Summary
The histone H2A variant H2AX is rapidly phosphorylated in response to DNA double-stranded breaks to produce γ-H2AX. γ-H2AX stabilizes cell cycle checkpoint proteins and DNA repair factors at the break site. We previously found that the protein phosphatase PP2A is required to resolve γ-H2AX foci and complete DNA repair after exogenous DNA damage. Here we describe a three-protein PP4 phosphatase complex in mammalian cells, containing PP4C, PP4R2 and PP4R3β, that specifically dephosphorylates ATR-mediated γ-H2AX generated during DNA replication. PP4 efficiently dephosphorylates γ-H2AX within mononucleosomes in vitro. The effect of PP4 on γ-H2AX is independent of ATR and checkpoint kinase activity. When the PP4 complex is silenced, repair of DNA replication mediated breaks is inefficient, and cells are hypersensitive to DNA replication inhibitors, but not radiomimetic drugs. Therefore γ-H2AX elimination at DNA damage foci is required for DNA damage repair, but accomplishing this task involves distinct phosphatases with potentially overlapping roles.
Introduction
DNA breaks occur constantly from endogenous (e.g. reactive oxygen species, metabolic byproducts, DNA replication and recombination) and exogenous (e.g. genotoxic chemicals, ionizing radiation (IR), UV irradiation) sources. Each type of DNA damage elicits a specific cellular repair response (Harrison and Haber, 2006). One of the earliest events in the double stranded DNA break (DSB) response is the phosphorylation of the histone H2A variant, H2AX, at Ser139 by members of the PI(3)K (phosphatidyl-inositol-3-OH kinase)-like kinases, ATM (ataxia telangiectasia mutated), ATR (ATM and Rad3-related) and DNA-PK (DNA-dependent protein kinase) (Fernandez-Capetillo et al., 2004). The three kinases have significant functional redundancy, but they are activated in a stress-specific manner. ATM and DNA-PK redundantly phosphorylate H2AX induced by ionizing radiation and radiomimetic drugs, whereas ATR seems to respond to endogenous or exogenous agents that interfere with DNA replication (Shiloh, 2003).
Phosphorylated H2AX (γ-H2AX) has a role in repair, replication, recombination of DNA and cell cycle regulation (Fernandez-Capetillo et al., 2004). The large γ-H2AX domains generated at each DSB, visualized as nuclear foci, stabilize cell cycle and DNA repair factors (cohesins, MDC1, Mre11, BRCA1, 53BP1 etc.) at the break site (Petrini and Stracker, 2003; Stucki and Jackson, 2006). Recent studies in mouse B cells suggest that γ-H2AX stabilizes the broken DNA ends during class switching, giving the repair machinery sufficient time to make appropriate joins (Franco et al., 2006; Ramiro et al., 2006). Importantly, loss of a single H2AX allele compromises genomic integrity and enhances cancer susceptibility in mice. The H2AX gene maps to a cytogenetic region frequently altered in human cancers, implicating similar functions in man (Bassing et al., 2003; Celeste et al., 2003a). Therefore the formation of γ-H2AX is of utmost importance for DNA repair. Although the kinases and stimuli involved in γ-H2AX formation have been intensely investigated, how γ-H2AX is eliminated in mammalian cells and the functional consequences of having constitutively phosphorylated H2AX remain unclear.
