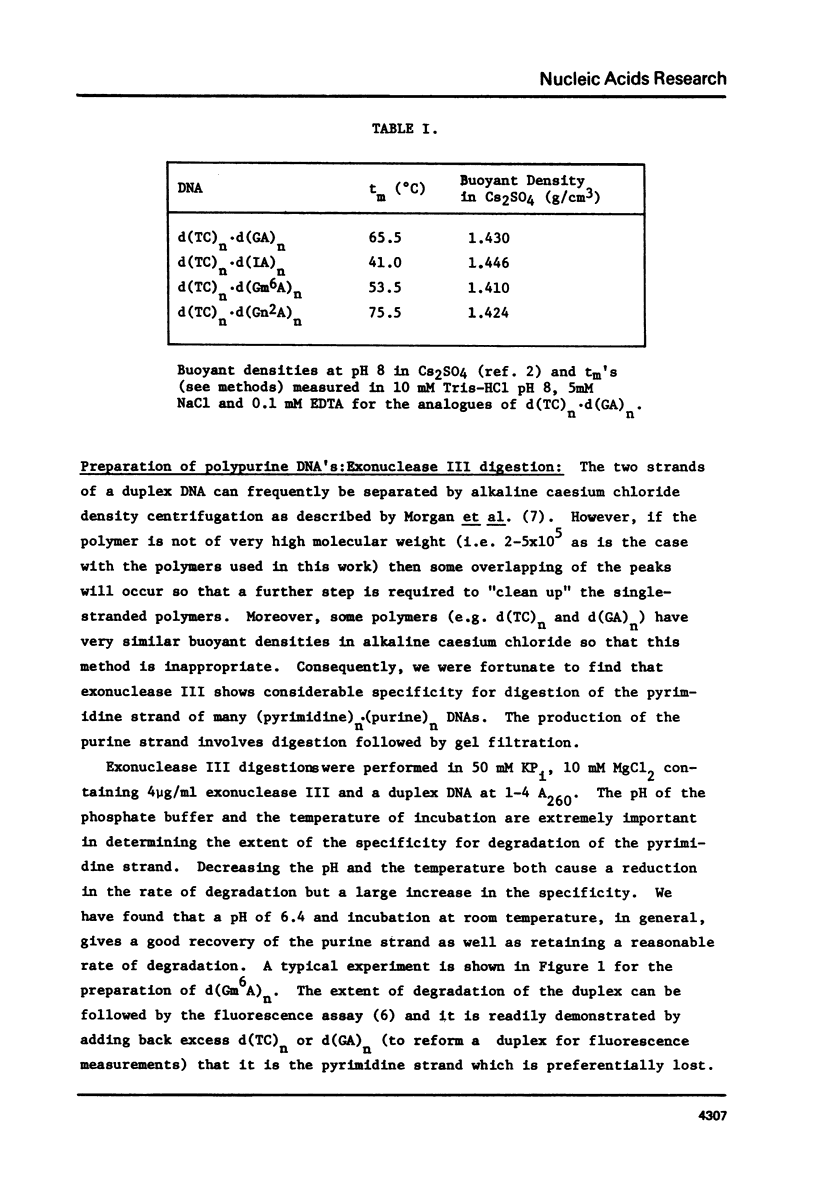
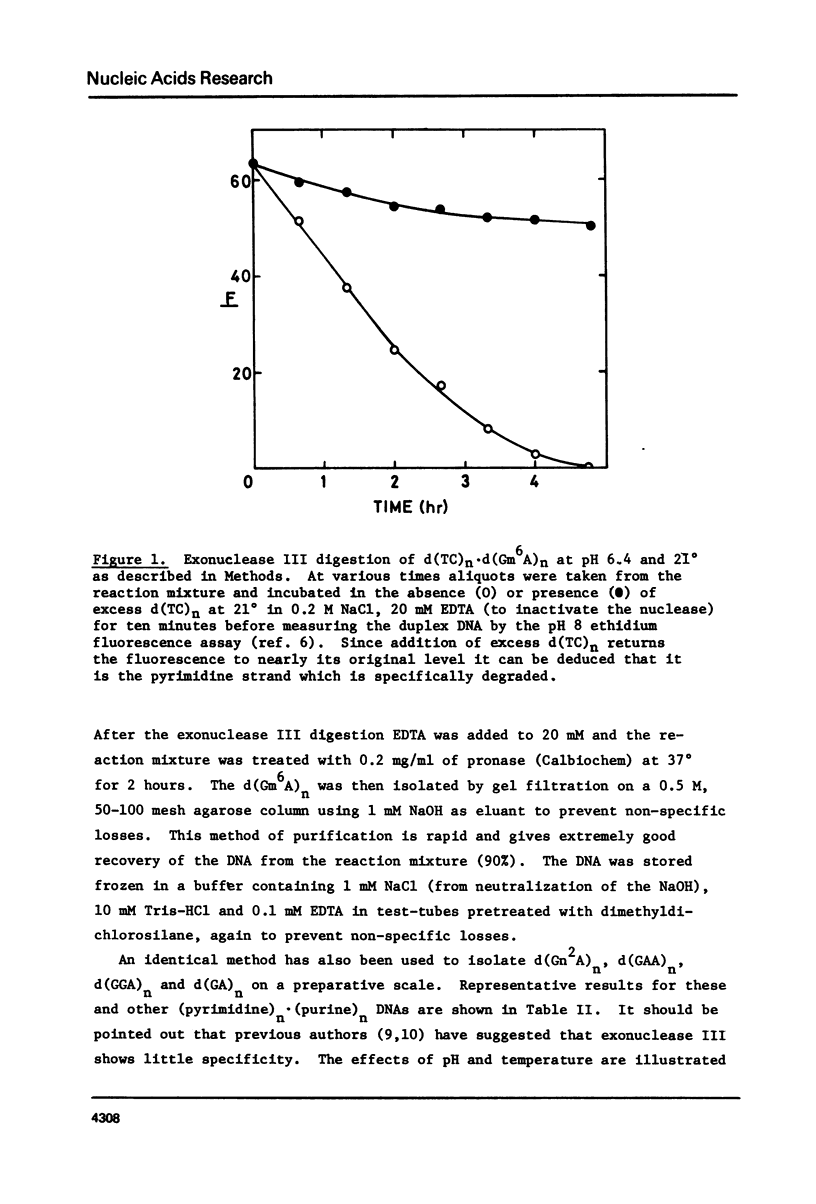
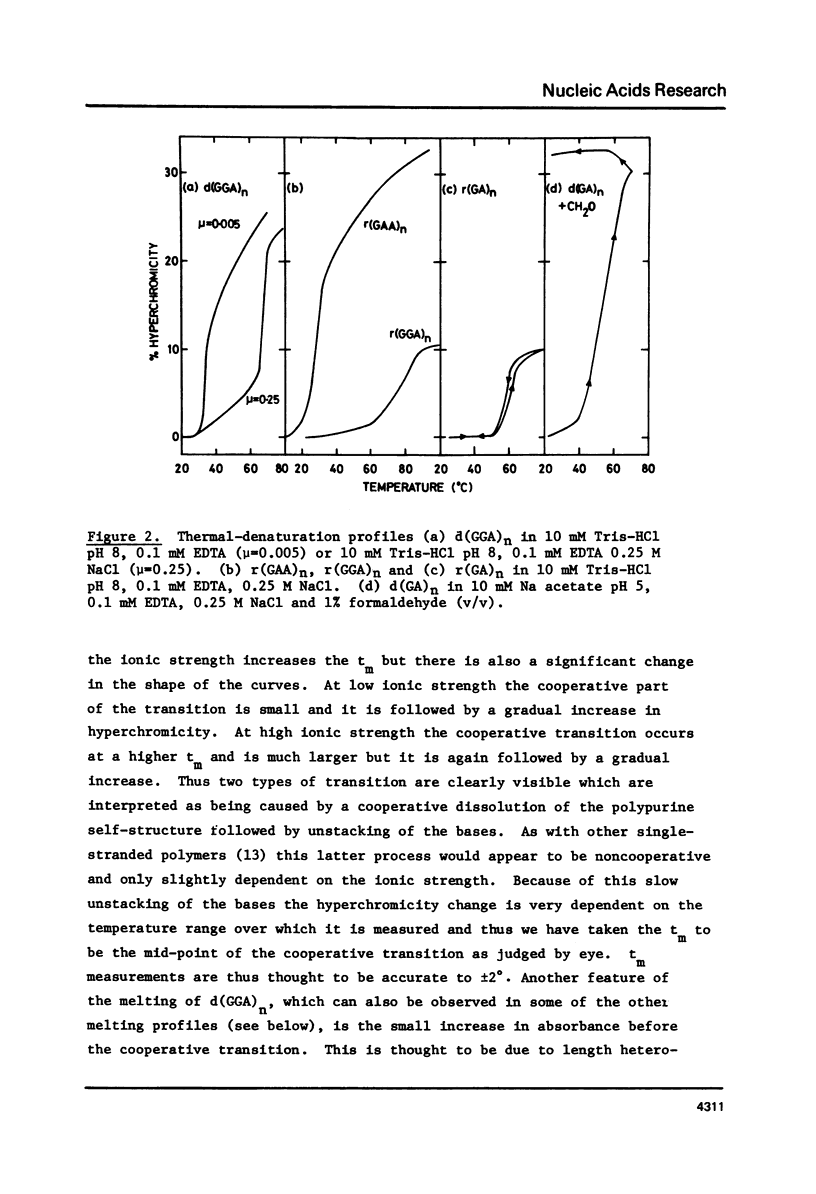
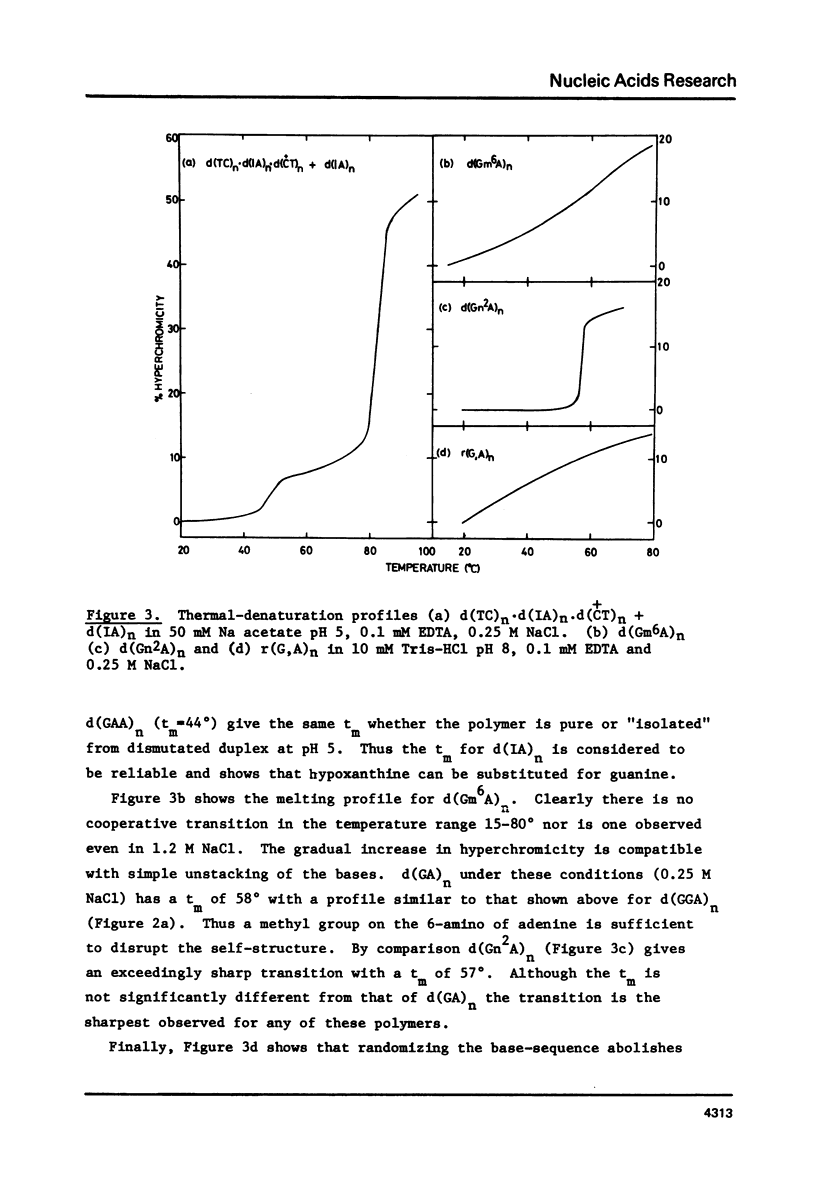
Abstract
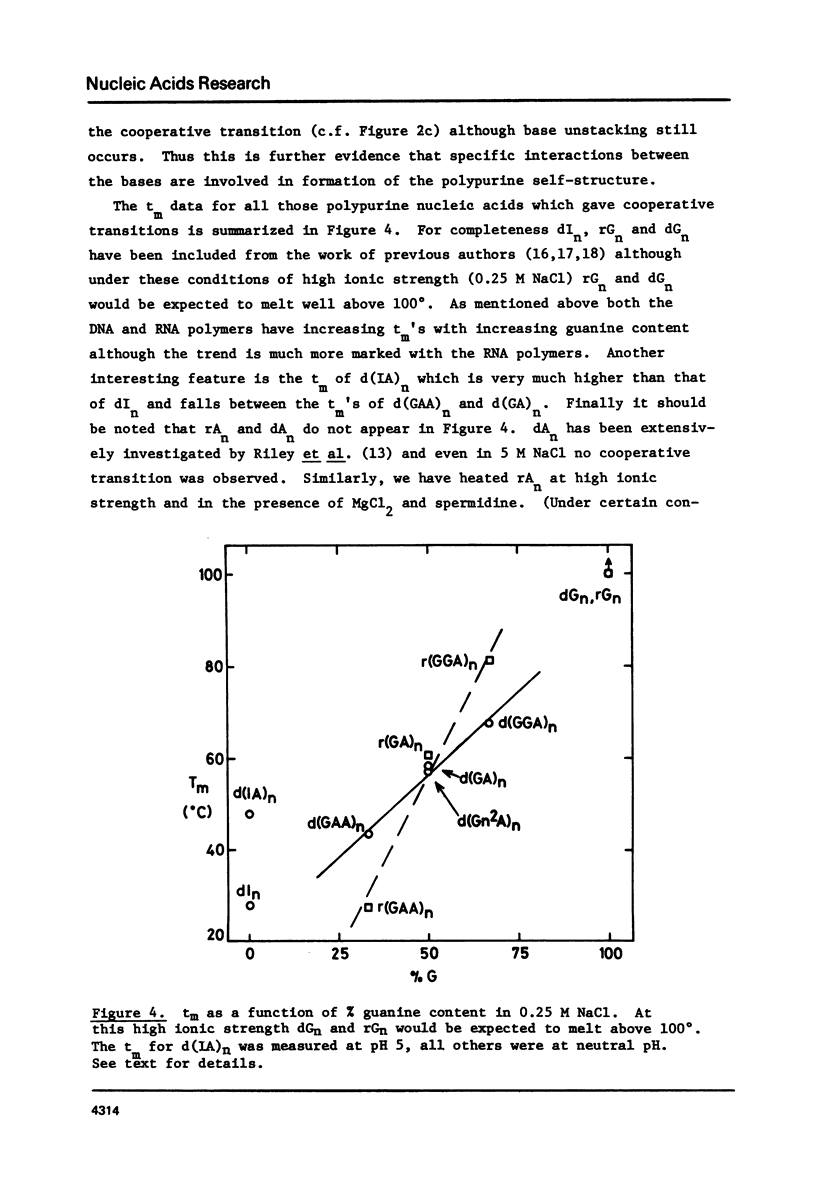
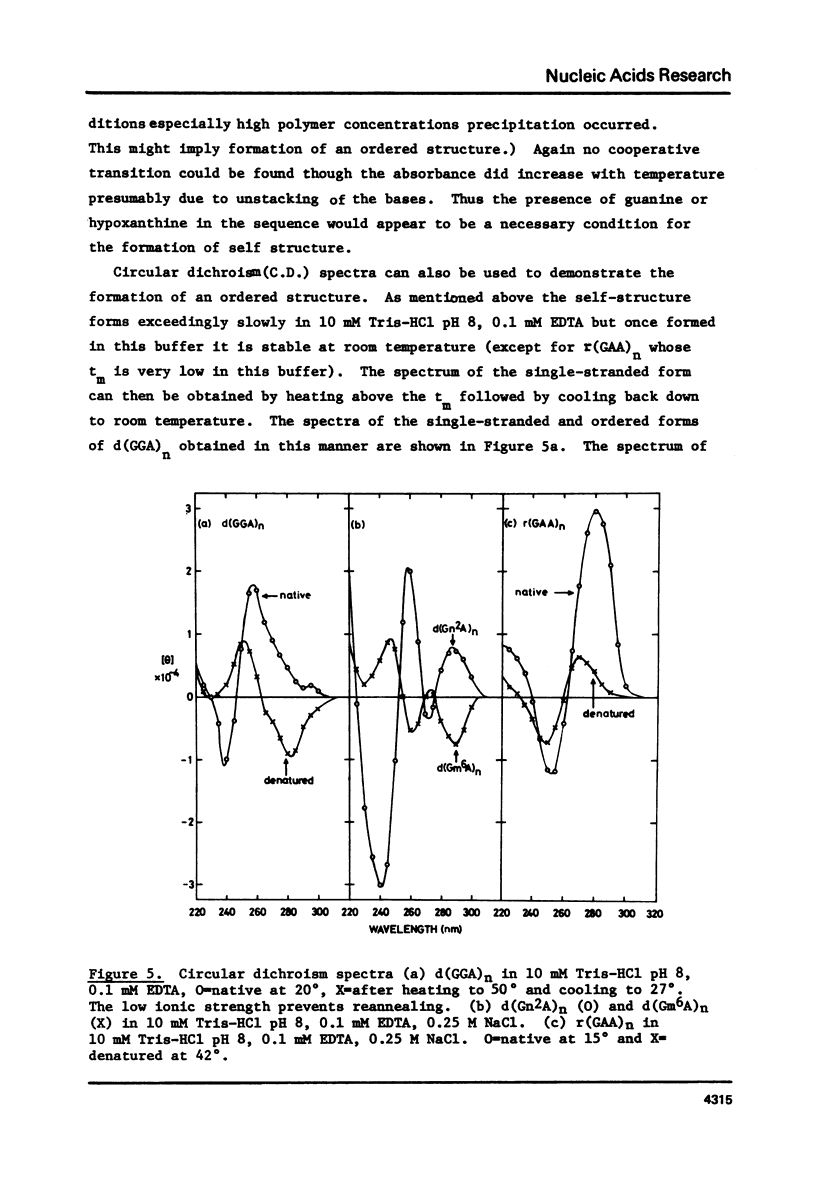
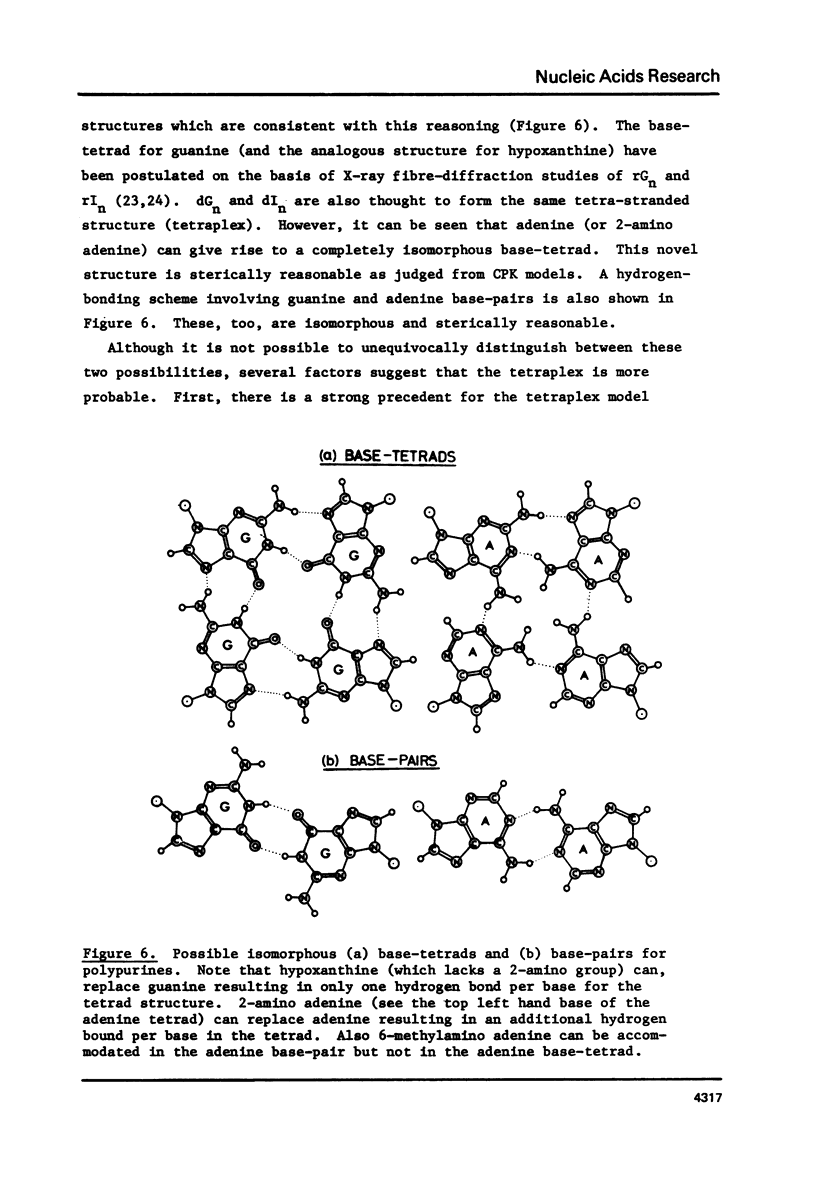
Polypurine DNAs and RNAs containing at least 33% guanine form a stable secondary structure at neutral pH and moderate ionic strengths. The tm's of the polymers increase with increasing guanine content. To eliminate possible structures three novel polymers, d(Gn2A)n, d(Gm6A)n and d(IA)n, as well as the random copolymer rr(G,A)n were were studied. Both d(Gn2A)n and d(IA)n can form a secondary structure whereas d(Gm6A)n and r(G,A)n cannot. Model building suggested two possible structures, one a duplex and the other a tetra-stranded polymer. The latter is considered to be the more likely, since previous X-ray diffraction studies have shown that rGn and rIn are tetra-stranded. Circular dichroism spectra are also consistent with such an interpretation.