in Table 1. The demographic and clinical characteristics of the population, as well as methods of tissue harvest, preparation, and storage, have been described in detail elsewhere (Torrey et al, 2000). We find no significant diagnostic differences in post-mortem interval, pH, age, or RIN.
Table 1. Demographic Characteristics of Brain Samples.
| N | Sex | Age | PMI | pH | RINa | Suicide | AD | MS | AP | |
|---|---|---|---|---|---|---|---|---|---|---|
| Fresh frozen samples | ||||||||||
| Stanley foundation neuropathology consortium | ||||||||||
| Parietal cortex (BA39-40) | ||||||||||
| Nonpsychotic subjects | 15 | 6F/9M | 48±10.6 | 23.7±9.9 | 6.3±0.24 | 4.9±0.97 | 0 | 0 | 0 | 0 |
| Psychotic subjects | 26 | 11F/15M | 44±12.4 | 30.4±14.0 | 6.2±0.23 | 5.5±0.90 | 11 | 11 | 10 | 20 |
| Formaldehyde fixed samples | ||||||||||
| Harvard tissue resource center | ||||||||||
| Prefrontal cortex (BA9) | ||||||||||
| Nonpsychotic subjects | 25 | 7F/18M | 58.4±17.9 | 20.2±6.1 | 6.5±0.32 | 0 | 0 | 0 | 0 | |
| Psychotic subjects | 30 | 9F/21M | 58.8±18.7 | 19.9±7.9 | 6.4±0.23 | 6 | 13 | 14 | 21 | |
Abbreviations: AD, antidepressants; AP, antipsychotics; MS, mood stabilizers; RIN, RNA integrity.
Subjects with RIN values below three were excluded for mRNA analyses. For mRNA analyses, 19 psychotic and 13 nonpsychotic subjects were included.
We found no significant differences in age, post-mortem interval (PMI), pH, or RIN whether subjects were separated based on the absence or presence of psychosis or based on diagnosis. Numbers represent mean±SD.
Prefrontal (BA9) cortical brain specimens fixed in 4% formaldehyde were obtained from the Harvard Brain Tissue Resource Center (HBTRC) (Belmont, MA). Each sample was transferred into 30% sucrose in 0.1 M PBS 72 h before histological preparation. The psychiatric diagnoses were established by two senior psychiatrists based on clinical and family histories and according to Diagnostic and Statistical Manual of Mental Disorders IV criteria. The demographic data are summarized in Table 1.
Quantitative Reverse Transcriptase-Polymerase Chain Reaction
Total RNA from the SFNC-obtained parietal cortices were isolated using TRIZOL reagent (Life Technologies, Grand Island, NY). DNA removal was performed using Ambion DNase (Ambion, Austin, TX). RNA was further purified using Qiagen RNeasy minikit (Qiagen, Valencia, CA). RNA integrity (RIN) was assessed with an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA). RNA was converted to cDNA using Applied Biosystems High Capacity Archive Kit (ABI, Foster City, CA).
We measured transcripts of several putative DNA demethylation genes and a potential target gene, BDNF IXabcd (Figure 6d). The potential DNA demethylating genes include: GADD45 genes (GADD45a, GADD45b, and GADD45g), DNA glycosylases (MBD4 and TDG), and an endonuclease (XPG). There are three known splice variants for GADD45a (Figure 2a). We chose to design primers for the full-length transcript, GADD45a, isoform 1. GADD45a, isoform 3 is transcribed but not translated, and isoform 2 has been noted to not bind other proteins in the same manner as isoform 1 and may be antagonistic to the actions of isoform 1 (Zhang et al, 2009). GADD45b primers were designed to include both CRA_a and CRA_b transcripts (Figure 2b). The third transcript, which we have designated ‘Alt' is missing much of the known protein binding sites, which extend from the middle portion of the protein to the N-terminus. The function of this isoform is unknown (Ying et al, 2005).
For qRT-PCR, cDNA samples were analyzed using a Stratagene Mx3005P QPCR System (Stratagene, La Jolla, CA) and Brilliant SYBR Green QPCR Master Mix (Stratagene). To confirm amplification specificity, the PCR products were subject to a melting curve analysis, in which only one peak was observed. Crossing point values were measured with the Stratagene MxPro QPCR analysis software. Each gene per individual analyzed was performed in duplicate and performed twice on two separate days. Primers were designed to span at least one intron–exon boundary. We evaluated four housekeeping genes (GAPDH, NSE, β-actin, and transferrin receptor (p90, CD71) (TFRC)) that represent accurate controls for mRNA expression analysis of post-mortem brain samples (Barrachina et al, 2006; Silberberg et al, 2009). For each housekeeping gene, we measured the gene stability and ranked it using the geNorm algorithm (Vandesompele et al, 2002). Stepwise exclusion of the gene with the highest gene stability value allowed ranking of the tested genes according to their expression stability. On the basis of this analysis, we normalized using geometric averaging of multiple housekeeping genes, TFRC and β-actin (Vandesompele et al, 2002). The following cycling conditions were used: 10 min 95 °C then 40 cycles at 95 °C for 30 s, 60 °C for 1 min, and 72 °C for 30 s. See Table 2 for primers.
Table 2. Primer Sequences Used in this Study.
| Genes | 5′ primer | 3′ primer |
|---|---|---|
| mRNA expression primers | ||
| GADD45a | AGAGCAGAAGACCGAAAGGATGGA | GCAGGATGTTGATGTCGTTCTCGC |
| GADD45b | ATTGCAACATGACGCTGGAAGAGC | GGATGAGCGTGAAGTGGATT |
| GADD45g | GACACAGTTCCGGAAAGCAC | TCAAGACTTTGGCTGACTCG |
| MBD4 | AAAACGTGGCTCTGAAATGG | TCTGTGTTCGTGGGATGGTA |
| TDG | AACATGGTGGAAAGGACCAC | ACACTGCTATTCGTGGCTGA |
| XPG | CTTTGTTTCATCGGCTCTGC | CGTTTTCCTGGAGTCACTGG |
| BDNF IXabcd | AACCTTGACCCTGCAGAATG | TGGTCATCACTCTTCTCACCTG |
| NSE | TGTCTCTGGCCGTGTGTAAG | GCATGAGAGCCACCATTGAT |
| GAPDH | CGAGATCCCTCCAAAATCAA | TTCACACCCATGACGAACAT |
| ACTB | TCCCTGGAGAAGAGCTACGA | TGAAGGTAGTTTCGTGGATGC |
| TFRC | AAAATCCGGTGTAGGCACAG | CACCAACCGATCCAAAGTCT |
| Chromatin immunoprecipitation primers | ||
| BDNFIXabcd −60 to +50 | CCTCTGGCAAACAGGAAGAG | CGCGCTCTGAGTTTATCCTA |
| BDNFIXabcd +1185 to +1305 | AAACATCCGAGGACAAGGTG | TTCGAAAGTGTCAGCCAATG |
|
Genes |
Probe |
|
| In situ mRNA probes | ||
| BDNFIXabcd +514 to +537 | AGCAGTTGGGGAACTATAAGGAG | |
| BDNFIXabcd +761 to +784 | AATGATCTCTGCTCATGCTGTC | |
| BDNFIXabcd +714 to +740 | GTTCTCCTTCTTCCCACTTTAGC | |
We correlated RIN scores with ΔCT values of each of the genes studied normalized to either β-actin or TFRC. We find no significant correlation between RIN and ΔCT for RIN scores >3. We have eliminated all mRNA expression data from samples with RIN scores <3. Further, prior studies indicate that expression data are independent of RIN values for amplicons <250 bp (Fleige and Pfaffl, 2006). In this study, no amplicon exceeds this length.