Two recent studies – one in mammals, the other in S. cerevisiae – identified roles for PP2A family phosphatases in γ-H2AX dephosphorylation (Chowdhury et al., 2005; Keogh et al., 2006). The PP2A family of serine/threonine phosphatases includes 4 distinct catalytic components in mammals – two closely related PP2A enzymes (PP2ACα, PP2ACβ), PP4C and PP6C (Honkanen and Golden, 2002). The most closely homologous yeast enzymes are Pph21 and Pph22, Pph3 and Sit4, respectively (Zabrocki et al., 2002). The catalytic components of these enzymes form dimeric or trimeric complexes with regulatory subunits that confer substrate specificity, tissue/cell type-specific targeting and control the extremely vigorous activity of the catalytic subunits. PP2A plays an important role in countering oncogenic kinases in cell cycle control and is the target of the SV40 small T antigen (Janssens et al., 2005); (Janssens and Goris, 2001). Little is known about the function of mammalian PP4 and PP6, although their yeast and fly homologues have been implicated in centrosome maturation and microtubule organization, resistance to apoptosis induced by UV irradiation and cisplatin, and recovery from the DNA damage checkpoint (PP4) (Cohen et al., 2005; Gingras et al., 2005; Hastie et al., 2006) and G1-S cell cycle progression (PP6) (Stefansson and Brautigan, 2007). We previously identified PP2A as a phosphatase that removes γ-H2AX foci formed in mammalian cells in response to DNA damage by the topoisomerase I inhibitor camptothecin (CPT) (Chowdhury et al., 2005). PP2AC colocalizes at γ-H2AX foci, suggesting that PP2A dephosphorylates γ-H2AX near a DSB. Importantly, when PP2AC is inhibited or silenced by RNA interference, γ-H2AX levels following DNA damage increase, γ-H2AX foci persist and DSB repair is impaired (Chowdhury et al., 2005). In a parallel study in yeast a deletion screen revealed a trimeric complex (histone H2A phosphatase complex) containing Pph3, Psy2 and Ybl046w, that regulates basal γ-H2AX levels (Keogh et al., 2006). Although both PP2A and Pph3 efficiently dephosphorylate γ-H2AX in vitro, their roles in cells appear to be distinct. Pph3 does not accumulate at the site of an engineered DSB and therefore was hypothesized to dephosphorylate γ-H2AX only after it has been removed from chromatin, whereas PP2A likely works on chromatin-bound γ-H2AX. Pph3 deletion has no effect on repair kinetics and the rate of γ-H2AX loss at a single exogenously induced DSB. Pph3 mutants show a constitutive increase in γ-H2AX levels even in the absence of exogenous DNA damage, while silencing PP2A in mammalian cells does not affect basal γ-H2AX in the absence of an exogenous insult (Chowdhury et al., 2005);(Keogh et al., 2006). Although the differences between PP2A and Pph3 mediated removal of γ-H2AX might be due to different regulatory mechanisms in yeast and mammals, we speculated that more than one phosphatase might be involved in γ-H2AX removal in mammalian cells and that another mammalian phosphatase might be functionally analogous to Pph3. PP4C is the closest human homolog of yeast Pph3 (Fig. 1A). Although several putative PP4C-containing complexes have been identified, their biological function is not well defined (Gingras et al., 2005). Genetic deletion of the PP4 catalytic subunit PP4C in mice results in early embryonic lethality. Conditional deletion of PP4C in thymocytes leads to an early block in thymic development at the DN3 stage and enhanced apoptosis (Shui et al., 2007). Few physiological substrates of mammalian PP4C have been identified. PP4 has been implicated in TNFα signaling (Mihindukulasuriya et al., 2004; Zhou et al., 2002) and NF-κb regulation (Hu et al., 1998; Yeh et al., 2004). Recently, histone deacetylase 3 (HDAC3) and a mitotic regulatory protein NDEL1 were shown to be regulated by a specific PP4 complex (Toyo-oka et al., 2008; Zhang et al., 2005).