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Marttila C. M. Structures for polyinosinic acid and polyguanylic acid. Biochem J. 1974 Aug;141(2):537–543. doi: 10.1042/bj1410537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahms J., Michelson A. M., Van Holde K. E. Adenylate oligomers in single- and double-strand conformation. J Mol Biol. 1966 Feb;15(2):467–488. doi: 10.1016/s0022-2836(66)80122-5. [DOI] [PubMed] [Google Scholar]
- CHAMBERLIN M. J., PATTERSON D. L. PHYSICAL AND CHEMICAL CHARACTERIZATION OF THE ORDERED COMPLEXES FORMED BETWEEN POLYINOSINIC ACID, POLYCYTIDYLIC ACID AND THEIR DEOXYRIBO-ANALOGUES. J Mol Biol. 1965 Jun;12:410–428. doi: 10.1016/s0022-2836(65)80264-9. [DOI] [PubMed] [Google Scholar]
- CHAMBERLIN M., BERG P. Deoxyribo ucleic acid-directed synthesis of ribonucleic acid by an enzyme from Escherichia coli. Proc Natl Acad Sci U S A. 1962 Jan 15;48:81–94. doi: 10.1073/pnas.48.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor C. R., Warshaw M. M., Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9(9):1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- Chan A., Kilkuskie R., Hanlon S. Correlations between the duplex winding angle and the circular dichroism spectrum of calf thymus DNA. Biochemistry. 1979 Jan 9;18(1):84–91. doi: 10.1021/bi00568a013. [DOI] [PubMed] [Google Scholar]
- Donelson J. E., Wu R. Nucleotide sequence analysis of deoxyribonucleic acid. VII. Characterization of Escherichia coli exonuclease 3 activity for possible use in terminal nucleotide sequence analysis of duplex deoxyribonucleic acid. J Biol Chem. 1972 Jul 25;247(14):4661–4668. [PubMed] [Google Scholar]
- Engel J. D., von Hippel P. H. Effects of methylation on the stability of nucleic acid conformations. Studies at the polymer level. J Biol Chem. 1978 Feb 10;253(3):927–934. [PubMed] [Google Scholar]
