Abstract
Herpesviruses, a large family of double-stranded DNA enveloped viruses, accomplish infection of their host cells through viral membrane fusion with either the plasma membrane or endocytic vesicle membranes. Efficient herpesvirus infection of cells requires the concerted effort of multiple glycoproteins and involves multiple host receptors, making it a remarkably complex system of viral fusion relative to the majority of enveloped viruses, which generally require only one or two viral glycoproteins. The structures of the major glycoproteins and receptors involved in the entry of the prototypical herpesviruses herpes simplex virus (HSV) and Epstein-Barr virus (EBV) are now known. These structural studies have accelerated our understanding of HSV and EBV binding and fusion by revealing conformational changes that occur upon receptor binding, depicting potential sites of functional protein and lipid interactions, and identifying the likely viral fusogen.
Herpesviruses form a large, diverse family of double-stranded DNA enveloped viruses. There are eight human herpesviruses (HHV): herpes simplex viruses type 1 and 2 (HSV-1, HSV-2), Epstein Barr virus (EBV), varicella zoster virus (VZV), cytomegalovirus (CMV), HHV-6, HHV-7 (roseolovirus), and Kaposi’s sarcoma-associated herpesvirus (KSHV)1. HHVs are ubiquitous in the human population and establish latent infections in a wide range of hosts that can be reactivated to cause various diseases. The prototypical herpesviruses HSV-1 and HSV-2, members of the alphaherpesvirus subfamily, most commonly cause localized mucocutaneous lesions known as oral herpes (HSV-1) or genital lesions (HSV-2) but can also cause meningitis and encephalitis. EBV, a member of the gammaherpesvirus subfamily, is the most common cause of infectious mononucleosis and is causally associated with several malignancies, including Burkitt’s lymphoma and Hodgkin’s lymphoma.
Although the pathogenesis of infection with herpesviruses differs, they enter host cells through a similar mechanism and use a conserved set of viral glycoproteins for membrane fusion. Entry of herpesviruses into cells proceeds in two distinct steps (Fig. 1A). First, the virus binds to the host cell through specific receptors. This brings the viral fusion apparatus in close proximity to the membrane of the cell. Some of these receptor-binding events trigger fusion, whereas others simply serve to tether the virus to the cell and are dispensable for fusion. Second, the viral membrane fuses with either the host plasma membrane or endocytic membrane through the action of a viral fusion protein that perturbs the host membrane. Herpesviruses generally require a larger number of envelope proteins to accomplish entry than most other enveloped viruses, which often accomplish target cell binding and membrane fusion using a single viral envelope protein. Recent structural studies of herpesvirus envelope glycoproteins have shed light on the complex process of herpes virus binding and fusion. In this review, we will focus on the requirements for viral binding and fusion of two prototypic HHVs, HSV and EBV, with a particular emphasis on the structural properties of the viral glycoproteins responsible for these functions, including details of the conformational changes that may trigger fusion and the functional domains present in the primary viral fusion protein and its regulators.
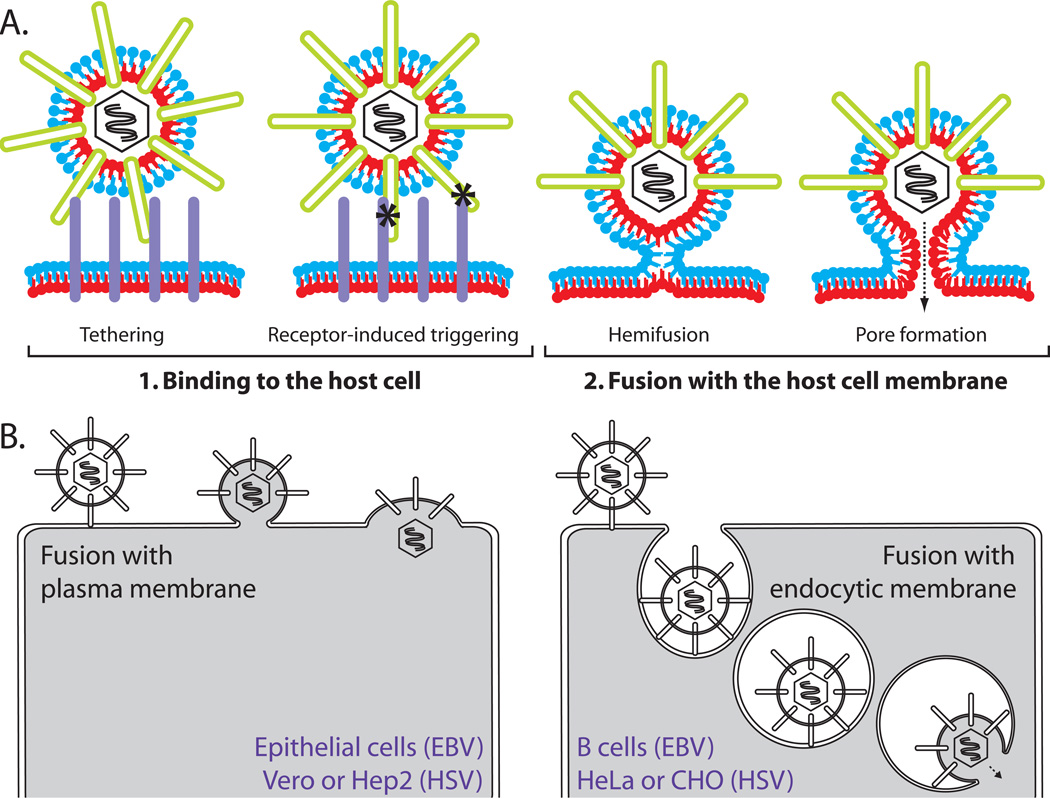
Figure 1. Herpesvirus entry.
(A) Routes of entry. Depending upon cell type, both HSV and EBV can enter cells by fusion at the plasma membrane or fusion with an endocytic membrane after endocytosis. Examples of common cells for which each entry route is used are noted. (B) Two steps of virus entry. 1. Virus binds to cellular receptors (purple) via envelope glycoproteins (green). Some receptor binding events serve simply to tether the virus to the cell. Other specific receptor binding events trigger conformational changes in the entry glycoproteins that mediate membrane fusion (asterisks). 2. Fusion of the viral and cellular membranes progresses through a hemifusion intermediate, in which the outer membrane leaflets (blue) mix. This is followed by full fusion, where the inner membrane leaflets (red) mix and a fusion pore is formed. This figure is not drawn to scale.
Cell tropism
HSV infects a variety of host cells, including lymphocytes, epithelial cells, fibroblasts, and neurons. The pathways of HSV entry are varied as well and depend upon the target cell type. Although initial studies indicated that HSV enters cells by direct fusion with the plasma membrane at neutral pH, further analyses revealed that HSV entry also can occur by endocytosis in either a pH-dependent or a pH-independent manner (Fig. 1B)2, 3. The entry route differs among cell lines and the reason that endocytosis is sometimes required is unclear. In fact, for some cells in which endocytosis is the required route of entry, cell surface expression of the viral glycoproteins that mediate entry causes cell-cell fusion 2. None of the HSV entry glycoproteins require low pH to function, although low pH has been shown to result in minor conformational differences in glycoprotein B (gB)4, 5. The ability to enter cells using multiple pathways may facilitate the productive infection of the multiple cell types encountered by HSV.
EBV, in contrast, predominately infects epithelial cells and B cells and, under some circumstances can infect other cell types, such as monocytes6. The route of entry used by EBV is also cell-type dependent; entry into epithelial cells occurs by direct fusion at the cell surface, whereas entry into B cells occurs via endocytosis in a pH-independent manner7.
Entry machinery
At least a dozen different glycoproteins are expressed on the herpesvirus envelope and a subset of these is necessary and sufficient for viral entry. The herpesvirus entry process is complex because not only do the viruses employ multiple glycoproteins to mediate entry, but also each of these glycoproteins is multifunctional (Table 1). Each entry glycoprotein may bind multiple receptors, interact with other glycoproteins, and/or undergo conformational changes to induce membrane fusion. Some glycoproteins are required for entry, whereas others simply enhance entry. EBV and HSV share a set of three conserved glycoproteins that are required for fusion with all cell types: gB and a heterodimer composed of glycoprotein H (gH) and glycoprotein L (gL), referred to as gH/gL. HSV fusion also requires the receptor binding activity of glycoprotein D (gD). Similarly, EBV fusion with B cells, but not epithelial cells, requires the receptor binding activity of glycoprotein 42 (gp42). Glycoproteins that tether the virus to the cell but are not essential for entry include HSV glycoprotein C (gC) and EBV glycoproteins 350/220 (gp350/220) and BMRF-2.
Table 1.
Glycoprotein functions and receptor interactions
| Glycoproteins | Function | Cellular Receptor(s) | |
|---|---|---|---|
| EBV | BMRF2 | Binds epithelial cells (dispensable for fusion) | β1 or α5β1 integrins |
| gp350 | Binds cells (dispensable for fusion) | CR2/CD21 | |
| gp42 | Binds B cells, triggers fusion | HLA class II | |
| gH/gL | Binds epithelial cells Triggers/regulates fusion with epithelial and B cells | αvβ6 & αvβ8 integrins | |
| gB | Catalyze membrane fusion | ||
| HSV | gC | Binds cells (dispensable for fusion) | Heparan sulfate |
| gD | Binds cells, triggers fusion | HVEM, nectin-1, nectin-2, 3-O-sulfated heparan sulfate | |
| gH/gL | Triggers/regulates fusion | unknown | |
| gB | Catalyze membrane fusion | heparan sulfate, DC-SIGN PILRα, MAG, NMHC-IIA |
Tethering to cells
Both HSV and EBV employ glycoproteins for viral tethering to host cells. In the absence of these glycoproteins viral entry can still occur, albeit at lower efficiency. Tethering serves to concentrate the virus at the cell surface but does not specifically trigger fusion. HSV tethers to host cells through the interactions of gB and gC with heparan sulfate on cell surface proteoglycans. Although gB is required for HSV fusion, its heparan sulfate-binding activity is not essential for fusion 8. gC has no required role in HSV fusion and is dispensable for viral attachment in vitro, however, gC deletion decreases the efficiency of virus binding9, 10. gB and gC also bind to dendritic cell-specific C-type lectin (DC-SIGN), which facilitates attachment during infection of dendritic cells 11.