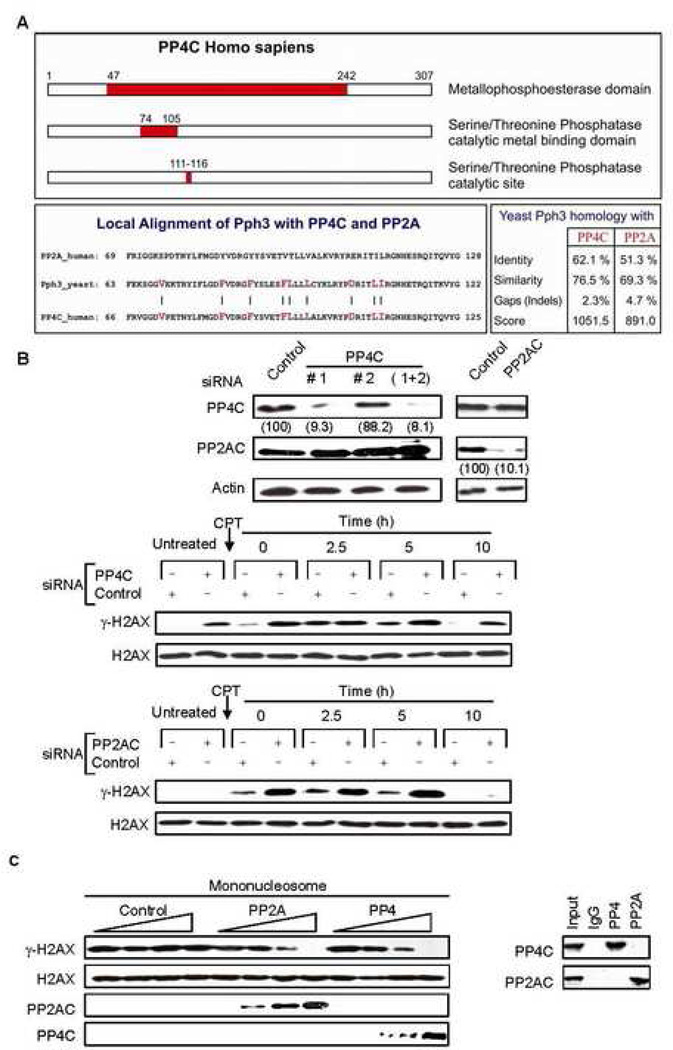
Fig. 1. PP4C dephosphorylates γ–H2AX in vitro, and silencing PP4C increases cellular γ–H2AX in the presence and absence of exogenous DNA damage.
A) Schematic depiction of the PP4C catalytic domain and sequence comparison of the respective catalytic domains of human PP4C and PP2AC with S. cerevisiae Pph3. Alignment of Pph3 with either PP4C or PP2AC shows relatively stronger homology between Pph3 and PP4C. The bars and highlighted residues represent the amino acids which were identical between PP4C and Pph3 but different for PP2AC. The percentage of identity and similarity between yeast Pph3 and PP4C or PP2AC is represented in a table in the lower right.
B) Silencing PP4C increases basal γ-H2AX. HeLa cells were transfected with control or PP4C siRNAs and harvested after 48 h. Two different siRNA duplexes to PP4C were used individually or in combination, and one siRNA complex was used for PP2AC. Immunoblots were probed for PP4C, PP2AC or β-actin. Numbers in parentheses indicate signal intensity as a percent of the control. Subsequent experiments were performed using PP4C siRNA duplex #1, which efficiently silenced PP4C. PP4C (upper panel), PP2AC (lower panel) or control siRNA-transfected HeLa cells, treated or not with CPT (2 µM, 1 h) were harvested at indicated times and immunoblots of whole cell extracts were probed for γ–H2AX or H2AX. Relative to control, PP4C-silenced cells have higher levels of γ-H2AX in untreated cells and after CPT treatment, whereas PP2AC-silenced cells show a relative increase in γ-H2AX only after CPT treatment.
C) PP4 dephosphorylates γ–H2AX in vitro as efficiently as PP2A. HA-tagged PP4C and PP2AC were in vitro transcribed and translated and the respective proteins were immunopurified using anti-HA beads without cross contamination (right). Immunoprecipitated PP4C and PP2AC were serially diluted in the phosphatase reaction. PP4 dephosphorylates human γ–H2AX assembled in mononucleosomes as efficiently as PP2A (left).