- Englander J. J., Kallenbach N. R., Englander S. W. Hydrogen exchange study of some polynucleotides and transfer RNA. J Mol Biol. 1972 Jan 14;63(1):153–169. doi: 10.1016/0022-2836(72)90527-x. [DOI] [PubMed] [Google Scholar]
- Fry K., Brutlag D. Detection and resolution of closely related satellite DNA sequences by molecular cloning. J Mol Biol. 1979 Dec 15;135(3):581–593. doi: 10.1016/0022-2836(79)90165-7. [DOI] [PubMed] [Google Scholar]
- Harwood S. J., Wells R. D. Micrococcus luteus deoxyribonucleic acid polymerase. Studies on the initiation of deoxyribonucleic acid synthesis in vitro. J Biol Chem. 1970 Nov 10;245(21):5625–5634. [PubMed] [Google Scholar]
- Johnson D., Morgan A. R. Unique structures formed by pyrimidine-purine DNAs which may be four-stranded. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1637–1641. doi: 10.1073/pnas.75.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovin T. M., Englund P. T., Bertsch L. L. Enzymatic synthesis of deoxyribonucleic acid. XXVI. Physical and chemical studies of a homogeneous deoxyribonucleic acid polymerase. J Biol Chem. 1969 Jun 10;244(11):2996–3008. [PubMed] [Google Scholar]
- Lee J. S., Johnson D. A., Morgan A. R. Complexes formed by (pyrimidine)n . (purine)n DNAs on lowering the pH are three-stranded. Nucleic Acids Res. 1979 Jul 11;6(9):3073–3091. doi: 10.1093/nar/6.9.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomant A. J., Fresco J. R. Structural and energetic consequences of noncomplementary base oppositions in nucleic acid helices. Prog Nucleic Acid Res Mol Biol. 1975;15(0):185–218. doi: 10.1016/s0079-6603(08)60120-8. [DOI] [PubMed] [Google Scholar]
- MILES H. T., FRAZIER J. INFRARED SPECTRA IN 2H2O SOLUTION OF GUANYLIC ACID HELICES AND OF POLY-G. Biochim Biophys Acta. 1964 Jan 27;79:216–220. doi: 10.1016/0926-6577(64)90057-9. [DOI] [PubMed] [Google Scholar]