EBV tethers to target B cells primarily through the interaction of gp350/220, one of the most abundant EBV envelope glycoproteins, with complement receptor type 2 (CR2/CD21), a B cell-specific surface receptor12–16. Although gp350/220 is not strictly required for entry into B cells, its absence reduces infection efficiency17. gp350/220 is a single-pass membrane protein, which as a result of alternative splicing is made in two forms with approximates masses of 350 and 220 kDa. The N-terminal residues (1–470) of either form of the protein can bind CR2/CD2118 and no specific roles for two forms of the protein have been determined. The structure of gp350 has been solved 19 and the CR2/CD21 binding site maps to a glycan-free patch on its surface.
gp350/220 is also an important mediator of EBV tethering to epithelial cells (for CR2/CD21-expressing epithelial cells), as is BMRF-2 (for integrin-expressing oral epithelial cells). EBV BMRF-2 is a transmembrane envelope glycoprotein with an integrin-binding RGD motif capable of interacting with the β1 and α5 family integrins expressed on oral epithelial cells20, 21. Recombinant EBV lacking BMRF-2 shows a 50% reduction in attachment to oral epithelial cells, suggesting that BMRF-2 plays a significant role at the portal of entry for human EBV infection in vivo22. BMRF-2 has also been shown to facilitate lateral cell-to-cell spread of EBV within polarized oral epithelial cells23.
EBV triggers of fusion
EBV gp42 receptor binding on B cells
The tethering of EBV to cells is not sufficient to trigger fusion. EBV gp42 is the receptor-binding protein required to trigger fusion with B cells. gp42 triggers membrane fusion after binding human leukocyte antigen type II (HLA class II) on target B cells. EBV entry can be triggered by all three HLA class II isotypes (HLA-DR, HLA-DP, and HLA-DQ), however some HLA-DQ alleles are nonfunctional24. gp42 belongs to the C-type lectin superfamily and contains a C-terminal C-type lectin domain (CTLD). gp42 has sequence homologs among the closely related primate lymphocryptoviruses and functional homologs in other herpesviruses such as HSV gD, discussed later in this review. gp42 is a type II membrane protein with a N-terminal transmembrane (TM) domain. It is cleaved adjacent to the TM domain, to generate a functional soluble form of gp42 in vivo25. Soluble gp42 can trigger viral fusion with B cells in the absence of membrane-bound gp42 and a mutation of the gp42 cleavage site that blocks production of soluble gp42 inhibits B cell fusion26.
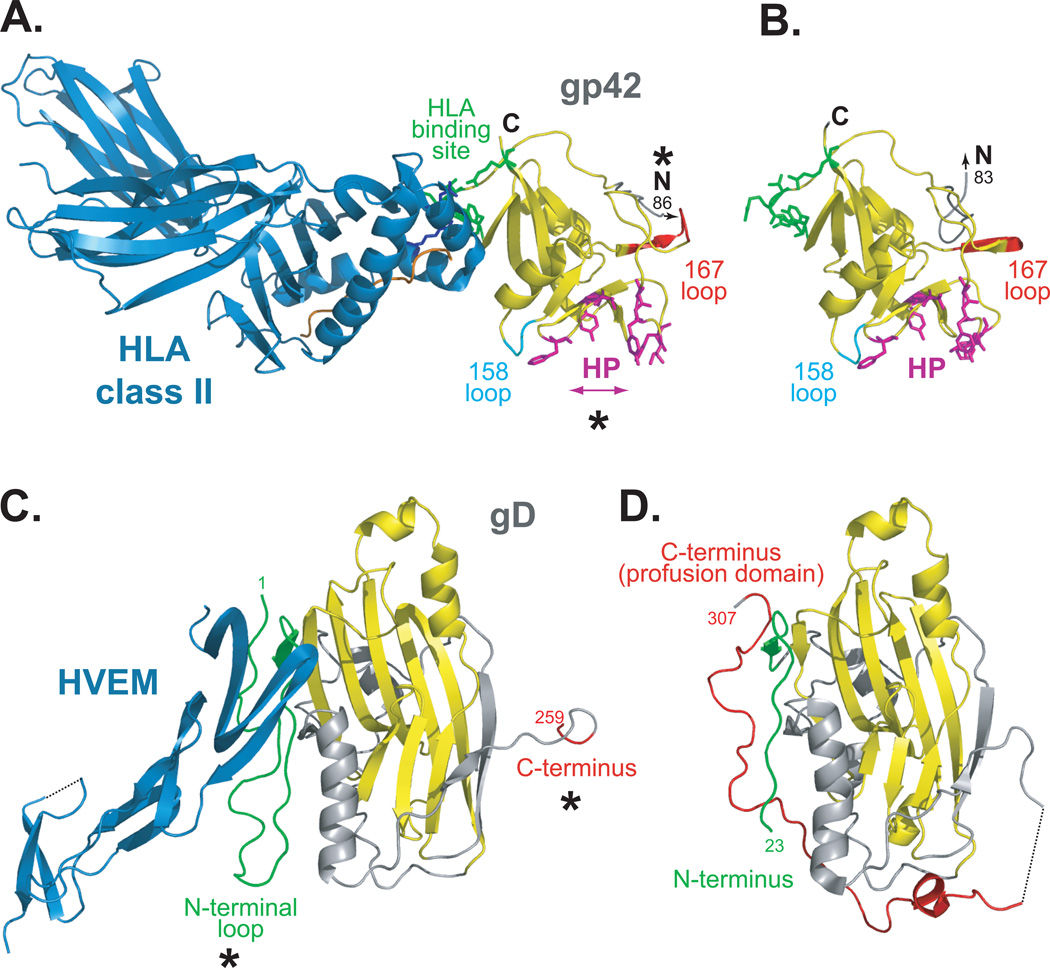
The structures of both HLA class II-bound gp42 and unbound gp42 have been determined (Fig. 2A, 2B)27, 28. The structure of gp42 bound to HLA class II revealed three key structural features that relate well to mutagenic studies of gp42 28. First, HLA class II binds within the gp42 CTLD (residues 94–221) at a site distinct from the canonical HLA-interactive site present in other CTLDs, such as that of the natural killer cell receptor Ly49A, which forms complexes with HLA class I29. Only two key HLA class II β chain side chains make extensive interactions with gp42: HLA class II glutamic acid at position β46 interacts with gp42 residue 220, and HLA class II arginine β72 interacts with gp42 residues 104–107. Second, the N-terminal portion of gp42, known to interact with gH/gL via residues 36–8130, is flexible and not resolved in the structure. Lastly, a hydrophobic pocket is present at the canonical ligand-binding site of gp42 Residues within this hydrophobic pocket are essential for B cell entry31, indicating that this pocket may interact with another ligand during B cell entry. Crystallographic dimerization of HLA-bound gp42 via its N-terminal region was observed 28, although dimerization does not occur with high affinity in solution32; hence, the oligomeric state of native gp42 at the viral membrane surface is not yet clear.
Figure 2. EBV and HSV receptor binding proteins, in bound and unbound states.
(A) EBV gp42 bound to class II HLA-DR1 (PDB ID 1KG0). (B) Unbound gp42 (PDB ID 3FD4). gp42 binds HLA class II (blue) at a non-canonical site within the gp42 CTLD (yellow, residues 94–221). HLA class II residues E46 and R72 (blue sticks) make extensive contacts with gp42 residues 104–107 and R200 (green sticks). The canonical CTLD interaction site lies on an opposite face of gp42 at a pocket lined with hydrophobic residues (magenta sticks). Asterisks denote conformational differences seen in gp42 in the presence or absence of HLA class II. When bound to HLA, gp42 loops at residue 158 (cyan) and 167 (red) are shifted and the hydrophobic pocket (HP) widens compared to unbound gp42. In the absence of HLA, the gp42 N-terminus projects outward in a path distinct from that observed for HLA-bound gp42. The N-terminus of gp42 is flexible and the residues that bind to gH/gL (gp42 residues 36–81) were not resolved. Peptide loaded in the HLA class II is colored orange. (C) HSV-1 gD bound to HVEM (PDB ID 1JMA). A truncated form of the gD ectodomain (gD285) with high affinity for HVEM was crystallized in complex with HVEM (blue). (D) Unbound gD (PDB ID 2C3A). A cysteine was added to the C-terminus of the gD ectodomain [gD(23–306)307C] which stabilized a gD dimer. A monomer is shown. The core of gD forms an Ig fold (yellow) that is flanked by N-terminal (green) and C-terminal (red) extensions. Asterisks denote conformational differences seen in gD in the presence or absence of receptor. In the presence of HVEM, the N-terminus of gD (green) forms a loop that serves as the receptor binding site and is stabilized by receptor binding. The C-terminal region, residues 260–285, is disordered in the crystal. In the absence of HVEM, the C-terminal region (red), which includes the pro-fusion domain, is anchored near the N-terminal region (green). The C-terminus masks the receptor binding site and the N-terminal loop conformation is not present. The structure of gD285 (PDB ID 1L2G, not shown) in the absence of receptor demonstrates a similar loss of the N-terminal loop and its N-terminus is disordered past residue 14 59.
In the more recently solved structure of unbound gp4227, 140 two regions of gp42 displayed noticeably different conformations in the absence of HLA class II, which may indicate a mechanism by which gp42 triggers membrane fusion upon receptor binding. First, the hydrophobic pocket of gp42 is widened in response to HLA class II-binding, emphasizing its critical and seemingly dynamic role in EBV B cell entry. It has been hypothesized that the gp42 hydrophobic pocket may contain a second, higher-affinity binding site for gH/gL or a binding site for gB or a cellular receptor27. Second, in the unbound structure, the flexible N-terminal region of one gp42 molecule (residues 87–93, between the gH/gL binding region and the CTLD) contacts the HLA-binding site of another gp42 molecule. The N-terminal region also projects outward from the CTLD in a path distinct from that observed in the HLA-bound structure. Changes in the gp42 N-terminal region could occur in response to receptor binding and could contribute to fusion triggering.
gp42 as a viral tropism switch
Whereas a three-part complex of gp42/gH/gL is required along with gB for B cell entry, gH/gL and gB are necessary and sufficient for epithelial cell entry. Soluble gp42 can rescue the B cell infectivity of gp42-null EBV; however the presence of soluble gp42 is inhibitory to epithelial cell membrane fusion and entry33, 34. This gp42-mediated inhibition of epithelial cell entry is due to the binding of gp42 to gH/gL, since an N-terminal peptide of gp42 (residues 36–81) that is sufficient for gH/gL binding also inhibits epithelial cell entry30, 32. This gp42 peptide inhibits B cell fusion as well when fusion is triggered in vitro with soluble gp42, however when gp42 and gH/gL are co-expressed in the same cell, the peptide exhibits only minimal B cell fusion inhibition, likely because gp42/gH/gL complexes are preformed during biosynthesis prior to exposure to inhibitory peptide at the cell surface. Virions synthesized in B cells contain lower amounts of gp42 in the virion envelope due to sequestration by cellular HLA class II, whereas EBV synthesized in infected epithelial cells contains higher amounts of gp42 in its envelope35. Hence, EBV appears to utilize gp42 as a switch of cell tropism. Interestingly, some other herpesviruses, including the betaherpesviruses CMV36 and HHV637, also encode glycoproteins that redirect cell tropism by forming stable complexes with gH/gL. No such stable complexes with HSV gH/gL have been reported.