Here we show that PP4C, like yeast Pph3, controls basal levels of γ-H2AX. PP4C dephosphorylates γ-H2AX incorporated in mononucleosomes in vitro with comparable efficiency as PP2AC. Silencing any component of a PP4C complex, containing mammalian homologues of Psy2 and Ybl046w, increases basal γ-H2AX foci that associate with MRE11. The increase in γ-H2AX when PP4 is knocked down is ATR-mediated and requires DNA replication. Unresolved γ-H2AX foci occur only in dividing cells and bind PCNA, suggesting they form at stalled replication forks. However, PP4-deficiency does not alter ATR kinase activity or checkpoint protein phosphorylation, suggesting that γ-H2AX is a direct substrate of PP4. Dividing cells with silenced PP4 are hypersensitive to DNA-replication inhibitors, but not other DNA damaging agents. Moreover, resolution of endogenous DNA damage that occurs during DNA replication requires PP4.
Results
Silencing PP4C increases γ-H2AX basal levels
To identify a Pph3 functional analog in mammalian cells, we first looked at whether PP4C might fulfill that role. Although all the PP2A family catalytic subunits are highly homologous, PP4 is the closest human homolog of the yeast phosphatase Pph3, especially in the catalytic metal binding domain and active site (residues 74–125) (Fig. 1A). To investigate a possible role of PP4C in regulating γ-H2AX, small interfering RNAs (siRNA) were designed to down-regulate PP4C expression specifically without altering expression of the related PP2A family members (Pandey et al., 2003). HeLa cells transfected with two PP4C siRNAs duplexes were assayed for phosphatase proteins by immunoblot three days later. Only PP4C siRNA duplex #1 efficiently silenced PP4C, and this duplex was used in subsequent experiments. PP4C siRNA had no effect on PP2AC and vice versa (Fig. 1B, upper panel). We next investigated how PP4C deficiency affected γ-H2AX levels (Fig. 1B, middle panel). HeLa cells transfected with control or PP4C siRNAs (Fig. 1B, middle panel) or PP2AC specific siRNAs (Fig. 1B, lower panel) were treated with CPT for 1 h and γ–H2AX levels analyzed by immunoblot (Fig. 1B, middle panel). In the absence of PP4C or PP2AC siRNAs, γ–H2AX was not detected in cells not treated with CPT, began to be detected immediately after adding CPT, peaked two and a half hours after CPT removal and returned to background by 10 h. However, in the presence of either PP4C or PP2AC siRNA, γ–H2AX levels were significantly higher and were only diminished 10 h after removing CPT. However, the pattern of γ–H2AX, but not total H2AX, protein varied in PP2AC and PP4C silenced cells. The most striking result was a significant increase in basal γ-H2AX in PP4C-deficient cells before adding CPT, which was not observed when PP2AC was silenced. However, γ-H2AX levels increased in response to CPT most dramatically in PP2A-deficient cells compared to control or PP4-silenced cells and this increase lasted for at least 5 h after DNA damage. Ten hours after CPT treatment γ-H2AX was detected only in PP4-silenced cells and was comparable to basal γ–H2AX levels in PP4-deficient cells that were not subjected to exogenous DNA damage. To confirm the specificity of the PP4C siRNA effect, we co-transfected HeLa cells with the PP4C siRNA that effectively silenced PP4C and a Flag-tagged PP4C expression plasmid containing silent mutations in the siRNA recognition site. Expressing the siRNA-resistant PP4C eliminated the PP4C siRNA-mediated increase in basal γ-H2AX (Suppl. Fig. 1). These results suggest that although both PP2A and PP4 regulate the cellular pool of γ–H2AX, there are key kinetic and contextual differences in the regulatory pathways.
PP4C dephosphorylates γ-H2AX as efficiently as PP2AC in vitro
To determine whether PP4C can dephosphorylate γ-H2AX directly and compare the efficiency of dephosphorylation with PP2AC, we compared the in vitro activity of the two enzymes against Ribosomal S6 kinase 1 (Rsk1)-phosphorylated human γ-H2AX reconstituted in mononucleosomes (Fig. 1C). The enzymes were obtained by immunoprecipitating HA-tagged PP4C and PP2AC from in vitro transcribed and translated rabbit reticulocyte lysates. Both PP2AC and PP4C dephosphorylated nucleosomal γ-H2AX in a dose-dependent manner and importantly, their in vitro activities on γ-H2AX were comparable.