- Marck C., Thiele D. Poly(dG).poly(dC) at neutral and alkaline pH: the formation of triple stranded poly(dG).poly(dG).poly(dC). Nucleic Acids Res. 1978 Mar;5(3):1017–1028. doi: 10.1093/nar/5.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. R., Coulter M. B., Flintoff W. F., Paetkau V. H. Enzymatic synthesis of deoxyribonucleic acids with repeating sequences. A new repeating trinucleotide deoxyribonucleic acid, d(T-C-C)n-d(G-G-A)n. Biochemistry. 1974 Apr 9;13(8):1596–1603. doi: 10.1021/bi00705a007. [DOI] [PubMed] [Google Scholar]
- Morgan A. R., Evans D. H., Lee J. S., Pulleyblank D. E. Review: ethidium fluorescence assay. Part II. Enzymatic studies and DNA-protein interactions. Nucleic Acids Res. 1979 Oct 10;7(3):571–594. doi: 10.1093/nar/7.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. R., Lee J. S., Pulleyblank D. E., Murray N. L., Evans D. H. Review: ethidium fluorescence assays. Part 1. Physicochemical studies. Nucleic Acids Res. 1979 Oct 10;7(3):547–569. doi: 10.1093/nar/7.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. R., Wells R. D. Specificity of the three-stranded complex formation between double-stranded DNA and single-stranded RNA containing repeating nucleotide sequences. J Mol Biol. 1968 Oct 14;37(1):63–80. doi: 10.1016/0022-2836(68)90073-9. [DOI] [PubMed] [Google Scholar]
- RICHARDSON C. C., LEHMAN I. R., KORNBERG A. A DEOXYRIBONUCLEIC ACID PHOSPHATASE-EXONUCLEASE FROM ESCHERICHIA COLI. II. CHARACTERIZATION OF THE EXONUCLEASE ACTIVITY. J Biol Chem. 1964 Jan;239:251–258. [PubMed] [Google Scholar]
- Riley M., Maling B. Physical and chemical characterization of two- and three-stranded adenine-thymine and adenine-uracil homopolymer complexes. J Mol Biol. 1966 Sep;20(2):359–389. doi: 10.1016/0022-2836(66)90069-6. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Sures I., Lowry J., Kedes L. H. The DNA sequence of sea urchin (S. purpuratus) H2A, H2B and H3 histone coding and spacer regions. Cell. 1978 Nov;15(3):1033–1044. doi: 10.1016/0092-8674(78)90287-8. [DOI] [PubMed] [Google Scholar]
- Thiele D., Guschlbauer W. Protonated polynucleotide structures. IX. Disproportionation of poly (G)-poly (C) in acid medium. Biopolymers. 1971;10(1):143–157. doi: 10.1002/bip.360100111. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Larson J. E., Grant R. C., Shortle B. E., Cantor C. R. Physicochemical studies on polydeoxyribonucleotides containing defined repeating nucleotide sequences. J Mol Biol. 1970 Dec 28;54(3):465–497. doi: 10.1016/0022-2836(70)90121-x. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Cohen G. H., Davies D. R. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J Mol Biol. 1975 Feb 25;92(2):181–192. doi: 10.1016/0022-2836(75)90222-3. [DOI] [PubMed] [Google Scholar]