EBV gH/gL receptor binding on epithelial cells
Compared to EBV B cell entry, epithelial cell entry has only recently begun to be understood. Integrins, broadly expressed receptors that mediate attachment of cells to surrounding tissue, seem to be intimately involved in EBV epithelial cell attachment and fusion. The minimal required viral glycoproteins for EBV attachment to epithelial cells are gH/gL38. Although it has been noted for some time that EBV gH/gL can bind epithelial, but not B cells39–41, the first epithelial receptor for gH/gL was determined only recently42. Binding of a soluble form of gH/gL to epithelial cells is reduced by siRNA-downregulation of αv integrins. Soluble gH/gL is also capable of co-precipitating with αvβ6 and αvβ8, but not αvβ3 integrins. Furthermore, the addition of soluble αvβ6 or αvβ8 integrin induced fusion of cells expressing gH/gL and gB42. Taken together, these lines of evidence strongly suggest EBV gH/gL binds these integrins and this receptor-binding is sufficient to trigger EBV entry into epithelial cells.
HSV triggers of fusion
HSV gD receptor binding
gD is the main HSV receptor-binding protein. It binds three classes of receptors: herpes virus entry mediator (HVEM), a member of the TNF receptor family; nectin-1 and nectin-2, cell adhesion molecules of the immunoglobulin superfamily; and 3-O-sulfated heparan sulfate43, 44. Soluble forms of gD and its receptors can trigger fusion, indicating that the gD-receptor interaction does more than tether the virus to the cell 45–47. Which receptors are important during in vivo infection is not clear, however in mouse models of both genital herpes and herpes simplex encephalitis, nectin-1 is the dominant receptor utilized by virus48–50. Notably, nectin-1 is expressed in neurons, the site of HSV latency. Interestingly, insertion of heterologous ligands into the gD sequence can be used to retarget HSV to novel receptors, such as IL13Rα251 or HER252.
The structure of a truncated soluble form of gD in complex with HVEM has been solved, revealing a V-like immunoglobulin fold at the gD core flanked by a large C-terminal extension and an N-terminal loop. The C-terminal extension wraps around the core domain and anchors the protein to the viral membrane53 (Fig. 2C). The N-terminal loop contains all of the HVEM contact sites within gD at residues 7–15 and 24–3253–55. These N-terminal gD residues are critical for interaction with all of the gD receptors. Mutagenesis studies have shown that gD residues 7–32 are required for fusion triggered by 3-O-sulfated heparan sulfate binding56 and that the nectin-1 and nectin-2 interaction also involves residues at the gD N-terminus, most notably Y38, as well as underlying residues within the core of gD57, 58.
Although the N-terminal region of gD contains the receptor-binding site, the C-terminal region of gD is necessary for its the ability to trigger fusion and has been termed the “pro-fusion domain”45. The structure of unliganded gD revealed that in the absence of receptor, the gD N-terminus is flexible and the gD C-terminus is anchored near the N-terminus, masking the receptor-binding site 59 (Fig. 2D). When the gD C-terminus was locked in a ‘receptor-unliganded state’ by introducing an intramolecular disulfide bond, receptor binding and virus entry were abrogated. This suggests that the C-terminus of unbound gD autoinhibits the N-terminal receptor-binding site59, 60. Upon receptor binding, the C-terminus moves and this movement is necessary for both binding receptor and triggering fusion61. Consistent with an autoinhibitory model, deletion of portions of the gD C-terminus increases binding affinity for both HVEM and nectin-162.
HSV gB receptor binding
gB can bind to paired immunoglobulin-like type 2 receptor alpha (PILRα) and this interaction can trigger viral fusion in the presence of gD63. The relatively new finding that gB receptor-binding can trigger fusion is of particular interest because it is a departure from the dominant model in which gD acts as the receptor-binding glycoprotein and gB acts strictly as a fusion protein. PILRα is likely an immune system regulator and the in vivo significance of the gB-PILRα interaction has yet to be explored, but it mediates the infection of several cell lines in vitro63, 64, and may play a minor role in HSV-2 entry into human retinal pigment epithelial cells65.
In addition to PILRα, two recently-identified receptors also have been shown to interact with gB and trigger HSV-1 entry: non-muscle myosin heavy chain IIA (NMHC-IIA)66 and myelin-associated glycoprotein (MAG)67, a protein expressed on neural tissues.
HSV gH/gL receptor binding
Although gH/gL receptor binding has not been shown to be a critical trigger for HSV entry, some studies have indicated a receptor-binding role for gH/gL. HSV gH contains an RGD motif, which may mediate attachment to integrins but is not essential for virus entry 68. A soluble form of gH/gL has been shown to bind to αvβ3 integrin-expressing cells in an RGD motif-specific manner69 and engagement of αvβ3 integrin by gH/gL in the virion may direct the route of entry of HSV70. In addition, soluble gH/gL has been shown to bind cells independent of αvβ3 integrin, and to inhibit virus entry into a number of cell types in vitro71. Similarly, expression in target cells of gH/gL from HSV71or CMV72 inhibits infection, possibly by sequestering cellular receptors.
Conserved core fusion machinery
The HSV and EBV receptor binding activities described above act to trigger fusion, yet are not sufficient for the completion of fusion. To mediate membrane fusion, HSV and EBV employ the conserved proteins gB and gH/gL, collectively considered the core fusion machinery of herpesviruses. The structures of both HSV-1 and EBV gB demonstrate that it has structural homology to viral fusion proteins and is likely a key fusion protein in herpesviruses73, 74. Fusion proteins can insert into target membranes and refold through large conformational changes to draw viral and target cell membranes together, resulting in fusion pore formation. The role of gH/gL during viral fusion is less clear. Recently, the gH/gL structures of HSV-2, EBV, and a partial structure of pseudorabies virus (PRV) gH were solved74–76, helping to define their roles as regulators of fusion.
The role of gH/gL during membrane fusion
The specific role gH/gL plays in fusion has been the most elusive among the required entry glycoproteins. Several studies previously suggested that gH/gL acts as a fusion protein, possibly mediating the hemifusion step77 (Fig. 1A). Peptides corresponding to HSV gH heptad repeats, common features of fusion proteins, were shown to inhibit fusion and peptides corresponding to gH hydrophobic regions, candidate “fusion peptides”, were shown to bind lipids78–83. VZV gH/gL expressed alone using recombinant vaccinia virus was shown be sufficient for cell-cell fusion84, 85. Lastly, 255 HSV gH/gL was shown to promote fusion of the virion envelope with the outer nuclear membrane during viral egress 86.
While these studies suggested that gH/gL has fusogenic capability, a more recent study was unable to demonstrate the ability of gH/gL to induce hemifusion87 and transfection of cells in a vaccinia virus-free setting demonstrated that a combination of VZV gH/gL and gB was required for fusion67. Most importantly, the recently solved crystal structures of the gH/gL ectodomains demonstrate that gH/gL has no structural homology with any known fusion protein74–76 and the candidate ‘fusion peptides’ of gH are buried76. Thus, the structural data support a model in which gH/gL acts not as a fusogen, but primarily as a regulator of fusion through interactions with gB.
HSV-2 gH/gL has an unusual “boot-shaped” structure composed of three domains, with the N-terminal domain formed by both gH and gL76 (Fig. 3A). The interface of gH and gL within this N-terminal domain is extensive and the proteins likely require one another for proper folding. Interestingly, the EBV gH/gL structure has a more linear domain arrangement with significantly different interdomain packing arrangements and modified domain designations74 (Fig. 3B). This difference in the orientation of the N-terminal domains may represent static structural variation between the two protein complexes. Alternatively, the structural differences may reflect that gH/gL domains can undergo dynamic rearrangement.
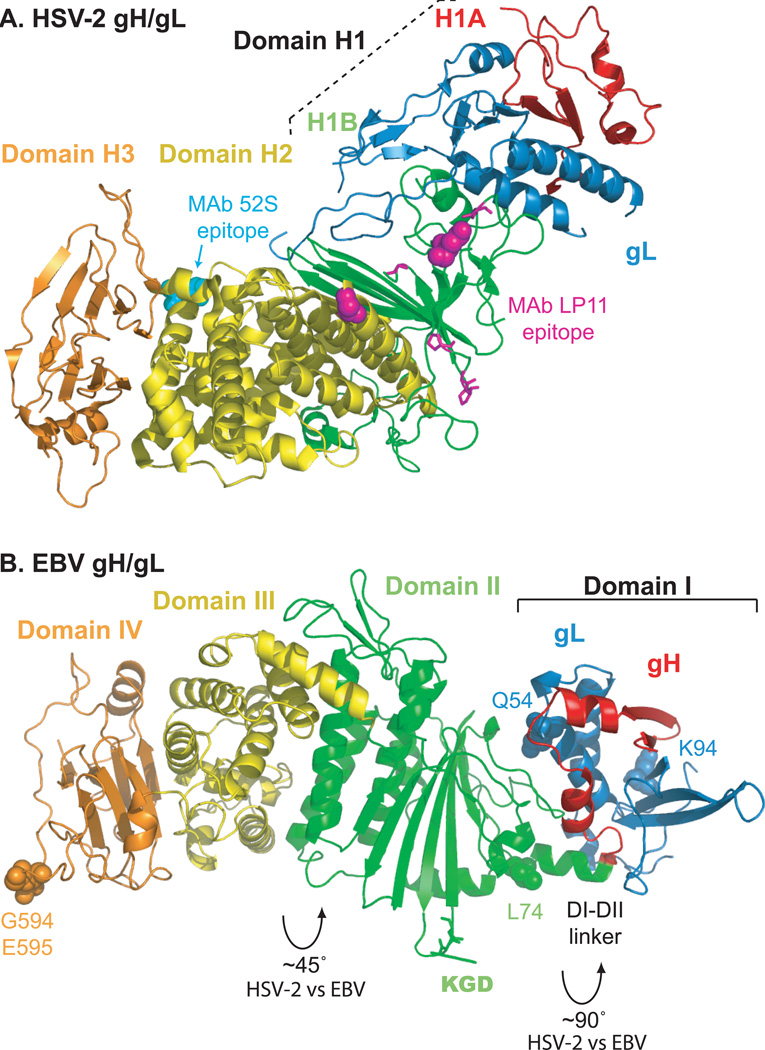
Figure 3. Conserved entry glycoproteins gH/gL.
(A) HSV-2 gH/gL (PDB ID 3M1C). The HSV gH/gL heterodimer adopts a boot-like configuration. The N-terminal gH domain H1, comprised of subdomains H1A (red) and H1B (green), clamps gL (blue) and make extensive contacts. The C-terminal gH domains H2 (yellow) and H3 (orange) are shown. Substitution mutations at gH residues 168 or 329 (magenta spheres) or insertion mutations at gH residues 300, 313, 316, or 317 (magenta sticks) prevent binding of the neutralizing MAb LP11. On the opposite face of the complex, substitution mutations at gH 536 or 537 (cyan spheres) prevent binding of the neutralizing MAb 52S. (B) EBV gH/gL (PDB ID 3PHF). EBV gH/gL domain designations differ somewhat from HSV-2. Domain I is comprised of gL (blue) and the N-terminus of gH (red). Domain II (green) contains an eight-stranded β-sheet that forms a ‘picket fence’ separating domain I from a helical bundle within domain II that lies parallel to the fence. Domain III (yellow) is mainly helical and domain IV (orange) forms a β-sandwich. The EBV gH/gL heterodimer is more linear and elongated than the HSV-2 complex because the interdomain packing arrangements of EBV gH/gL differ significantly. Packing angles differ by ~90° between domains I and II and ~45° between domains II and III. EBV gL residues Q54 and K94 (blue spheres) contribute to a functional interaction with EBV gB. A KGD motif (green sticks) is located in a prominent loop of domain II and lies adjacent to the helical linker between domains I and II. A mutation at residue gH-L74 (green spheres) near the linker results in increased fusion promotion. Mutations at gH residues G594 or E595 (orange spheres) differentially affect fusion promotion.