Multiple PP4C-containing complexes
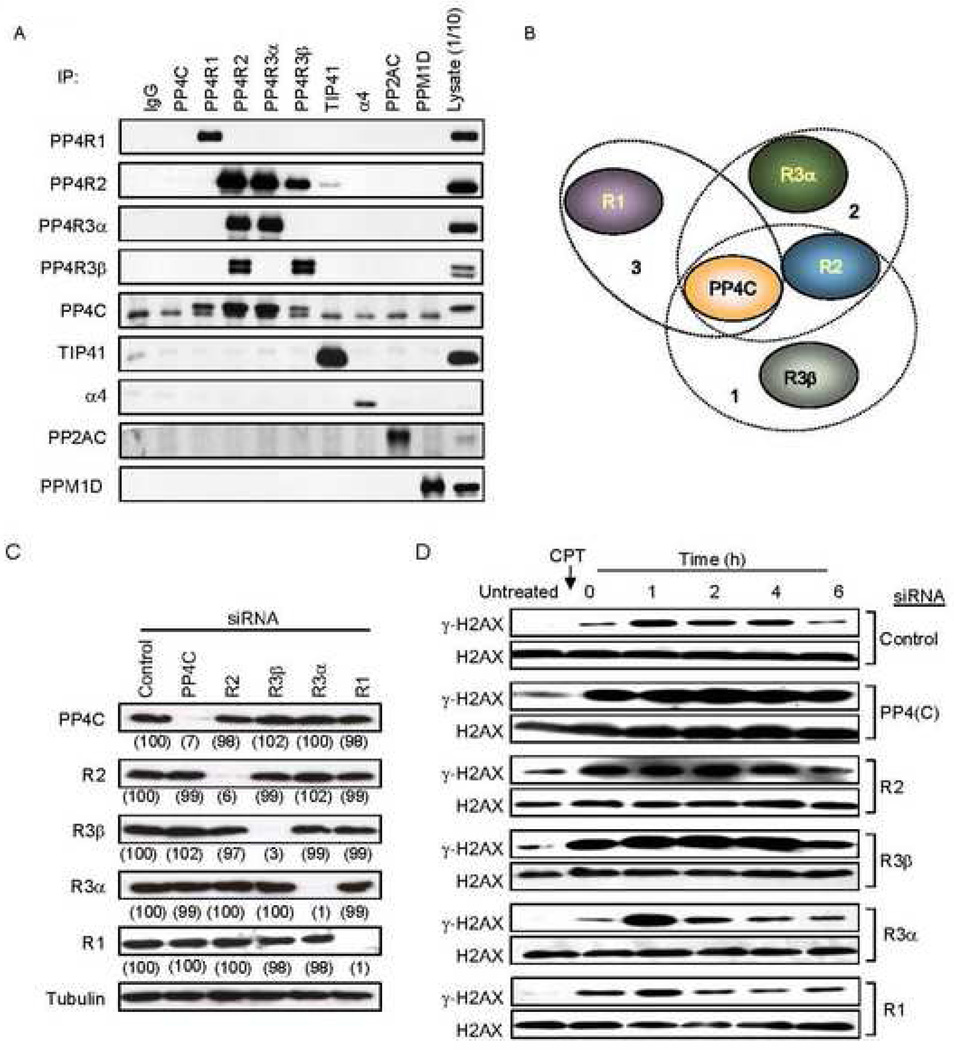
PP4C forms several multimeric complexes with proteins that have homology to the subunits of the γ-H2AX regulating yeast phosphatase complex, HTP-C (Cohen et al., 2005). However, the subunit composition and molecular architecture, cellular functions and subcellular localization of the different PP4-complexes are not well characterized. A recent study using mass spectrometry analysis of TAP-tagged proteins systematically analyzed yeast Pph3 and mammalian PP4C interacting proteins and identified several distinct PP4C-containing protein complexes (Gingras et al., 2005). To confirm these results and identify which of the PP4 regulatory subunits are involved in the regulation of γ-H2AX in mammalian cells, we produced antibodies against the predicted PP4 subunits, R1, R2, R3α, R3β and PP4C, and for 3 other proteins, TIP41, α4, and Gemin 4, which have also been reported to associate with PP4C (Carnegie et al., 2003; Gingras et al., 2005). We analyzed the association of these proteins by reciprocal immunoprecipitation/immunoblot assays using 293T cell lysates. Immunoprecipitation with non-specific IgG controlled for specificity. Antibodies against three other phosphatases, PP2AC, PPM1D (also known as PP2Cδ or WIP1) (Fig. 2A), PP6C (data not shown) served as additional controls (Fig. 2A). As in the earlier study (Gingras et al., 2005), we identified three distinct PP4C-containing complexes (Fig. 2B): 1) a complex containing PP4R2 and PP4R3β, 2) a complex containing PP4R2 and PP4R3α, and 3) a binary complex with PP4R1. None of these PP4 complex subunits interacted with PP2AC, PP6C or PPM1D. Although PP4R3α and PP4R3β are close homologs, we failed to detect any physical interaction between them. Contrary to earlier results, we did not detect any interaction between PP4C and TIP41 or α4 (Fig. 2A) or Gemin 4 (data not shown). This discrepancy may be due to the difference in cell or the method (i.e. immunoprecipitating over-expressed tagged proteins versus immunoprecipitating endogenous protein).
Fig. 2. Knocking down the PP4C-R2-R3β complex alters basal γ-H2AX levels.