GADD45b Immunoblotting
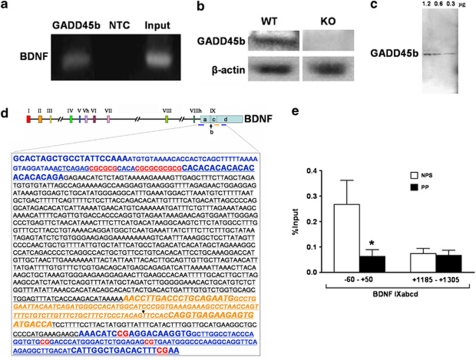
Protein was extracted from samples using TRIZOL reagent protocol (Life Technologies). Proteins were separated on 10–20% gradient Tris-Glycine gels (Invitrogen) with constant voltage (125 V) for 1.5 h and transferred overnight to PVDF membranes (Bio-Rad #162–0181). Membranes were incubated with either an anti-GADD45b polyclonal antibody (Santa Cruz, SC-133606) for 48 h at 4 °C or a β-actin monoclonal antibody (Sigma; #A5316) for 1 h, in blotto buffer. Enhanced chemiluminescence (ECL, Amersham) was detected using a STORM fluorescent imager (Molecular Dynamics) and quantified with IMAGEQUANT 5.0 software (Figure 3c). For immunoblotting, each subject was analyzed across a linear range of GADD45b normalized to β-actin 1.2, 0.6, and 0.3 μg total protein. One band at the correct molecular weight was detected for the antibody used. All further mention of GADD45b protein reflects the normalized levels to β-actin.
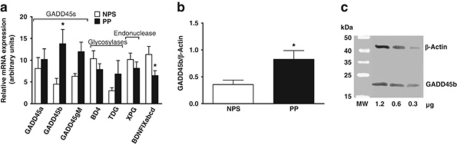
Figure 3.
GADD45b mRNA and protein expression are increased in psychosis. In post-mortem parietal cortical sections obtained from the Stanley Foundation Neuropathology Consortium (SFNC) mRNA was measured using qRT-PCR with β-actin and transferrin receptor (p90, CD71) (TFRC) as controls. Expression was calculated via geometric averaging of multiple internal control genes (Vandesompele et al, 2002). GADD45b (p=0.034) and BDNF IXabcd (p=0.016) mRNA significantly differ between nonpsychotic subjects (NPS) (n=13) and psychotic patients (PP) (n=19) (a). In SFNC samples, GADD45b protein (Santa Cruz, SC-133606) normalized to β-actin (Sigma-Aldrich #A5316) is higher in PP (n=26) compared with NPS (n=15) (p=0.04) (b). Representative immunoblot (c). Measurements were made for each subject in the linear range of 1.2, 0.6, and 0.3 μg of total protein with good agreement at all protein concentrations (molecular weight markers, MW). *p<0.05.
GADD45b Chromatin Immunoprecipitation (ChIP) and Methylated DNA Immunoprecipitation (MeDIP)
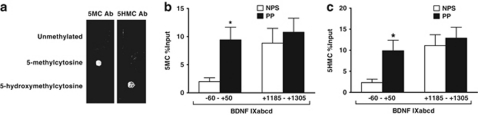
ChIP and MeDIP experiments were conducted using a previously published protocol (Gavin et al, 2009a; Dong et al, 2008). We used rabbit anti-GADD45b polyclonal antibody (Aviva, ARP48346_P050). Antibody specificity was tested in a western blot of human tissue that revealed one band, and in a western blot using wild-type (WT) mouse and GADD45b knockout (KO) mouse brain tissue (Figures 6a–c). Mouse monoclonal anti-5MC (Diagenode) and rabbit polyclonal 5HMC (Active Motif) antibodies were used in MeDIP, which had no detectable cross-reactivity based on a dot blot (Figure 7a). We decided to use the MeDIP procedure because bisulfite and most enzyme-dependent methods are incapable of distinguishing 5MC from the approximately 14% of methylcytosines in the brain that are 5HMC (Globisch et al, 2010). Recent studies demonstrate that these two cytosine modifications may have very different functions and genomic locations (Valinluck et al, 2004; Jin et al, 2011; Guo et al, 2011). Results are expressed as percent input minus negative control (adapted from MagMeDIP kit instruction manual; Diagenode).
Light Microscopy Immunohistochemistry
Twenty-five micron slices were obtained from sucrose immersed samples using a cryostat (Microm HM 550; Thermo Scientific). Sections were first incubated in 0.25% Tween in PBS for 30 min at room temperature. For immunohistochemistry experiments, they were then incubated with 5% NGS, 2% BSA in PBS for 30 min, then 48 h at 4 °C and 1 h at room temperature with rabbit anti-GADD45b polyclonal antibody (Santa Cruz, #SC-133606) in 1% NGS and 0.4% BSA in PBS.
After addition of the secondary antibody, sections were stained following a previously described protocol (Rodriguez et al, 2002), and sections were reacted with 3,3-diaminobenzidine (DAB) with nickel-ammonium sulfate to obtain a dark-brown reaction product (Rodriguez et al, 2002). For each brain, five to six sections were taken (one every fourth slice), and cells were counted using a bi-dimensional cell counting method (Veldic et al, 2007). The counts were performed blindly in three randomly selected squares in each of five to six sections; thus, a total of 15–18 squares per sample were counted. All analyses were carried out in comparable areas (100 × 100 μm) under the same optical and illumination conditions (for example, light intensity was set to 8 on a scale of 1–10, and exposure time was set to 1.5 ms) in a Zeiss MicroImager. Black and white images were digitized and viewed on a computer by using AxioVision software. To count strongly stained cells and exclude weakly stained or nonspecifically labeled cells from the analysis, the threshold intensity of staining was established at 3 × the background (measured by Zeiss Software).