The gH domains proximal to the viral membrane are more conserved among viruses and adopt similar arrangements in the HSV and EBV structures. In contrast, the N-terminal domains of gH/gL are the most divergent in sequence among herpesviruses, suggesting that this membrane distal portion of the complex might interact with other virus- or cell-specific proteins. For example, EBV gH domain II contains an integrin-binding KGD motif within an exposed loop. In addition, the EBV gH/gL domain arrangement creates a prominent interdomain groove, near the KGD motif and a domain I-II linker, which could serve as an interaction site for gp42 or an epithelial cell receptor. Using chimeric gL molecules comprised of EBV and rhesus lymphocrytovirus sequences, a species-specific functional interaction between gH/gL and gB was mapped to EBV gL residues 54 and 9488. This gB interaction site on EBV gH/gL is distinct from that proposed for HSV gH/gL. The HSV gB interaction site is proposed to lie further down the complex, just below the epitope for antibody LP11 (Fig, 3A), on the surface of domain H2 in a region that is conserved between HSV-1 and HSV-276.
Neutralizing antibodies (Ab) bind multiple regions of gH/gL, possibly reflecting the fact that gH/gL must interact with several partners for function. For example, the anti-HSV gH/gL neutralizing Ab LP11 binds at the border of domains H1B and H2, whereas the neutralizing Ab 52S maps to the opposite face of the complex in domain H2 (Fig. 3A). Similarly, the anti-EBV gH/gL neutralizing Ab E1D1 binds in domain I near residues 65 and 69, whereas the neutralizing Ab E1D1 maps to domains III and IV 40, 41.
Mutagenesis studies also indicate that both the membrane distal and membrane proximal domains of the gH/gL ectodomain contribute to fusion promotion, consistent with gH/gL having multiple roles during entry. HSV gH is relatively tolerant of insertion mutations, however insertions in multiple domains reduce fusion efficiency and insertions at membrane proximal HSV gH residues 791 and 799, downstream of the solved structure but prior to the TM, abrogate fusion68, 89. Similarly, mutation of membrane proximal EBV gH residue 594 abrogates epithelial and B cell fusion, whereas mutation of residue 595 enhances B cell fusion and decreases epithelial cell fusion41. At the other end of the complex, mutation of EBV gH residue 74, which lies in the domain I-II linker near the interdomain groove, moderately enhances fusion promotion90.
The solved structures depict only the ectodomains of gH/gL, however mutagenesis studies indicate that the gH TM domain and cytoplasmic tail also play a role in fusion promotion 89, 91–93. Although a soluble version of the HSV gH/gL lacking the TM and cytoplasmic tail was shown to promote fusion94, its efficiency was greatly reduced.
gB: A conserved herpesvirus fusion protein
The HSV-1 and EBV gB structures revealed surprising structural homology to both glycoprotein G from vesicular stomatitis virus (VSV G), the sole fusion protein of VSV, and baculovirus gp64, the major envelope protein of baculovirus that is necessary and sufficient for cell entry73, 95–97. This structural homology could not be predicted by sequence similarity and indicates that, despite being insufficient for herpesvirus fusion alone, gB has a similar fold to known fusion proteins and may undergo large conformational changes to bring about fusion. While evidence for gB refolding has yet to be obtained, a model for ‘prefusion gB’ has been proposed95 and it seems likely that gB undergoes a refolding transition during fusion. Together these proteins represent a newly defined class of fusogens, called class III, with structural characteristics distinct from the well described class I fusogens, such as influenza virus hemagglutinin, and class II fusogens, such as flavivirus E protein98.
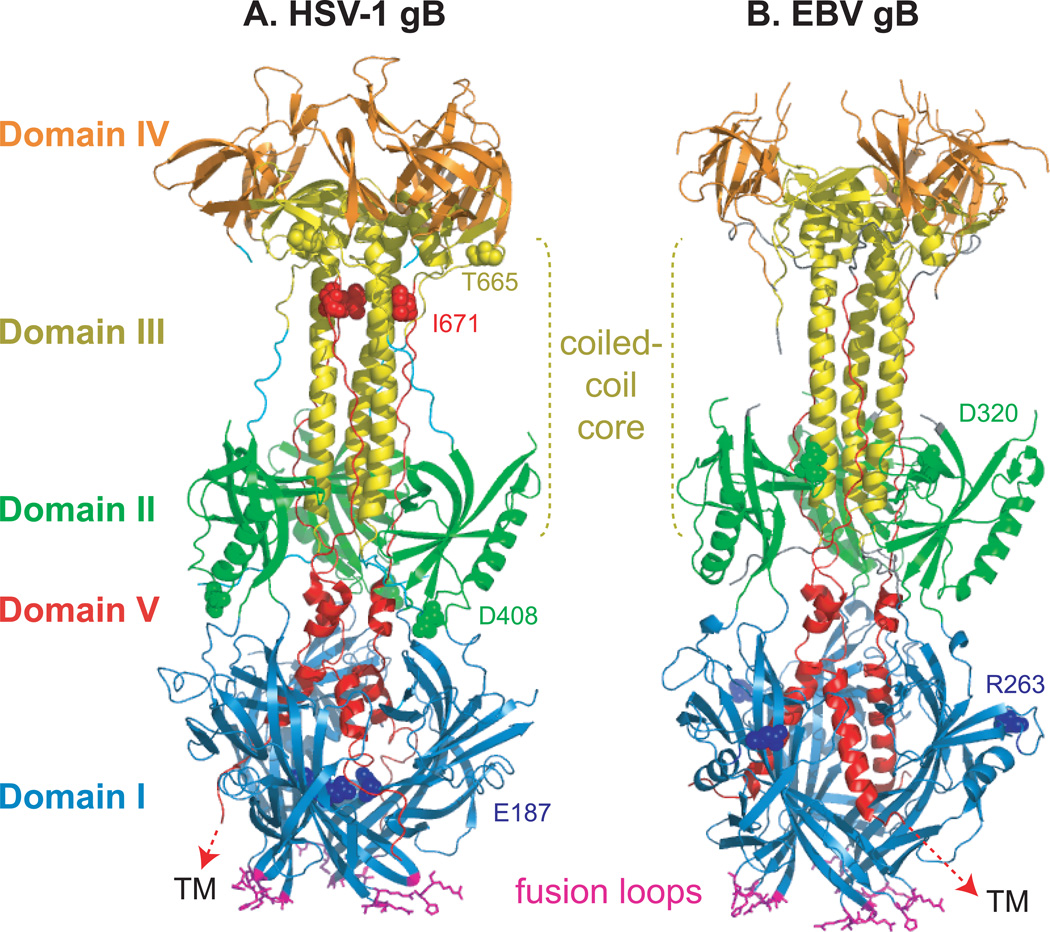
HSV and EBV gB are trimers and each protomer contains five distinct domains (Fig. 4). The fusion loops of gB are critical for membrane fusion99, 100, and are located in domain I, structurally close to the expected location of the TM region. The core of the protein is composed of an alpha-helical coiled-coil, a feature typical of many viral fusion proteins including those of classes I and III. Long linker regions leading into and out of domains I and II are hypothesized to permit a large-scale rotation necessary for the transition from pre- to post-fusion conformations. Mutational and antigenic analyses highlight that multiple domains of gB are critical for function, oligomerization is critical for function, and in the case of EBV, gB may participate in the determination of cell tropism101–103. Insertion mutations in all five of the gB domains can abrogate fusion, consistent with the concept that gB undergoes a complex and ordered refolding process to drive fusion102, 103 (Fig. 4).
Figure 4. Conserved fusion protein gB.
(A) HSV-1 gB (PDB ID 2GUM). (B) EBV gB (PDB ID 3FVC). Domain I (blue) contains hydrophobic fusion loops (magenta) that insert into the target cell membrane. For crystallization, the hydrophobic residues in the EBV gB fusion loops were replaced with corresponding residues from HSV-1 gB. The C-terminal domain V (red) packs against the coiled-coil core formed by domain III (yellow) and proceeds through domain I, headed towards the transmembrane (TM) region. This creates a hairpin-like organization of the structure wherein the fusion loops and TM lie at the same end of the trimer. The location of insertion mutations that reduce fusion without preventing gB expression are noted (spheres colored according to domain).
Overall, the HSV and EBV structures are remarkably similar. The most noticeable difference between the HSV-1 and EBV gB structures is a shift in the position of domain IV, the crown domain that contains epitopes for neutralizing anti-HSV antibodies. The fusion loop sequences and electrostatic surfaces of the trimers also differ, perhaps indicating that they interact with target phospholipid membranes in differently95.
It seems likely that the gB structures represent the post-fusion conformation of the protein. Both pre- and post-fusion forms of the structurally homologous VSV G protein have been solved97, 104 and the gB structures more closely resemble the VSV G post-fusion or low pH structure. Furthermore, the gB fusion loops sit in close proximity to where the C-terminal TM domain would lie, a feature shared with other post-fusion fusion protein structures, including influenza virus hemagglutinin and paramyxovirus F protein98, 105. However, nearly 50 residues at the C-terminus of the gB ectodomain are absent from both gB structures and these could interrupt the perceived proximity between the fusion loops and TM domain. Additionally, the epitopes for neutralizing anti-gB antibodies map to the surface of the solved structure of HSV gB. Neutralizing antibodies could reasonably be expected to recognize the pre-fusion structure of the protein, and at least some of these epitopes would likely be obscured 345 in the post-fusion form of the protein. How gB undergoes substantial folding changes to adopt distinct pre-fusion and post-fusion conformations remains to be understood.
The gB cytoplasmic tail is not included in the solved structures, however many studies have highlighted its role in gB function. Mutations in the HSV gB cytoplasmic tail can disrupt its transport to the cell surface, reduce fusion, or enhance fusion, depending on the mutation location106, 107. The HSV gB cytoplasmic tail expressed alone can interact with membranes, possibly providing a mechanism for negative regulation of fusion108. Close but distinct regions within the EBV gB tail positively and negatively regulate fusion109. Furthermore, distinct mutations within the EBV gB tail can cause increased cell-surface expression and fusion independent of gH/gL38. Collectively, these studies highlight that in addition to the structurally complex gB ectodomain, several functional regions exist within the intracellular cytoplasmic tail of gB.