A) Reciprocal immunoprecipitation/immunoblot of PP4 complex proteins in 293T cell lysates. The PP4C antibody worked for immunoblot, but failed to immunoprecipitate detectable amounts of PP4C. The lower band in the PP4C blot is a nonspecific cross-reacting band. The PP4 complex subunits R1, R2, R3α and R3β associated with PP4C and did not interact with the other phosphatases, PP2AC and PPM1D.
B) Schematic representation of the distinct PP4 complexes.
C) siRNAs targeting specific PP4 subunits reduce their expression without altering other PP4 subunits or tubulin. HeLa cells transfected with the indicated siRNAs were harvested after 40 h and analyzed by immunoblot. Experiments described henceforth were performed using these siRNAs. Numbers in parentheses indicate signal intensity as a percent of control.
D) Knocking down certain PP4 subunits increases basal γ-H2AX levels. Control or PP4 subunit specific siRNA-transfected HeLa cells, treated or not with CPT (2 µM, 1 h), were harvested at indicated times and whole cell extracts were probed for γ–H2AX or H2AX. PP4R3β, PP4R2 and PP4C deficient cells have increased γ-H2AX both in untreated cells and after CPT treatment.
A PP4C-R2-R3β complex regulates basal γ-H2AX
To determine which PP4-complex controls cellular γ-H2AX, we designed siRNA duplexes against all the subunits. Three days after HeLa cells were transfected with siRNA duplexes targeting each subunit, PP4 complex subunit levels were assessed by immunoblot (Fig. 2C). siRNAs against each subunit specifically reduced expression of the respective subunit by >90%. More importantly, it did not affect the expression of any other PP4 subunit or tubulin. We next investigated how knockdown of the different PP4 subunits affected γ-H2AX. HeLa cells transfected with control or PP4-subunit siRNAs were treated with CPT for 1 h and γ–H2AX levels analyzed by immunoblot (Fig. 2D). After knockdown of PP4C, PP4R2 or PP4R3β, γ–H2AX levels were significantly higher at all times examined. Consistent with Fig. 1B, even in the absence of exogenous DNA damage, basal γ-H2AX levels were increased specifically in PP4C, PP4R2 or PP4R3β deficient cells. Knocking down the two other subunits, PP4R1 or PP4R3α, had no consistent effect on γ-H2AX. Together these results suggest that a PP4 complex, comprised of the catalytic subunit and the two regulatory subunits PP4R2 and PP4R3β, controls cellular levels of γ-H2AX, independently of external DNA damage.