Confocal Fluorescence Microscopy
Immunofluorescence labeling was performed following a modification of the procedure described by Veldic et al. (2007) and Agis-Balboa et al. (2006). For colocalization experiments, 25 μm sections were incubated for 48 h at 4 °C and 1 h at room temperature with rabbit anti-GADD45b polyclonal antibody (Santa Cruz, #SC-133606) and either a GABAergic cell marker, mouse anti-GAD65/67 monoclonal antibody (Millipore, AB1511), pyramidal cell marker, mouse anti-VGLUT2 monoclonal antibody (Abcam, ab79157), or glial cell marker, mouse anti-S100β monoclonal antibody (Sigma, S2532). After labeling with the primary antibodies, slices were incubated with Cy5-labeled goat anti-mouse or anti-rabbit IgG (diluted 1 : 1000; Amersham Biosciences, Piscataway, NJ) to produce red fluorescent staining or Cy2-labeled streptavidin (diluted 1 : 1000; Amersham Biosciences) to produce green fluorescent staining, as indicated in the Figure legends. The reactions were carried out in 1% normal goat serum and 0.4% BSA in PBS for 1 h.
The numbers of cells in which green and red fluorescence colocalize compared with the numbers of cells that express only green or only red fluorescence was quantified using confocal microscopy (Leica, Bannockburn, IL) at a magnification of × 40 in a counting box of 100 × 100 × 20 μm. To test the specificity of the immunological detection, the primary antibody was omitted and no fluorescent staining was detected.
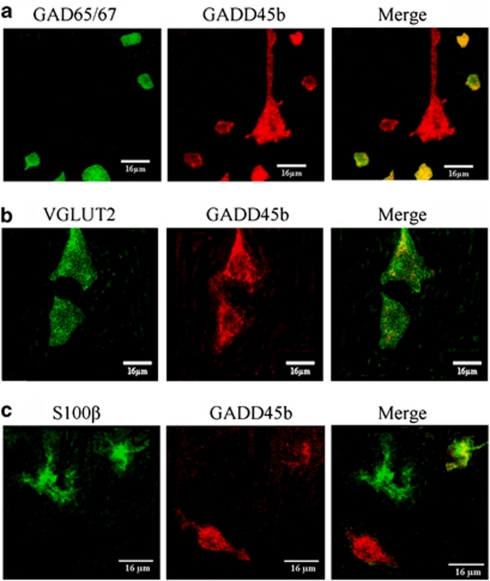
The choice of VGLUT2 was made based on our experience with this protein in prior studies in which we demonstrate its abundance in cortical pyramidal cells (Agis-Balboa et al, 2006, 2007). We have documented its presence in layer V of the cortex (Agis-Balboa et al, 2007). Others indicate that it is also present in layer II, III, and IV pyramidal neurons (Uezato et al, 2009; McCullumsmith and Meador-Woodruff, 2003; Fremeau et al, 2001; Nakamura et al, 2005).
In situ Hybridization
Three oligoprobes complementary to the human BDNF IXabcd transcript were used for in situ hybridization experiments conducted based on a previously published protocol (Rodriguez et al, 2002; Veldic et al, 2004). A BLAT search of each probe sequence against the UCSC Genome Bioinformatics group (http://genome.ucsc.edu/) detected no sequence identity with other genomic sequences. The oligonucleotide 3′ terminals were labeled with a digoxigenin kit (Roche Diagnostics). Twenty micron sections were incubated for 48 h with 50 pmol/ml of antisense oligonucleotides using three separate probes complementary to the BDNF IXabcd transcript with similar results for all three (see Table 2 for probe sequences). After the hybridization reactions, tissue sections were incubated with sheep anti-digoxigenin antibody (Roche Diagnostics) at 4 °C in 1% normal goat serum and 0.4% BSA in PBS for 24 h. Samples were then processed and visualized identically to light microscopy immunohistochemistry samples as above.
Digital Photomicrography
DAB (Sigma) staining images were captured AxioVision 3.1. (Zeiss) and confocal immunofluorescence by using a Leica Confocal Microscope (Leica Microsystems). The final composites were processed using Photoshop (Adobe Systems) and PowerPoint (Microsoft).
Statistical Analysis
We used SPSS version 15.0 for Windows (SPSS) for all statistical analyses. For post-mortem experiments, we compared mRNA expression, protein expression, ChIP, and MeDIP data between groups using multiple linear regression analysis. Spearman's correlations were also calculated. All significance levels are two-tailed.
RESULTS
Post-Mortem Expression Studies
In the SFNC cohort, psychotic patients include: 11 samples from patients with BP and 15 from patients with SZ. Nonpsychotic subjects include: 15 nonpsychiatric subjects (NPS) (Table 1). We included only samples with RIN scores >3 for mRNA analyses. For mRNA experiments, this left 19 psychotic subjects (8 BP and 11 SZ) and 13 NPS. We performed a multiple linear regression with each gene as the dependent variable and sex, age, psychosis, pH, and RIN as explanatory variables. We find that psychosis is a significant predictor for GADD45b mRNA expression (β=−0.409, F(1, 31)=5.020, p=0.034) with psychotic patients having significantly higher levels compared with controls (mean±SEM, 13.8±3.3 vs 4.5±1.3) (Figure 3a). We do not find age, sex, pH, or RIN to be significant predictors. A lack of significant correlation between GADD45b mRNA and these potential confounds verified this result. To establish whether there exist differences in GADD45b among psychotic patients taking antipsychotics (n=14) and those who were not (n=5), we performed a second multiple linear regression analysis, this time limiting the analysis to patients with psychosis. Use of antipsychotic medications is not a significant predictor of GADD45b mRNA expression in these patients. We found no differences in expression in the other GADD45 genes or the thymidine glycosylases (Figure 3).
As shown in Figure 3a, we find that psychosis is a significant predictor also for decreased BDNF IXabcd mRNA expression (β=0.459, F(1, 31)=6.661, p=0.016; mean±SEM, 6.5±1.1 vs 12.1±2.1) (Figure 3a). We do not find age, sex, pH, RIN, or antipsychotics to be significant predictors, nor significant correlations between this transcript and these variables.
To establish whether the increase in GADD45b mRNA expression in psychosis translates into an increase in the corresponding protein, we performed immunoblotting experiments on these same subjects from the SFNC cohort. We performed similar multiple linear regression analyses as in the mRNA experiments except excluding RIN as a potential explanatory variable and with GADD45b protein as the dependent variable. We find that psychosis is a significant predictor for GADD45b protein increase (β=−0.320, F(1, 40)=4.447, p=0.041; mean±SEM, 0.82±0.16 vs 0.36±0.08) (Figures 3b and c). We do not find age, sex, pH, or antipsychotic use to be significant predictors, nor significant correlations between GADD45b protein and these variables.