Interaction of viral proteins
Triggering virus fusion with the right cells at the right time requires coordinated interactions among multiple entry glycoproteins (Fig. 5). The most well defined interactions are those of gp42 and gD with their receptors, which are stable enough for co-crystallization. EBV gp42 and gH/gL also have a high affinity interaction and a future structure of gH/gL bound to gp42 or the previously mentioned gp42 peptide should reveal how the complex forms. Details of the other glycoprotein interactions are not completely understood because they are likely low affinity and/or transient and difficult to capture using wild-type proteins. Upon binding receptor, gD and gp42 must transmit a signal to gH/gL and/or gB. Several studies suggest that a critical step in fusion is the interaction between gH/gL and gB, with the latter glycoprotein being required for a committed and expanding fusion pore110–113.
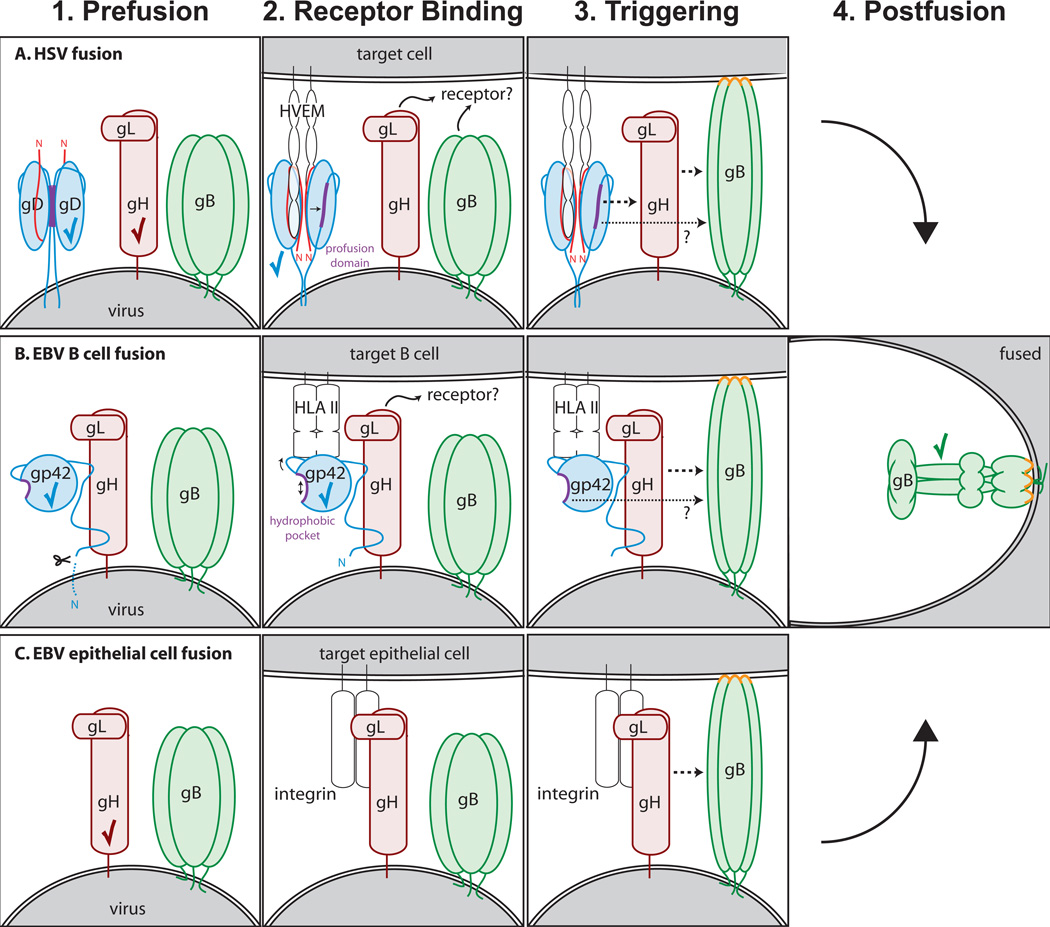
Figure 5. Models of fusion.
(A) HSV fusion with a target cell at the plasma or endosomal membrane. 1. gD is expressed as a dimer with its C-termini occluding its receptor binding site. 2. Upon binding HVEM, the N-terminus of gD (red) forms a loop and displaces the gD C-terminus that contains the profusion domain (purple). Nectin-1 binds to an overlapping site on gD and displaces its C-terminus in a similar manner. The gB trimer also possesses receptor-binding activity and the gH/gL heterodimer may bind to a cellular receptor as well. 3. Different sites within the gD profusion domain can interact with gH/gL and gB. In addition, gH/gL and gB may interact with one another in response to gD binding receptor. These interactions trigger gB to insert its fusion loops (orange) into the target membrane. 4. gB then refolds to its postfusion conformation to mediate fusion of the viral and cellular membranes. (B) EBV fusion with B cell endosomal membrane. 1. gp42, a type II glycoprotein, undergoes N-terminal cleavage and is retained on the virion by binding gH/gL via the gp42 N-terminus. 2. When HLA class II binds to gp42, a hydrophobic pocket (purple) on gp42 widens. gH/gL may also bind a receptor. 3. Conformational changes within gp42 and/or gH/gL trigger gB fusion loop (orange) insertion. The hydrophobic pocket in gp42 may serve as an interaction site to promote fusion. 4. gB refolds to a postfusion conformation, thereby mediating membrane fusion. (C) EBV fusion with epithelial cell plasma membrane. 1. gp42 functions as a tropism switch and inhibits entry into epithelial cells. Virus produced in B cells is deficient in gp42 and thus able to infect epithelial cells. 2. gH/gL binds to receptor such as integrin. 3. After receptor binding, gH/gL interacts with gB and triggers fusion loop (orange) insertion. 4. gB refolds to a postfusion conformation, thereby driving membrane fusion. Checkmarks denote structures that have been solved. Note that gp42 and gH/gL were not solved in complex. Structures for the other protein complexes and conformations, such as receptor-bound gH/gL and pre-fusion gB, remain to be determined.
EBV gp42 and gH/gL form a stable complex via the N-terminus of gp4230, 114. Since gp42 inhibits epithelial cell entry33, 34, it is likely that gp42 binding on gH/gL overlaps with an epithelial receptor binding site, possibly near the loop that contains an integrin-binding KGD motif (Fig. 3B). EBV gL residues 54 and 94 contribute to a functional interaction with gB88, suggesting that the N-terminal region of EBV gH/gL may physically interact with gB.
HSV gD has been reported to associate with both gH/gL and gB. gD mutations mapped a gD-gH/gL interactive site to the gD pro-fusion domain between residues 260–310 and a gD-gB interactive site to gD residues 240–260 and 304–305115. A gD interaction with both gH/gL and gB also has been detected using bimolecular complementation (BiMC). In BiMC, two inactive halves of a fluorescent protein are fused onto separate glycoproteins of interest. The resulting proteins are transiently expressed and their interaction restores a fluorescent signal.
An association of HSV gH/gL with gB has also been shown using BiMC, however it is unclear whether the gH/gL-gB interaction requires that gD first bind a receptor110, 112. A gB interaction site on HSV gH/gL has been proposed to overlap with the epitope for the neutralizing antibody LP11, since that antibody inhibits the gH/gL-gB BiMC signal76 (Fig. 3A). The gH/gL-gB interaction likely occurs prior to fusion and not as a result of fusion, since some anti-HSV gB antibodies can block fusion without disrupting gH/gL-gB BiMC116. In addition, mutations within the gB fusion loops blocked both gB-gH/gL BiMC and fusion, perhaps suggesting that gB must insert its fusion loops into the membrane prior to its interaction with gH/gL.
Whether the associations detected by BiMC represent functional interactions remains to be determined. A more recent study suggests that gD binding to receptor activates gH/gL and this activated gH/gL triggers gB in a cascading fashion to drive fusion 94. The interaction of gH/gL with gB does not appear to require coexpression on the same membrane, since cell-cell fusion has been reported when gB and gH/gL were expressed on distinct cells for both HSV and CMV 94, 117.
Concluding remarks and key questions
With the recently solved gH/gL crystal structures, the structures for each glycoprotein required for virus entry (HSV gD, gB, gH/gL and EBV gp42, gB, gH/gL), as well as two of the cellular receptors that bind virus and trigger fusion (HVEM and HLA class II), are now known. The structures illustrate conformational changes that may trigger fusion and identify the viral fusogen. These viral glycoproteins serve as targets for neutralizing antibodies and each represents a potential target for anti-viral therapy. Inhibitors of entry that target either the viral or the receptor side of the interaction have been successful for HIV therapy118 and dissection of the EBV gH/gL/gp42 interactions have identified a high affinity peptide inhibitor of EBV fusion30, 114. These herpesvirus structures can be used both for rational design of novel attachment and/or fusion inhibitors and to analyze inhibitors identified via drug screens. The ability to model the conserved glycoproteins gB and gH/gL from the other human and animal herpesviruses using these structures further expands their significance.
The structural examination of the proteins that mediate virus entry is ongoing and current working models of HSV and EBV fusion will continue to be refined (Fig. 5). The structures of the receptors nectin-1, PILRα, MAG, NMHC-IIA, and integrin αvβ6/8 remain to be solved, both alone and in complex with glycoproteins. Furthermore, it is likely that multiple entry receptors remain to be identified, since soluble forms of both HSV gB and gH/gL bind to cells and inhibit virus entry71, 119. On the viral side, the structures of the tethering proteins HSV gC and EBV BMRF2 and the tripartite gp42/gH/gL complex have not been solved. In addition, a structural comparison of the prefusion versus postfusion conformations of gB would shed light on how this fusion protein refolds to mediate membrane fusion during virus entry. Analysis of gB conformational changes will contribute to our understanding of entry of other viruses that employ class III fusogens, including VSV and baculovirus. Although many viruses enter cells using a single protein for both attachment and fusion, herpesviruses are not alone in their complexity. Understanding how herpesvirus entry proteins interact may provide insight into the entry of paramyxoviruses, which use separate binding and fusion glycoproteins105 and poxviruses, which probably use even more entry proteins than herpesviruses120.
Box 1: Therapeutic benefits of structural studies.
Currently, no entry inhibitors for herpesvirus entry are routinely used clinically. Studies focused on herpesvirus entry are valuable for the design of such inhibitors. Two types of inhibitors could emerge from such studies. First, inhibitors that block virus glycoprotein-glycoprotein or virus glycoprotein-cellular receptor interactions essential for receptor binding and the triggering of fusion may result. As an example, gp42 peptides have been identified that prevent B cell and epithelial cell fusion by interfering with the essential interaction of gH/gL with gp42 for B cell infection or possibly with receptor for epithelial infection30. High throughput screening approaches using biochemical assays developed from this observation114 may result in small molecule inhibitors that interfere with normal gH/gL function in EBV entry. Similar interactions between gD, gB, and gH/gL may also be exploited. Second, inhibitors that prevent the fusion function of gB may be developed. A large number of peptides have been designed that recognize heptad repeat regions in herpesvirus gB, some of which are able to block fusion and entry possibly by preventing conformational changes that occurs as gB refolds to its post-fusion form121, 122. Solving the gB prefusion structure would simplify the design and understanding of how such inhibitors function. This class of drug has shown previous therapeutic success in the form of Fuzeon, a drug that targets the HIV fusion protein. Finally, the determination of the structure of herpesvirus protein complexes and the mapping functional domains will allow a better understanding of why certain antibodies are neutralizing while others are not. This may lead to the development of more effective vaccines to prevent herpesvirus infections.
Supplementary Material
Acknowledgments
Research in the Jardetzky (AI076183 and CA117794) and Longnecker (AI076183, CA117794, and AI067048) laboratories was supported by Public Health Service grants. The authors would like to thank current and former members of their laboratories for their contributions to the work described, as well as our many colleagues throughout the world who have contributed to understanding the entry of EBV, HSV, and other herpesviruses.
Glossary
- Enveloped virus
Viruses that when outside of the cell, contain a phospholipid membrane surrounding the viral protein coat.
- Endocytic vesicle
A membrane-bound vesicle participating in the endocytic membrane transport pathway from the plasma membrane (outer membrane of a cell) to the lysosome (a membrane-bound organelle containing digestive enzymes that can, among other things, degrade invading bacteria and viruses).
- Latent virus infection
A persistent viral infection that can occur in some cell types, during which the virus ceases to replicate but limited expression of some viral genes (a “latent phase transcriptional program”) still occurs.
- Heterodimer
A compound consisting of two non-identical subunits (in this case, a dimer composed of one gH molecule and one gL molecule).
- C-type lectin superfamily
A large family of functionally diverse proteins containing conserved structures of double-loops stabilized by disulfide bridges, termed C-type lectin-like domains.
- Hydrophobic pocket
A groove or pocket in a protein that may serve as a binding site and is composed of mostly hydrophobic amino acid residues.
- Cell tropism
The preferential targeting of specific cell types within a host by a pathogen.
- Proteoglycans
Proteins that contain an abundance of sugars including one or more glycosaminoglycan chains on their surface.
- Fusion peptide or loop
A sequence of hydrophobic amino acid residues within a fusion protein, sometimes in the shape of a loop, that inserts into a target cell membrane during the fusion event.
- Antibody epitope
The portion of an antigen (a protein capable of eliciting an immune response) recognized by a specific antibody directed against that antigen.
- Heptad repeat
A structural motif in the amino acid sequence of proteins that can signal the formation of a coiled-coil, a structure common to viral fusion proteins.
Biographies
Richard Longnecker: Dr. Longnecker earned his bachelor’s degree in Cellular and Molecular Biology from the University of Michigan and his Ph.D. from the University of Chicago in Virology. At Northwestern, Dr. Longnecker is a Professor in the Department of Microbiology-Immunology, Director of the Viral Oncogenesis Basic Sciences at The RHLCCC, and the John Edward Porter Professor in Biomedical Research. Dr. Longnecker’s research focuses on mechanism and pathogenesis of herpesvirus infections in humans with a focus on HSV and EBV.
Theodore Jardetzky: Dr. Jardetzky was a Professor in the Department of Biochemistry, Molecular Biology and Cell Biology at Northwestern University from 1994–2007, after which he joined the Department of Structural Biology in the School of Medicine at Stanford University. Dr. Jardetzky’s laboratory focuses on elucidating the structures and mechanisms of biomolecular complexes important in human health and development, with the goal of developing novel approaches to intervening in disease processes.
Sarah A. Connolly: Dr. Connolly is a postdoctoral researcher in the lab of Richard Longnecker at Northwestern University, Chicago, IL, USA. She received her Ph.D. in Cell and Molecular Biology at the University of Pennsylvania while studying HSV entry. She then completed postdoctoral studies on paramyxovirus entry and is currently focusing on the mechanism of refolding of herpesvirus gB.
Julia O. Jackson: Julia Jackson is a Ph.D. candidate in the lab of Richard Longnecker at Northwestern University, Chicago, IL, USA. She is pursuing a Ph.D. in Microbiology and Immunology and a Masters in Public Health through the Northwestern University Integrated Graduate Program. Her current research investigates the functional domains of glycoprotein H, an essential herpesvirus entry glycoprotein.
References
- 1.Pellet PE, Roizman B. In: Fields' Virology. 5th Edition. Knipe DM, Howley PM, editors. New York, N.Y: Lippincott-Williams and Wilkins; 2007. pp. 2479–2499. [Google Scholar]
- 2.Milne RS, Nicola AV, Whitbeck JC, Eisenberg RJ, Cohen GH. Glycoprotein D receptor-dependent, low-pH-independent endocytic entry of herpes simplex virus type 1. J Virol. 2005;79:6655–6663. doi: 10.1128/JVI.79.11.6655-6663.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicola AV, McEvoy AM, Straus SE. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and Chinese hamster ovary cells. J Virol. 2003;77:5324–5332. doi: 10.1128/JVI.77.9.5324-5332.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dollery SJ, Delboy MG, Nicola AV. Low pH-induced conformational change in herpes simplex virus glycoprotein B. J Virol. 2010;84:3759–3766. doi: 10.1128/JVI.02573-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stampfer SD, Lou H, Cohen GC, Eisenberg RJ, Heldwein EE. Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus 1. J Virol. 2010 doi: 10.1128/JVI.01750-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutt-Fletcher L. Epstein-Barr virus entry. J Virol. 2007;81:7825–7832. doi: 10.1128/JVI.00445-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller N, Hutt-Fletcher LM. Epstein-Barr virus enters B cells and epithelial cells by different routes. J Virol. 1992;66:3409–3414. doi: 10.1128/jvi.66.6.3409-3414.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laquerre S, et al. Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread. J Virol. 1998;72:6119–6130. doi: 10.1128/jvi.72.7.6119-6130.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herold BC, WuDunn D, Soltys N, Spear PG. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J Virol. 1991;65:1090–1098. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shukla D, Spear PG. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J Clin Invest. 2001;108:503–510. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Jong MA, de Witte L, Bolmstedt A, van Kooyk Y, Geijtenbeek TB. Dendritic cells mediate herpes simplex virus infection and transmission through the C-type lectin DC-SIGN. J Gen Virol. 2008;89:2398–2409. doi: 10.1099/vir.0.2008/003129-0. [DOI] [PubMed] [Google Scholar]
- 12.Fingeroth JD, et al. Epstein-Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc Natl Acad Sci U S A. 1984;81:4510–4514. doi: 10.1073/pnas.81.14.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frade R, Barel M, Ehlin-Henriksson B, Klein G. gp140, the C3d receptor of human B lymphocytes, is also the Epstein-Barr virus receptor. Proc Natl Acad Sci U S A. 1985;82:1490–1493. doi: 10.1073/pnas.82.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nemerow GR, Mold C, Schwend VK, Tollefson V, Cooper NR. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J Virol. 1987;61:1416–1420. doi: 10.1128/jvi.61.5.1416-1420.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanner J, Weis J, Fearon D, Whang Y, Kieff E. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell. 1987;50:203–213. doi: 10.1016/0092-8674(87)90216-9. [DOI] [PubMed] [Google Scholar]
- 16.Tanner J, Whang Y, Sample J, Sears A, Kieff E. Soluble gp350/220 and deletion mutant glycoproteins block Epstein-Barr virus adsorption to lymphocytes. J Virol. 1988;62:4452–4464. doi: 10.1128/jvi.62.12.4452-4464.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janz A, et al. Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J Virol. 2000;74:10142–10152. doi: 10.1128/jvi.74.21.10142-10152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nemerow GR, Houghten RA, Moore MD, Cooper NR. Identification of an epitope in the major envelope protein of Epstein-Barr virus that mediates viral binding to the B lymphocyte EBV receptor (CR2) Cell. 1989;56:369–377. doi: 10.1016/0092-8674(89)90240-7. [DOI] [PubMed] [Google Scholar]
- 19.Szakonyi G, et al. Structure of the Epstein-Barr virus major envelope glycoprotein. Nat Struct Mol Biol. 2006;13:996–1001. doi: 10.1038/nsmb1161. [DOI] [PubMed] [Google Scholar]
- 20.Tugizov SM, Berline JW, Palefsky JM. Epstein-Barr virus infection of polarized tongue and nasopharyngeal epithelial cells. Nat Med. 2003;9:307–314. doi: 10.1038/nm830. [DOI] [PubMed] [Google Scholar]
- 21.Xiao J, Palefsky JM, Herrera R, Berline J, Tugizov SM. The Epstein-Barr virus BMRF-2 protein facilitates virus attachment to oral epithelial cells. Virology. 2008;370:430–442. doi: 10.1016/j.virol.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao J, Palefsky JM, Herrera R, Tugizov SM. Characterization of the Epstein-Barr virus glycoprotein BMRF-2. Virology. 2007;359:382–396. doi: 10.1016/j.virol.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 23.Xiao J, Palefsky JM, Herrera R, Berline J, Tugizov SM. EBV BMRF-2 facilitates cell-to-cell spread of virus within polarized oral epithelial cells. Virology. 2009;388:335–343. doi: 10.1016/j.virol.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haan KM, Longnecker R. Coreceptor restriction within the HLA-DQ locus for Epstein-Barr virus infection. Proc Natl Acad Sci U S A. 2000;97:9252–9257. doi: 10.1073/pnas.160171697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ressing ME, et al. Epstein-Barr virus gp42 is posttranslationally modified to produce soluble gp42 that mediates HLA class II immune evasion. J Virol. 2005;79:841–852. doi: 10.1128/JVI.79.2.841-852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sorem J, Jardetzky TS, Longnecker R. Cleavage and secretion of Epstein-Barr virus glycoprotein 42 promote membrane fusion with B lymphocytes. J Virol. 2009;83:6664–6672. doi: 10.1128/JVI.00195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kirschner AN, Sorem J, Longnecker R, Jardetzky TS. Structure of Epstein-Barr virus glycoprotein 42 suggests a mechanism for triggering receptor-activated virus entry. Structure. 2009;17:223–233. doi: 10.1016/j.str.2008.12.010. Offers a comparison between the HLA-bound gp42 and unbound gp42, prompting a structurally-based model of receptor-activated EBV fusion.
- 28. Mullen MM, Haan KM, Longnecker R, Jardetzky TS. Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol Cell. 2002;9:375–385. doi: 10.1016/s1097-2765(02)00465-3. Reports the first form of gp42 successfully crystallized, revealing that gp42 binds HLA using a surface site distinct from other C-type lectin superfamily members.