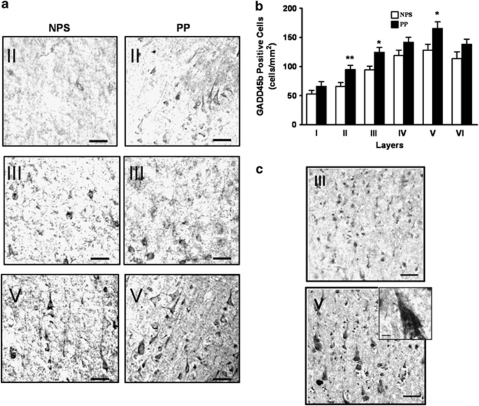
Using a different cohort (HBTRC), brain region (prefrontal cortex (BA9)), and assay (immunohistochemistry) from the one used for the mRNA and immunoblotting experiments (SFNC) we sought to confirm the increase in GADD45b protein among psychotic subjects. In this cohort, patients with psychosis include, 16 BP subjects and 14 SZ subjects. We performed a multiple linear regression with each layer as the dependent variable and sex, age, psychosis, pH as explanatory variables. We find that psychosis is a significant predictor for layers II (β=−0.361, F(1,53)=7.790, p=0.007; mean±SEM, 66.0±6.5 vs 94.7±7.4), III (β=−0.337, F(1,53)=6.647, p=0.013; mean±SEM, 94.3±6.4 vs 124.2±8.7), and V (β=−0.307, F(1,53)=5.422, p=0.024; mean±SEM, 128.2±10.1 vs 165.0±11.4) with psychotic patients having significantly higher levels compared with NPS (Figures 4a and b). We do not find age, sex, or pH to be significant predictors in these layers nor significant correlations between GADD45b stained cells and these variables. To establish whether there exist differences in GADD45b among psychotic patients taking antipsychotics (n=21) and those who were not (n=8), we performed a second multiple linear regression analysis, this time limiting the analysis to patients with psychosis. Use of antipsychotic medications is not a significant predictor of GADD45b staining in these layers.
Figure 4.
Immunohistochemistry and in situ hybridization results. In (a), is a representative immunohistochemistry with specific GADD45b antibodies using samples from the Harvard Brain Tissue Resource Center cohort, prefrontal cortex (BA9). Note more stained cells in cortical layers II, III, and V in psychotic patients (PP) compared with nonpsychotic subjects (NPS) (scale bar: 100 μm). In (b), we find significant increases in GADD45b positive neurons among PP (n=30) vs NPS (n=25) in layers II (p=0.007), III. (p=0.01), and V (p=0.02) (b). In (c), is a representative in situ hybridization experiment showing presence of BDNF IXabcd transcript in layer III and V cortical pyramidal neurons. Inset is a magnified cortical pyramidal neuron. (Scale bar: 100 μm; inset scale bar 10 μm). Error bars represent SEMs. *p<0.05; **p<0.01.
GADD45b is Highly Expressed in Pyramidal Neurons
Confocal colocalization experiments were conducted to determine which cell types primarily express GADD45b in the human brain (Figure 5). We find that about 60% of the GABAergic cells, approximately 80% of pyramidal cells, and a minority of glial cells expresses GADD45b.
Figure 5.
GADD45b is expressed in pyramidal and GABAergic cells, and not highly expressed in glial cells, in the post-mortem prefrontal cortex. In (a), is immunofluorescence showing colocalization of GADD45b (red) with a marker for GABAergic cells, GAD65/67 (Millipore, AB1511) (green). In (b), is immunofluorescence showing colocalization of GADD45b (red) with a marker for pyramidal cells, VGLUT2 (Abcam, ab79157) (green). In (c), is immunofluorescence showing colocalization of GADD45b (red) with a marker for glial cells, S100β (Sigma, S2532) (green).
GADD45b Protein is Less Bound to BDNF IXabcd in Psychosis
We chose to study the BDNF IXabcd promoter because Ma et al. (2009) documented that BDNF IXa is demethylated by GADD45b in mice. Although there are regulatory differences between human BDNF IXabcd and mouse BDNF IXa (Pruunsild et al, 2011), there also exist several similarities. In vivo and in vitro human BDNF IXabcd and mouse BDNF IXa are similarly induced by neuronal activity (Pruunsild et al, 2011; Koppel et al, 2009). In addition, both human and mouse transcripts are well-expressed widely throughout the body and the brain (Aid et al, 2007; Pruunsild et al, 2007). Finally, our in situ hybridization results indicate that BDNF IXabcd is well expressed in cortical pyramidal neurons, similar to GADD45b (Figure 4c).
We find significantly less GADD45b bound to the BDNF IXabcd gene at −60 to +50 (β=0.404, F(1,40)=6.833, p=0.013; mean±SEM, 0.06±0.03 vs 0.27±0.09) (Figure 6e) associated with an increase in 5MC (β=−0.370, F(1,40)=4.449, p=0.044; mean±SEM, 9.4±2.3 vs 1.99±0.68) and 5HMC (β=−0.349, F(1,40)=4.568, p=0.040; mean±SEM, 9.9±2.5 vs 2.3±0.81) in psychotic subjects (Figure 7). We do not find age, sex, pH, or antipsychotic use to be significant predictors, nor significant correlations between GADD45b binding, 5MC, 5HMC, and these variables. These differences in GADD45b binding (mean±SEM, 0.07±0.02 vs 0.07±0.02) (Figure 6e), 5MC (mean±SEM, 10.8±2.5 vs 8.8±2.6), or 5HMC (mean±SEM, 12.9±2.6 vs 11.1±2.6) (Figure 7) are not present at a position within the BDNF IXd exon (+1185 to +1305).
Figure 6.
GADD45b differentially binds to the BDNF IXabcd promoter in psychotic patients (PP) vs nonpsychotic subjects (NPS) in parietal cortical samples from the Stanley Foundation Neuropathology Consortium. In (a), is a representative gel from human brain chromatin immunoprecipitation (ChIP). Left gel lane reveals BDNF IXabcd PCR product in GADD45b (Aviva Systems Biology, ARP48346_P050) immunoprecipitated sample, center lane is negative control (NTC) (normal rabbit IgG; Santa Cruz, sc-2027) and right lane is input. In (b), is a western blot demonstrating that the antibody (Aviva Systems Biology, ARP48346_P050) used in ChIP experiments produces a band using protein from a wild-type (WT) mouse brain, but not from a GADD45b knockout (KO). In (c), is a western blot of human parietal tissue with 1.2, 0.6, and 0.3 μg of total protein indicating one major band for the GADD45b antibody. In (d), are the locations of human BDNF regions studied (Pruunsild et al, 2007). In ChIP experiments, we measured GADD45b binding and performed methylated DNA immunoprecipitation (MeDIP) experiments in the region surrounding the BDNF IXabcd transcription start site and within the exon BDNF IXd (blue lettering and blue lines below figure). Underlined is a transcribed portion of BDNF IXabcd. In red are the CpG sites in the ChIP areas studied. In italicized orange lettering is the location of the BDNF IXabcd transcript measured in RT-PCR experiments. The downward arrow represents the start of the shared 3′ exon used by other mRNA transcripts. Primer locations are in larger font size. In ChIP assays, GADD45b (Aviva, ARP48346_P050) is significantly more bound to a region of the BDNF IXabcd (p=0.013) promoter in NPS (n=15) compared with PP (n=26) (e). By contrast there is no difference in binding in a region of the BDNF IXd exon well downstream of the transcription start site (+1185 to +1305). Numbers represent pull down (ChIP) minus negative control as percent input. Error bars represent SEMs. *p<0.05.
Figure 7.
Increased 5-methylcytosine (5MC) and 5-hydroxymethylcytosine (5HMC) at the BDNF IXabcd promoter in parietal cortical samples from psychotic patients (PP) from the Stanley Foundation Neuropathology Consortium. In (a), is a dot blot indicating specificity of 5MC and 5HMC antibodies. 75 ng of either unmethylated, 5MC, or 5HMC standards (Active Motif, 55 008) were spotted onto two membranes. The membrane on the left was incubated with 5MC antibody (Ab) (Diagenode, MAb-081-100), while the right membrane was incubated with 5HMC Ab (Active Motif, 39769). In (b), methylated DNA immunoprecipitation (MeDIP) assays reveal significantly more 5MC (Diagenode, MAb-081-100) at the BDNF IXabcd (p=0.044) promoter in PP (n=26) compared with nonpsychotic subjects (NPS) (n=15). In (c), MeDIP experiments using anti-5HMC antibody reveal increased 5HMC at the BDNF IXabcd (p=0.040) promoter in psychotic patients. Numbers represent pull down (ChIP) minus negative control as percent input. Error bars represent SEMs. *p<0.05.
DISCUSSION
Our most significant and novel finding is the increased expression (Figure 3) and decreased promoter binding of GADD45b (Figure 6) in psychotic subjects. This increase in expression was confirmed using two separate cohorts (SFNC and HBTRC), in two different brain regions (IPL (BA39-40) and prefrontal cortex (BA9)), and using methods including qRT-PCR, immunoblotting, and immunohistochemistry. The publicly accessible Stanley Medical Research Institute (SMRI) online database, which includes 32 separate microarray studies also indicates an increase in GADD45b mRNA expression in BP (p=0.0004) with a trend for an increase in SZ (p<0.08) relative to controls (https://www.stanleygenomics.org/). We also find a decrease in BDNF IXabcd mRNA expression. We find no differences in other components of the demethylation pathway measured, such as GADD45a, GADD45g, MBD4, TDG, and XPG (Figures 3a). We also did not find a correlation between GADD45b and other immediate early genes such as FOS and JUN (unpublished data), indicating perhaps differences in their regulation.
To examine whether GADD45b induction may confer an epigenetic modulation in the expression of genes we focused on BDNF IXabcd. The presence of GADD45b in chromatin associated with a regulatory region of BDNF IXabcd points to a direct role of this protein in this gene's regulation and potentially in a demethylation process. An increase in BDNF IXabcd promoter 5MC and 5HMC connected with a decreased binding of GADD45b and reduced transcription in psychotic patients suggests this association (Figure 7).
Reconciling prior post-mortem studies of DNA methylation pathways is difficult as published reports indicate seemingly or occasionally genuinely contradictory results. While several studies from our group have pointed to a hypermethylating milieu in the brains of psychotic subjects (Grayson et al, 2005; Guidotti et al, 2011, 2007; Veldic et al, 2005, 2007; Zhubi et al, 2009; Ruzicka et al, 2007), others found decreased DNA methylation at particular genes (Abdolmaleky et al, 2006) or overall dysfunctional methylation networks comprised of both increases and decreases in methylation (Mill et al, 2008). Further, several studies have failed to replicate DNA methylation changes at particular genes (Tochigi et al, 2008; Mill et al, 2008; Dempster et al, 2006). These disparate results bespeak the increasingly recognized complexity of the DNA methyl mark.
Although CpG methylation was originally thought of as an on–off switch within promoter regions, the location of this mark has been shown to be integral to gene expression (Deaton et al, 2011; Irizarry et al, 2009). Further, the relationship between CpG methylation and transcription is not linear with transcription beginning at times before it is demethylated (Buschhausen et al, 1985; Sharma et al, 2010). Finally, recent studies indicate that 5HMC accounts for 14% of the methylcytosines in the brain (Globisch et al, 2010; Munzel et al, 2010). Most previous DNA methylation studies of clinical populations used methods incapable of distinguishing 5MC from 5HMC.
A limitation of the MeDIP method employed is that it is incapable of providing base-specific information regarding methylation status. It is possible that this is important as prior studies indicate that the methylation status of even a single CpG can determine whether a gene is transcribed (Sharma et al, 2010).
The increased expression of GADD45b in psychosis is somewhat surprising. This is because the majority of previous studies have reported increases in factors associated with a restrictive chromatin state, such as increased DNMT expression (Veldic et al, 2004, 2005, 2007; Zhubi et al, 2009; Ruzicka et al, 2007), DNA promoter methylation (Grayson et al, 2005; Abdolmaleky et al, 2005), and repressive histone modifications (Gavin et al, 2009a, 2009b; Benes et al, 2007; Huang et al, 2007) in psychotic patients. These abnormalities are hypothesized to cause the much-replicated reductions in GABAergic gene expression in psychosis (Benes et al, 2007; Akbarian et al, 1995; Fatemi et al, 2005; Hashimoto et al, 2008; Costa et al, 2004; Guidotti et al, 2000). Therefore, we expected GADD45b, which facilitates DNA demethylation, to be reduced in psychotic patients. One explanation for this discrepancy is suggested by our ChIP experiments. In these experiments, psychotic subjects, despite high levels of GADD45b, show reduced GADD45b binding to the BDNF IXabcd promoter compared with nonpsychotic subjects (Figure 6). This reduced binding is not found at a site well downstream of the transcription start site (TSS) of BDNF IXabcd (Figure 6), suggesting that this difference may be specific to gene promoter regions. In regards to the ChIP experiments, because GADD45b binding was examined at only certain genomic loci it is possible that this lack of binding is not generalizable to the genome as a whole. Additional genome-wide studies would be necessary to determine whether this is in fact the case. This may contribute to the understanding of the dyscoordination of promoter methylation processes reported in psychotic patients (Mill et al, 2008).