- 29.McShane MP, Mullen MM, Haan KM, Jardetzky TS, Longnecker R. Mutational analysis of the HLA class II interaction with Epstein-Barr virus glycoprotein 42. J Virol. 2003;77:7655–7662. doi: 10.1128/JVI.77.13.7655-7662.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirschner AN, Lowrey AS, Longnecker R, Jardetzky TS. Binding-site interactions between Epstein-Barr virus fusion proteins gp42 and gH/gL reveal a peptide that inhibits both epithelial and B-cell membrane fusion. J Virol. 2007;81:9216–9229. doi: 10.1128/JVI.00575-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silva AL, Omerovic J, Jardetzky TS, Longnecker R. Mutational analyses of Epstein-Barr virus glycoprotein 42 reveal functional domains not involved in receptor binding but required for membrane fusion. J Virol. 2004;78:5946–5956. doi: 10.1128/JVI.78.11.5946-5956.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirschner AN, Omerovic J, Popov B, Longnecker R, Jardetzky TS. Soluble Epstein-Barr virus glycoproteins gH, gL, and gp42 form a 1 : 1 : 1 stable complex that acts like soluble gp42 in B-cell fusion but not in epithelial cell fusion. J Virol. 2006;80:9444–9454. doi: 10.1128/JVI.00572-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Hutt-Fletcher LM. Epstein-Barr virus lacking glycoprotein gp42 can bind to B cells but is not able to infect. J Virol. 1998;72:158–163. doi: 10.1128/jvi.72.1.158-163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Kenyon WJ, Li Q, Mullberg J, Hutt-Fletcher LM. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J Virol. 1998;72:5552–5558. doi: 10.1128/jvi.72.7.5552-5558.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borza CM, Hutt-Fletcher LM. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat Med. 2002;8:594–599. doi: 10.1038/nm0602-594. [DOI] [PubMed] [Google Scholar]
- 36.Wang D, Shenk T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci U S A. 2005;102:18153–18158. doi: 10.1073/pnas.0509201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mori Y. Recent topics related to human herpesvirus 6 cell tropism. Cell Microbiol. 2009;11:1001–1006. doi: 10.1111/j.1462-5822.2009.01312.x. [DOI] [PubMed] [Google Scholar]
- 38.McShane MP, Longnecker R. Cell-surface expression of a mutated Epstein-Barr virus glycoprotein B allows fusion independent of other viral proteins. Proc Natl Acad Sci U S A. 2004;101:17474–17479. doi: 10.1073/pnas.0404535101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molesworth SJ, Lake CM, Borza CM, Turk SM, Hutt-Fletcher LM. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J Virol. 2000;74:6324–6332. doi: 10.1128/jvi.74.14.6324-6332.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu L, Borza CM, Hutt-Fletcher LM. Mutations of Epstein-Barr virus gH that are differentially able to support fusion with B cells or epithelial cells. J Virol. 2005;79:10923–10930. doi: 10.1128/JVI.79.17.10923-10930.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu L, Hutt-Fletcher LM. Point mutations in EBV gH that abrogate or differentially affect B cell and epithelial cell fusion. Virology. 2007;363:148–155. doi: 10.1016/j.virol.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chesnokova LS, Nishimura SL, Hutt-Fletcher LM. Fusion of epithelial cells by Epstein-Barr virus proteins is triggered by binding of viral glycoproteins gHgL to integrins alphavbeta6 or alphavbeta8. Proc Natl Acad Sci U S A. 2009;106:20464–20469. doi: 10.1073/pnas.0907508106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campadelli-Fiume G, Cocchi F, Menotti L, Lopez M. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev Med Virol. 2000;10:305–319. doi: 10.1002/1099-1654(200009/10)10:5<305::aid-rmv286>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 44.Spear PG, Eisenberg RJ, Cohen GH. Three classes of cell surface receptors for alphaherpesvirus entry. Virology. 2000;275:1–8. doi: 10.1006/viro.2000.0529. [DOI] [PubMed] [Google Scholar]
- 45.Cocchi F, et al. The soluble ectodomain of herpes simplex virus gD contains a membrane-proximal pro-fusion domain and suffices to mediate virus entry. Proc Natl Acad Sci U S A. 2004;101:7445–7450. doi: 10.1073/pnas.0401883101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwon H, et al. Soluble V domain of Nectin-1/HveC enables entry of herpes simplex virus type 1 (HSV-1) into HSV-resistant cells by binding to viral glycoprotein D. J Virol. 2006;80:138–148. doi: 10.1128/JVI.80.1.138-148.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsvitov M, et al. Characterization of soluble glycoprotein D-mediated herpes simplex virus type 1 infection. Virology. 2007;360:477–491. doi: 10.1016/j.virol.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kopp SJ, et al. Infection of neurons and encephalitis after intracranial inoculation of herpes simplex virus requires the entry receptor nectin-1. Proc Natl Acad Sci U S A. 2009;106:17916–17920. doi: 10.1073/pnas.0908892106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor JM, et al. Alternative entry receptors for herpes simplex virus and their roles in disease. Cell Host Microbe. 2007;2:19–28. doi: 10.1016/j.chom.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Y, et al. Durable protection from Herpes Simplex Virus-2 transmission following intravaginal application of siRNAs targeting both a viral and host gene. Cell Host Microbe. 2009;5:84–94. doi: 10.1016/j.chom.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou G, Ye GJ, Debinski W, Roizman B. Engineered herpes simplex virus 1 is dependent on IL13Ralpha 2 receptor for cell entry and independent of glycoprotein D receptor interaction. Proc Natl Acad Sci U S A. 2002;99:15124–15129. doi: 10.1073/pnas.232588699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Menotti L, Cerretani A, Hengel H, Campadelli-Fiume G. Construction of a fully retargeted herpes simplex virus 1 recombinant capable of entering cells solely via human epidermal growth factor 2. J Virol. 2008;82:10153–10161. doi: 10.1128/JVI.01133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Carfi A, et al. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol Cell. 2001;8:169–179. doi: 10.1016/s1097-2765(01)00298-2. Reports the structure of gD in complex with HVEM, revealing a V-like immunoglobulin fold at the gD core and a receptor-binding N-terminal extension.
- 54.Connolly SA, et al. Structure-based mutagenesis of herpes simplex virus glycoprotein D defines three critical regions at the gD-HveA/HVEM binding interface. J Virol. 2003;77:8127–8140. doi: 10.1128/JVI.77.14.8127-8140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Connolly SA, et al. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM) J Virol. 2002;76:10894–10904. doi: 10.1128/JVI.76.21.10894-10904.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoon M, Zago A, Shukla D, Spear PG. Mutations in the N termini of herpes simplex virus type 1 and 2 gDs alter functional interactions with the entry/fusion receptors HVEM, nectin-2, and 3-O-sulfated heparan sulfate but not with nectin-1. J Virol. 2003;77:9221–9231. doi: 10.1128/JVI.77.17.9221-9231.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manoj S, Jogger CR, Myscofski D, Yoon M, Spear PG. Mutations in herpes simplex virus glycoprotein D that prevent cell entry via nectins and alter cell tropism. Proc Natl Acad Sci U S A. 2004;101:12414–12421. doi: 10.1073/pnas.0404211101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Connolly SA, et al. Potential nectin-1 binding site on herpes simplex virus glycoprotein d. J Virol. 2005;79:1282–1295. doi: 10.1128/JVI.79.2.1282-1295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Krummenacher C, et al. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. Embo J. 2005;24:4144–4153. doi: 10.1038/sj.emboj.7600875. Reports the structure of HSV gD in the absence of receptor and reveals that the gD C-terminus masks the receptor-binding N-terminus and moves upon receptor binding.
- 60.Fusco D, Forghieri C, Campadelli-Fiume G. The pro-fusion domain of herpes simplex virus glycoprotein D (gD) interacts with the gD N terminus and is displaced by soluble forms of viral receptors. Proc Natl Acad Sci U S A. 2005;102:9323–9328. doi: 10.1073/pnas.0503907102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lazear E, et al. Engineered disulfide bonds in herpes simplex virus type 1 gD separate receptor binding from fusion initiation and viral entry. J Virol. 2008;82:700–709. doi: 10.1128/JVI.02192-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Willis SH, et al. Examination of the kinetics of herpes simplex virus glycoprotein D binding to the herpesvirus entry mediator, using surface plasmon resonance. J Virol. 1998;72:5937–5947. doi: 10.1128/jvi.72.7.5937-5947.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Satoh T, et al. PILRalpha is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell. 2008;132:935–944. doi: 10.1016/j.cell.2008.01.043. Establishes a novel role for gB-receptor binding that mediates HSV entry into at least some cell types.
- 64.Satoh T, Arase H. HSV-1 infection through inhibitory receptor, PILRalpha. Uirusu. 2008;58:27–36. doi: 10.2222/jsv.58.27. [DOI] [PubMed] [Google Scholar]
- 65.Shukla SY, Singh YK, Shukla D. Role of nectin-1, HVEM, and PILR-alpha in HSV-2 entry into human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2009;50:2878–2887. doi: 10.1167/iovs.08-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arii J, et al. Non-muscle myosin IIA is a functional entry receptor for herpes simplex virus-1. Nature. 2010;467:859–862. doi: 10.1038/nature09420. [DOI] [PubMed] [Google Scholar]
- 67.Suenaga T, et al. Myelin-associated glycoprotein mediates membrane fusion and entry of neurotropic herpesviruses. Proc Natl Acad Sci U S A. 2010;107:866–871. doi: 10.1073/pnas.0913351107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galdiero M, et al. Site-directed and linker insertion mutagenesis of herpes simplex virus type 1 glycoprotein H. J Virol. 1997;71:2163–2170. doi: 10.1128/jvi.71.3.2163-2170.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parry C, Bell S, Minson T, Browne H. Herpes simplex virus type 1 glycoprotein H binds to alphavbeta3 integrins. J Gen Virol. 2005;86:7–10. doi: 10.1099/vir.0.80567-0. [DOI] [PubMed] [Google Scholar]
- 70.Gianni T, Gatta V, Campadelli-Fiume G. {Alpha}V{beta}3-integrin routes herpes simplex virus to an entry pathway dependent on cholesterol-rich lipid rafts and dynamin2. Proc Natl Acad Sci U S A. 2010;107:22260–22265. doi: 10.1073/pnas.1014923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gianni T, et al. Herpes simplex virus glycoproteins H/L bind to cells independently of {alpha}V{beta} 3 integrin and inhibit virus entry, and their constitutive expression restricts infection. J Virol. 2010;84:4013–4025. doi: 10.1128/JVI.02502-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ryckman BJ, Chase MC, Johnson DC. HCMV gH/gL/UL128-131 interferes with virus entry into epithelial cells: evidence for cell type-specific receptors. Proc Natl Acad Sci U S A. 2008;105:14118–14123. doi: 10.1073/pnas.0804365105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Heldwein EE, et al. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. Reports the structure of HSV-1 gB, providing the first direct evidence that herpesvirus gB is a viral fusion protein.
- 74. Matsuura H, Kirschner AN, Longnecker R, Jardetzky TS. Crystal structure of Epstein-Barr virus glycoprotein H/glycoprotein L (gH/gL) complex. Proc Natl Acad Sci U S A. 2010;107:22641–22646. doi: 10.1073/pnas.1011806108. Reports the structure of EBV gH/gL and demonstrates that EBV gH/gL has different domain packing than HSV gH/gL.