Previous studies demonstrate that GADD45 proteins associate with markers of an ‘open' chromatin state such as acetylated histones (Carrier et al, 1999). It has been suggested that subjects with psychosis have histone marks characteristic of a restrictive chromatin state (Huang et al, 2007), such as reduced acetylated histone 3 (Gavin et al, 2008), increased dimethylated lysine 9 of histone 3 (Gavin et al, 2009b), reduced methylated lysine 4 of histone 3 in female schizophrenics (Huang et al, 2007), and increased HDAC1 expression (Benes et al, 2007; Sharma et al, 2008). Although, Akbarian et al. (2005) reported increases in an ‘open' histone mark associated with reduced metabolic gene expression in SZ (Akbarian et al, 2005). A restrictive chromatin state in psychosis could obstruct or fail to recruit GADD45b binding to specific promoters, and lead to a compensatory increase in its expression. Future cellular studies are necessary to determine if a restrictive chromatin state prevents GADD45b binding and DNA demethylation.
In this study, we cannot exclude the possibility that medications contributed to our findings. In our analyses, we find no significant differences in GADD45b expression based on the use of antipsychotic medications. However, few subjects were not taking antipsychotic medications at the time of death, which makes any assertions that detected abnormalities in psychosis are not the result of medications impossible from the data presented here. It should also be noted that mice treated with clozapine or valproic acid, but not haloperidol have increased GADD45b expression (Guidotti et al, 2011; Matrisciano et al, 2011).
As epigenetic mechanisms are amenable to pharmacological intervention the potential for developing agents, which target chromatin remodeling and DNA methylation is profound. A better understanding of these effects could lead to novel therapeutics that efficiently target hypermethylated candidate genes, and potentially reverse the abnormalities in coordinating histone modifications and DNA methylation (Huang and Akbarian, 2007). It remains to be determined whether the increase in GADD45b in psychosis is compensatory to a restrictive chromatin state or a hypermethylating cellular milieu, a contributor to psychosis, or a result of environmental factors caused by living with a psychotic illness. If the former hypothesis is correct then bolstering an individual's DNA demethylating capacity using chromatin ‘opening' drugs would be expected to increase GADD45b access to gene promoters and potentially result in favorable gene expression changes.
Acknowledgments
We thank Dr Ekrem Maloku for his excellent technical support. This work was supported in part by the APA AstraZeneca Young Minds in Psychiatry Award (DPG) and PHS Grants MH069839 (RPS) and MH070855 (AG).
The authors declare no conflict of interest.
References
- Abdolmaleky HM, Cheng KH, Faraone SV, Wilcox M, Glatt SJ, Gao F, et al. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2006;15:3132–3145. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, et al. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;134B:60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- Agis-Balboa RC, Pinna G, Pibiri F, Kadriu B, Costa E, Guidotti A. Down-regulation of neurosteroid biosynthesis in corticolimbic circuits mediates social isolation-induced behavior in mice. Proc Natl Acad Sci USA. 2007;104:18736–18741. doi: 10.1073/pnas.0709419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agis-Balboa RC, Pinna G, Zhubi A, Maloku E, Veldic M, Costa E, et al. Characterization of brain neurons that express enzymes mediating neurosteroid biosynthesis. Proc Natl Acad Sci USA. 2006;103:14602–14607. doi: 10.1073/pnas.0606544103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbarian S, Kim JJ, Potkin SG, Hagman JO, Tafazzoli A, Bunney WE, Jr, et al. Gene expression for glutamic acid decarboxylase is reduced without loss of neurons in prefrontal cortex of schizophrenics. Arch Gen Psychiatry. 1995;52:258–266. doi: 10.1001/archpsyc.1995.03950160008002. [DOI] [PubMed] [Google Scholar]
- Akbarian S, Ruehl MG, Bliven E, Luiz LA, Peranelli AC, Baker SP, et al. Chromatin alterations associated with down-regulated metabolic gene expression in the prefrontal cortex of subjects with schizophrenia. Arch Gen Psychiatry. 2005;62:829–840. doi: 10.1001/archpsyc.62.8.829. [DOI] [PubMed] [Google Scholar]
- Barrachina M, Castano E, Ferrer I. TaqMan PCR assay in the control of RNA normalization in human post-mortem brain tissue. Neurochem Int. 2006;49:276–284. doi: 10.1016/j.neuint.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Benes FM, Lim B, Matzilevich D, Walsh JP, Subburaju S, Minns M. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci USA. 2007;104:10164–10169. doi: 10.1073/pnas.0703806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks PJ, Marietta C, Goldman D. DNA mismatch repair and DNA methylation in adult brain neurons. J Neurosci. 1996;16:939–945. doi: 10.1523/JNEUROSCI.16-03-00939.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhausen G, Graessmann M, Graessmann A. Inhibition of herpes simplex thymidine kinase gene expression by DNA methylation is an indirect effect. Nucleic Acids Res. 1985;13:5503–5513. doi: 10.1093/nar/13.15.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrier F, Georgel PT, Pourquier P, Blake M, Kontny HU, Antinore MJ, et al. Gadd45, a p53-responsive stress protein, modifies DNA accessibility on damaged chromatin. Mol Cell Biol. 1999;19:1673–1685. doi: 10.1128/mcb.19.3.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Uchimura K, Stetler RA, Zhu RL, Nakayama M, Jin K, et al. Transient global ischemia triggers expression of the DNA damage-inducible gene GADD45 in the rat brain. J Cereb Blood Flow Metab. 1998;18:646–657. doi: 10.1097/00004647-199806000-00007. [DOI] [PubMed] [Google Scholar]
- Connor CM, Akbarian S. DNA methylation changes in schizophrenia and bipolar disorder. Epigenetics. 2008;3:55–58. doi: 10.4161/epi.3.2.5938. [DOI] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa E, Davis JM, Dong E, Grayson DR, Guidotti A, Tremolizzo L, et al. A GABAergic cortical deficit dominates schizophrenia pathophysiology. Crit Rev Neurobiol. 2004;16:1–23. doi: 10.1615/critrevneurobiol.v16.i12.10. [DOI] [PubMed] [Google Scholar]