- 75.Backovic M, et al. Structure of a core fragment of glycoprotein H from pseudorabies virus in complex with antibody. Proc Natl Acad Sci U S A. 2010;107:22635–22640. doi: 10.1073/pnas.1011507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chowdary TK, et al. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat Struct Mol Biol. 2010;17:882–888. doi: 10.1038/nsmb.1837. Reports the first gH/gL structure and provokes a shift in the model of herpesvirus membrane fusion by revealing that HSV-2 gH/gL does not resemble a fusion protein; additionally provides evidence that gH/gL may serve as a regulator of gB function.
- 77.Subramanian RP, Geraghty RJ. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc Natl Acad Sci U S A. 2007;104:2903–2908. doi: 10.1073/pnas.0608374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Galdiero S, et al. Evidence for a role of the membrane-proximal region of herpes simplex virus Type 1 glycoprotein H in membrane fusion and virus inhibition. ChemBioChem. 2007;8:885–895. doi: 10.1002/cbic.200700044. [DOI] [PubMed] [Google Scholar]
- 79.Galdiero S, et al. Analysis of a membrane interacting region of herpes simplex virus type 1 glycoprotein H. J Biol Chem. 2008;283:29993–30009. doi: 10.1074/jbc.M803092200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gianni T, Fato R, Bergamini C, Lenaz G, Campadelli-Fiume G. Hydrophobic alpha-helices 1 and 2 of herpes simplex virus gH interact with lipids, and their mimetic peptides enhance virus infection and fusion. J Virol. 2006;80:8190–8198. doi: 10.1128/JVI.00504-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gianni T, Martelli PL, Casadio R, Campadelli-Fiume G. The ectodomain of herpes simplex virus glycoprotein H contains a membrane alpha-helix with attributes of an internal fusion peptide, positionally conserved in the herpesviridae family. J Virol. 2005;79:2931–2940. doi: 10.1128/JVI.79.5.2931-2940.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gianni T, Menotti L, Campadelli-Fiume G. A heptad repeat in herpes simplex virus 1 gH, located downstream of the alpha-helix with attributes of a fusion peptide, is critical for virus entry and fusion. J Virol. 2005;79:7042–7049. doi: 10.1128/JVI.79.11.7042-7049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gianni T, Piccoli A, Bertucci C, Campadelli-Fiume G. Heptad repeat 2 in herpes simplex virus 1 gH interacts with heptad repeat 1 and is critical for virus entry and fusion. J Virol. 2006;80:2216–2224. doi: 10.1128/JVI.80.5.2216-2224.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Duus KM, Hatfield C, Grose C. Cell surface expression and fusion by the varicella-zoster virus gH:gL glycoprotein complex: analysis by laser scanning confocal microscopy. Virology. 1995;210:429–440. doi: 10.1006/viro.1995.1359. [DOI] [PubMed] [Google Scholar]
- 85.Maresova L, Pasieka TJ, Grose C. Varicella-zoster Virus gB and gE coexpression, but not gB or gE alone, leads to abundant fusion and syncytium formation equivalent to those from gH and gL coexpression. J Virol. 2001;75:9483–9492. doi: 10.1128/JVI.75.19.9483-9492.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Farnsworth A, et al. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc Natl Acad Sci U S A. 2007;104:10187–10192. doi: 10.1073/pnas.0703790104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jackson JO, Longnecker R. Re-evaluating Herpes Simplex Virus Hemifusion. J Virol. 2010 doi: 10.1128/JVI.01615-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Plate AE, Smajlovic J, Jardetzky TS, Longnecker R. Functional analysis of glycoprotein L (gL) from rhesus lymphocrytovirus in Epstein-Barr virus-mediated cell fusion indicates a direct role of gL in gB-induced membrane fusion. J Virol. 2009;83:7678–7689. doi: 10.1128/JVI.00457-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jackson JO, Lin E, Spear PG, Longnecker R. Insertion mutations in herpes simplex virus 1 glycoprotein H reduce cell surface expression, slow the rate of cell fusion, or abrogate functions in cell fusion and viral entry. J Virol. 2010;84:2038–2046. doi: 10.1128/JVI.02215-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Omerovic J, Lev L, Longnecker R. The amino terminus of Epstein-Barr virus glycoprotein gH is important for fusion with epithelial and B cells. J Virol. 2005;79:12408–12415. doi: 10.1128/JVI.79.19.12408-12415.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Browne HM, Bruun BC, Minson AC. Characterization of herpes simplex virus type 1 recombinants with mutations in the cytoplasmic tail of glycoprotein H. J Gen Virol. 1996;77(Pt 10):2569–2573. doi: 10.1099/0022-1317-77-10-2569. [DOI] [PubMed] [Google Scholar]
- 92.Jones NA, Geraghty RJ. Fusion activity of lipid-anchored envelope glycoproteins of herpes simplex virus type 1. Virology. 2004;324:213–228. doi: 10.1016/j.virol.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 93.Wilson DW, Davis-Poynter N, Minson AC. Mutations in the cytoplasmic tail of herpes simplex virus glycoprotein H suppress cell fusion by a syncytial strain. J Virol. 1994;68:6985–6993. doi: 10.1128/jvi.68.11.6985-6993.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Atanasiu D, Saw WT, Cohen GH, Eisenberg RJ. The cascade of events governing cell-cell fusion induced by HSV glycoproteins gD, gH/gL and gB. J Virol Sept. 2010 doi: 10.1128/JVI.01700-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Backovic M, Longnecker R, Jardetzky TS. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc Natl Acad Sci U S A. 2009;106:2880–2885. doi: 10.1073/pnas.0810530106. Reports the crystal structure of EBV gB, revealing differences in structural details between EBV gB and HSV-1 gB.
- 96.Kadlec J, Loureiro S, Abrescia NG, Stuart DI, Jones IM. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat Struct Mol Biol. 2008;15:1024–1030. doi: 10.1038/nsmb.1484. [DOI] [PubMed] [Google Scholar]
- 97.Roche S, Bressanelli S, Rey FA, Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 98.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Backovic M, Jardetzky TS, Longnecker R. Hydrophobic residues that form putative fusion loops of Epstein-Barr virus glycoprotein B are critical for fusion activity. J Virol. 2007;81:9596–9600. doi: 10.1128/JVI.00758-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hannah BP, et al. Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops. J Virol. 2009;83:6825–6836. doi: 10.1128/JVI.00301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bender FC, et al. Antigenic and mutational analyses of herpes simplex virus glycoprotein B reveal four functional regions. J Virol. 2007;81:3827–3841. doi: 10.1128/JVI.02710-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lin E, Spear PG. Random linker-insertion mutagenesis to identify functional domains of herpes simplex virus type 1 glycoprotein B. Proc Natl Acad Sci U S A. 2007;104:13140–13145. doi: 10.1073/pnas.0705926104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Reimer JJ, Backovic M, Deshpande CG, Jardetzky T, Longnecker R. Analysis of Epstein-Barr virus glycoprotein B functional domains via linker insertion mutagenesis. J Virol. 2009;83:734–747. doi: 10.1128/JVI.01817-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roche S, Rey FA, Gaudin Y, Bressanelli S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 2007;315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- 105.Lamb RA, Paterson RG, Jardetzky TS. Paramyxovirus membrane fusion: lessons from the F and HN atomic structures. Virology. 2006;344:30–37. doi: 10.1016/j.virol.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cai WH, Gu B, Person S. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol. 1988;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ruel N, Zago A, Spear PG. Alanine substitution of conserved residues in the cytoplasmic tail of herpes simplex virus gB can enhance or abolish cell fusion activity and viral entry. Virology. 2006;346:229–237. doi: 10.1016/j.virol.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 108.Chowdary TK, Heldwein EE. Syncytial phenotype of C-terminally truncated herpes simplex virus type 1 gB is associated with diminished membrane interactions. J Virol. 2010;84:4923–4935. doi: 10.1128/JVI.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Haan KM, Lee SK, Longnecker R. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology. 2001;290:106–114. doi: 10.1006/viro.2001.1141. [DOI] [PubMed] [Google Scholar]
- 110.Atanasiu D, et al. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc Natl Acad Sci U S A. 2007;104:18718–18723. doi: 10.1073/pnas.0707452104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Avitabile E, Forghieri C, Campadelli-Fiume G. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J Virol. 2007;81:11532–11537. doi: 10.1128/JVI.01343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Avitabile E, Forghieri C, Campadelli-Fiume G. Cross talking among the glycoproteins involved in herpes simplex virus entry and fusion: the interaction between gB and gH/gL does not necessarily require gD. J Virol. 2009 doi: 10.1128/JVI.01287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maurer UE, Sodeik B, Grunewald K. Native 3D intermediates of membrane fusion in herpes simplex virus 1 entry. Proc Natl Acad Sci U S A. 2008;105:10559–10564. doi: 10.1073/pnas.0801674105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu F, Marquardt G, Kirschner AN, Longnecker R, Jardetzky TS. Mapping the N-terminal Residues of EBV gp42 that Bind gH/gL Using Fluorescence Polarization and Cell-based Fusion Assays. J Virol. 2010 doi: 10.1128/JVI.00381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gianni T, Amasio M, Campadelli-Fiume G. Herpes simplex virus gD forms distinct complexes with fusion executors gB and gH/gL through the C-terminal profusion. J Biol Chem. 2009 doi: 10.1074/jbc.M109.005728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Atanasiu D, et al. Bimolecular complementation defines functional regions of Herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J Virol. 2010;84:3825–3834. doi: 10.1128/JVI.02687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vanarsdall AL, Ryckman BJ, Chase MC, Johnson DC. Human cytomegalovirus glycoproteins gB and gH/gL mediate epithelial cell-cell fusion when expressed in cis or in trans. J Virol. 2008;82:11837–11850. doi: 10.1128/JVI.01623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kuritzkes DR. HIV-1 entry inhibitors: an overview. Curr Opin HIV AIDS. 2009;4:82–87. doi: 10.1097/COH.0b013e328322402e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bender FC, Whitbeck JC, Lou H, Cohen GH, Eisenberg RJ. Herpes simplex virus glycoprotein B binds to cell surfaces independently of heparan sulfate and blocks virus entry. J Virol. 2005;79:11588–11597. doi: 10.1128/JVI.79.18.11588-11597.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Moss B. Poxvirus entry and membrane fusion. Virology. 2006;344:48–54. doi: 10.1016/j.virol.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 121.Galdiero S, et al. The identification and characterization of fusogenic domains in herpes virus glycoprotein B molecules. Chem Biochem. 2008;9:758–767. doi: 10.1002/cbic.200700457. [DOI] [PubMed] [Google Scholar]
- 122.Akkarawongsa R, Pocaro NE, Case G, Kolb AW, Brandt CR. Multiple peptides homologous to herpes simplex virus type 1 glycoprotein B inhibit viral infection. Antimicrob Agents Chemother. 2009;53:987–996. doi: 10.1128/AAC.00793-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.