- Craddock N, O'Donovan MC, Owen MJ. Genes for schizophrenia and bipolar disorder? Implications for psychiatric nosology. Schizophr Bull. 2006;32:9–16. doi: 10.1093/schbul/sbj033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Webb S, Kerr AR, Illingworth RS, Guy J, Andrews R, et al. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 2011;21:1074–1086. doi: 10.1101/gr.118703.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster EL, Mill J, Craig IW, Collier DA. The quantification of COMT mRNA in post mortem cerebellum tissue: diagnosis, genotype, methylation and expression. BMC Med Genet. 2006;7:10. doi: 10.1186/1471-2350-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Nelson M, Grayson DR, Costa E, Guidotti A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc Natl Acad Sci USA. 2008;105:13614–13619. doi: 10.1073/pnas.0805493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Stary JM, Earle JA, Araghi-Niknam M, Eagan E. GABAergic dysfunction in schizophrenia and mood disorders as reflected by decreased levels of glutamic acid decarboxylase 65 and 67 kDa and Reelin proteins in cerebellum. Schizophr Res. 2005;72:109–122. doi: 10.1016/j.schres.2004.02.017. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Frederikse M, Lu A, Aylward E, Barta P, Sharma T, Pearlson G. Sex differences in inferior parietal lobule volume in schizophrenia. Am J Psychiatry. 2000;157:422–427. doi: 10.1176/appi.ajp.157.3.422. [DOI] [PubMed] [Google Scholar]
- Fremeau RT, Jr, Troyer MD, Pahner I, Nygaard GO, Tran CH, Reimer RJ, et al. The expression of vesicular glutamate transporters defines two classes of excitatory synapse. Neuron. 2001;31:247–260. doi: 10.1016/s0896-6273(01)00344-0. [DOI] [PubMed] [Google Scholar]
- Gavin DP, Chase KA, Sharma RP. Enhancement of psychotherapy using epigenetic modulating drugs. Med Hypotheses. 2011;77:121–124. doi: 10.1016/j.mehy.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin DP, Kartan S, Chase K, Grayson DR, Sharma RP. Reduced baseline acetylated histone 3 levels, and a blunted response to HDAC inhibition in lymphocyte cultures from schizophrenia subjects. Schizophr Res. 2008;103:330–332. doi: 10.1016/j.schres.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin DP, Kartan S, Chase K, Jayaraman S, Sharma RP. Histone deacetylase inhibitors and candidate gene expression: An in vivo and in vitro approach to studying chromatin remodeling in a clinical population. J Psychiatr Res. 2009a;43:870–876. doi: 10.1016/j.jpsychires.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Gavin DP, Rosen C, Chase K, Grayson DR, Tun N, Sharma RP. Dimethylated lysine 9 of histone 3 is elevated in schizophrenia and exhibits a divergent response to histone deacetylase inhibitors in lymphocyte cultures. J Psychiatry Neurosci. 2009b;34:232–237. [PMC free article] [PubMed] [Google Scholar]
- Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JM, Goodman JM, Seidman LJ, Kennedy DN, Makris N, Lee H, et al. Cortical abnormalities in schizophrenia identified by structural magnetic resonance imaging. Arch Gen Psychiatry. 1999;56:537–547. doi: 10.1001/archpsyc.56.6.537. [DOI] [PubMed] [Google Scholar]
- Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, et al. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci USA. 2005;102:9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Chen Y, Davis JM, Dong E, Gavin DP, et al. Epigenetic GABAergic targets in schizophrenia and bipolar disorder. Neuropharmacology. 2011;60:1007–1016. doi: 10.1016/j.neuropharm.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Di-Giorgi-Gerevini V, Dwivedi Y, Grayson DR, et al. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch Gen Psychiatry. 2000;57:1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Ruzicka W, Grayson DR, Veldic M, Pinna G, Davis JM, et al. S-adenosyl methionine and DNA methyltransferase-1 mRNA overexpression in psychosis. Neuroreport. 2007;18:57–60. doi: 10.1097/WNR.0b013e32800fefd7. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Arion D, Unger T, Maldonado-Aviles JG, Morris HM, Volk DW, et al. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2008;13:147–161. doi: 10.1038/sj.mp.4002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- Huang HS, Akbarian S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PLoS One. 2007;2:e809. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Matevossian A, Whittle C, Kim SY, Schumacher A, Baker SP, et al. Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. J Neurosci. 2007;27:11254–11262. doi: 10.1523/JNEUROSCI.3272-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Yamada K, Takao H, Iwayama-Shigeno Y, Yoshikawa T, et al. DNA methylation status of SOX10 correlates with its downregulation and oligodendrocyte dysfunction in schizophrenia. J Neurosci. 2005;25:5376–5381. doi: 10.1523/JNEUROSCI.0766-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez JA, Mancini-Marie A, Lakis N, Rinaldi M, Mendrek A. Disturbed sexual dimorphism of brain activation during mental rotation in schizophrenia. Schizophr Res. 2010;122:53–62. doi: 10.1016/j.schres.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Jin SG, Wu X, Li AX, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Kondo T, Takada I, Youn MY, Yamamoto Y, Takahashi S, et al. DNA demethylation in hormone-induced transcriptional derepression. Nature. 2009;461:1007–1012. doi: 10.1038/nature08456. [DOI] [PubMed] [Google Scholar]
- Koppel I, Aid-Pavlidis T, Jaanson K, Sepp M, Pruunsild P, Palm K, et al. Tissue-specific and neural activity-regulated expression of human BDNF gene in BAC transgenic mice. BMC Neurosci. 2009;10:68. doi: 10.1186/1471-2202-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrisciano F, Dong E, Gavin DP, Nicoletti F, Guidotti A. Activation of group-Ii metabotropic glutamate receptors promotes DNA demethylation in the mouse brain. Mol Pharmacol. 2011;80:174–182. doi: 10.1124/mol.110.070896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullumsmith RE, Meador-Woodruff JH. Expression of transcripts for the vesicular glutamate transporters in the human medial temporal lobe. Ann NY Acad Sci. 2003;1003:438–442. doi: 10.1196/annals.1300.046. [DOI] [PubMed] [Google Scholar]
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, et al. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munzel M, Globisch D, Bruckl T, Wagner M, Welzmiller V, Michalakis S, et al. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew Chem Int Ed Engl. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- Naghavi HR, Nyberg L. Common fronto-parietal activity in attention, memory, and consciousness: shared demands on integration. Conscious Cogn. 2005;14:390–425. doi: 10.1016/j.concog.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Hioki H, Fujiyama F, Kaneko T. Postnatal changes of vesicular glutamate transporter (VGluT)1 and VGluT2 immunoreactivities and their colocalization in the mouse forebrain. J Comp Neurol. 2005;492:263–288. doi: 10.1002/cne.20705. [DOI] [PubMed] [Google Scholar]
- Niznikiewicz M, Donnino R, McCarley RW, Nestor PG, Iosifescu DV, O'Donnell B, et al. Abnormal angular gyrus asymmetry in schizophrenia. Am J Psychiatry. 2000;157:428–437. doi: 10.1176/appi.ajp.157.3.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Pope HG, Jr, Yurgelun-Todd D. Schizophrenic individuals with bipolar first-degree relatives: analysis of two pedigrees. J Clin Psychiatry. 1990;51:97–101. [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90:397–406. doi: 10.1016/j.ygeno.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruunsild P, Sepp M, Orav E, Koppel I, Timmusk T. Identification of cis-elements and transcription factors regulating neuronal activity-dependent transcription of human BDNF gene. J Neurosci. 2011;31:3295–3308. doi: 10.1523/JNEUROSCI.4540-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, Luu KV, et al. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci USA. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenberg A, Harvey PD, Bowie CR, Mojtabai R, Rabinowitz J, Heaton RK, et al. Neuropsychological function and dysfunction in schizophrenia and psychotic affective disorders. Schizophr Bull. 2009;35:1022–1029. doi: 10.1093/schbul/sbn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MA, Caruncho HJ, Costa E, Pesold C, Liu WS, Guidotti A. In Patas monkey, glutamic acid decarboxylase-67 and reelin mRNA coexpression varies in a manner dependent on layers and cortical areas. J Comp Neurol. 2002;451:279–288. doi: 10.1002/cne.10341. [DOI] [PubMed] [Google Scholar]
- Ruzicka WB, Zhubi A, Veldic M, Grayson DR, Costa E, Guidotti A. Selective epigenetic alteration of layer I GABAergic neurons isolated from prefrontal cortex of schizophrenia patients using laser-assisted microdissection. Mol Psychiatry. 2007;12:385–397. doi: 10.1038/sj.mp.4001954. [DOI] [PubMed] [Google Scholar]
- Schmitz KM, Schmitt N, Hoffmann-Rohrer U, Schafer A, Grummt I, Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell. 2009;33:344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Sharma RP, Gavin DP, Grayson DR. CpG methylation in neurons: message, memory, or mask. Neuropsychopharmacology. 2010;35:2009–2020. doi: 10.1038/npp.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma RP, Grayson DR, Gavin DP. Histone deactylase 1 expression is increased in the prefrontal cortex of schizophrenia subjects: analysis of the National Brain Databank microarray collection. Schizophr Res. 2008;98:111–117. doi: 10.1016/j.schres.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberberg G, Baruch K, Navon R. Detection of stable reference genes for real-time PCR analysis in schizophrenia and bipolar disorder. Anal Biochem. 2009;391:91–97. doi: 10.1016/j.ab.2009.05.026. [DOI] [PubMed] [Google Scholar]
- Szyf M. A dynamic methylome; implications of non-CG methylation/demethylation. Cell Cycle. 2010;9:3846–3847. doi: 10.4161/cc.9.19.13563. [DOI] [PubMed] [Google Scholar]
- Tochigi M, Iwamoto K, Bundo M, Komori A, Sasaki T, Kato N, et al. Methylation status of the reelin promoter region in the brain of schizophrenic patients. Biol Psychiatry. 2008;63:530–533. doi: 10.1016/j.biopsych.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Torrey EF. Schizophrenia and the inferior parietal lobule. Schizophr Res. 2007;97:215–225. doi: 10.1016/j.schres.2007.08.023. [DOI] [PubMed] [Google Scholar]
- Torrey EF, Webster M, Knable M, Johnston N, Yolken RH. The stanley foundation brain collection and neuropathology consortium. Schizophr Res. 2000;44:151–155. doi: 10.1016/S0920-9964(99)00192-9. [DOI] [PubMed] [Google Scholar]
- Tsuang MT. Morbidity risks of schizophrenia and affective disorders among first-degree relatives of patients with schizoaffective disorders. Br J Psychiatry. 1991;158:165–170. doi: 10.1192/bjp.158.2.165. [DOI] [PubMed] [Google Scholar]
- Uezato A, Meador-Woodruff JH, McCullumsmith RE. Vesicular glutamate transporter mRNA expression in the medial temporal lobe in major depressive disorder, bipolar disorder, and schizophrenia. Bipolar Disord. 2009;11:711–725. doi: 10.1111/j.1399-5618.2009.00752.x. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldic M, Caruncho HJ, Liu WS, Davis J, Satta R, Grayson DR, et al. DNA-methyltransferase 1 mRNA is selectively overexpressed in telencephalic GABAergic interneurons of schizophrenia brains. Proc Natl Acad Sci USA. 2004;101:348–353. doi: 10.1073/pnas.2637013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldic M, Guidotti A, Maloku E, Davis JM, Costa E. In psychosis, cortical interneurons overexpress DNA-methyltransferase 1. Proc Natl Acad Sci USA. 2005;102:2152–2157. doi: 10.1073/pnas.0409665102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldic M, Kadriu B, Maloku E, Agis-Balboa RC, Guidotti A, Davis JM, et al. Epigenetic mechanisms expressed in basal ganglia GABAergic neurons differentiate schizophrenia from bipolar disorder. Schizophr Res. 2007;91:51–61. doi: 10.1016/j.schres.2006.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Angyal L. Zur bedeutung des interparietalen syndroms bei der schizophrenie. Eur Arch Psychiatry Clin Neurosci. 1934;102:107–119. [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Ying J, Srivastava G, Hsieh WS, Gao Z, Murray P, Liao SK, et al. The stress-responsive gene GADD45G is a functional tumor suppressor, with its response to environmental stresses frequently disrupted epigenetically in multiple tumors. Clin Cancer Res. 2005;11:6442–6449. doi: 10.1158/1078-0432.CCR-05-0267. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Beezhold K, Castranova V, Shi X, Chen F. Characterization of an alternatively spliced GADD45alpha, GADD45alpha1 isoform, in arsenic-treated epithelial cells. Mol Carcinog. 2009;48:454–464. doi: 10.1002/mc.20483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu RL, Graham SH, Jin J, Stetler RA, Simon RP, Chen J. Kainate induces the expression of the DNA damage-inducible gene, GADD45, in the rat brain. Neuroscience. 1997;81:707–720. doi: 10.1016/s0306-4522(97)00205-4. [DOI] [PubMed] [Google Scholar]
- Zhubi A, Veldic M, Puri NV, Kadriu B, Caruncho H, Loza I, et al. An upregulation of DNA-methyltransferase 1 and 3a expressed in telencephalic GABAergic neurons of schizophrenia patients is also detected in peripheral blood lymphocytes. Schizophr Res. 2009;111:115–122. doi: 10.1016/j.schres.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]