γ–H2AX foci increase independently of exogenous DNA damage in cells deficient in PP4C, PP4R2 or PP4R3β
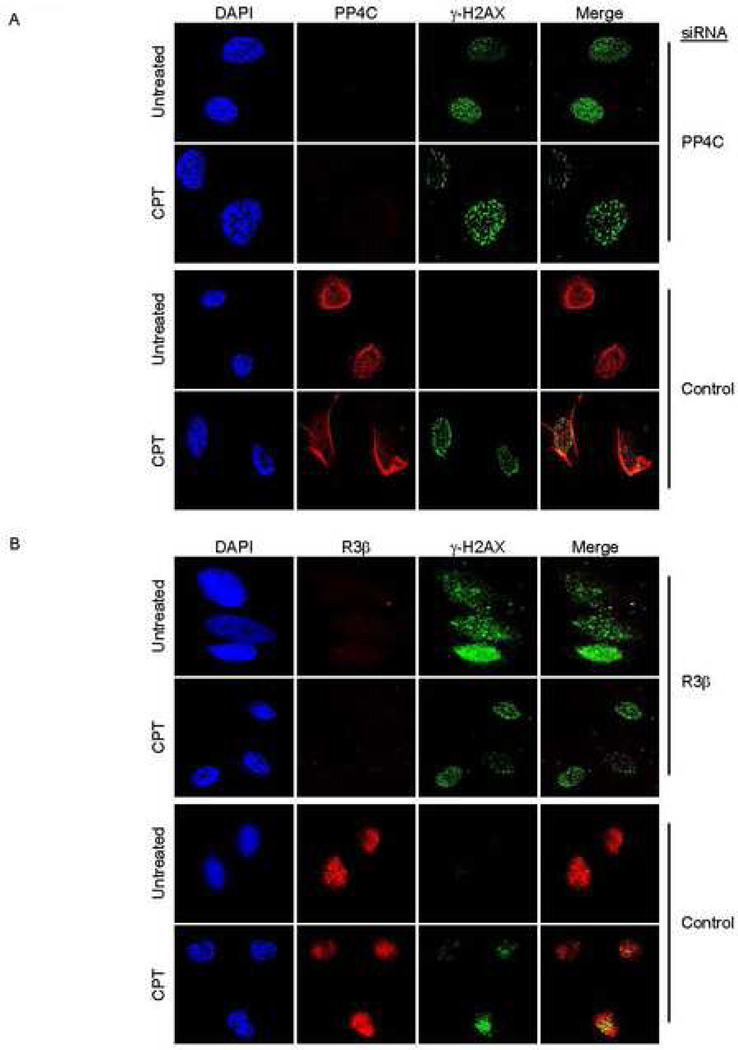
γ–H2AX accumulation at discrete nuclear foci is an early marker of DSB sites undergoing DNA repair. These foci, visible by immunofluorescence microscopy, can be seen at a low frequency in cycling cells, reflecting the occasional DSB that occurs as DNA is replicated. To investigate whether the PP4 complex regulates γ-H2AX focus formation, HeLa cells transfected with control or PP4C siRNAs were either untreated or treated with CPT for 1 h and observed for γ–H2AX foci 2 h later (Fig. 3A). In untreated cells γ-H2AX foci increased greatly in the absence of PP4C. Focus formation in response to CPT was not that different in control or PP4C-deficient cells. Of note, PP4C in control cells also did not appear to colocalize with CPT-induced γ–H2AX foci (see below). Therefore the basal increase in cellular γ-H2AX in PP4C-knocked down cells (as observed in Fig. 1B and 2D) also resulted in formation of γ-H2AX foci. The DNA repair factors, MRE11, which co-localizes with DSBs, and PCNA, which specifically co-localizes with DSB generated during DNA replication (Ward and Chen, 2001), also were recruited to γ-H2AX foci generated in PP4C deficient cells, suggesting that γ-H2AX is induced in response to DSB generated during DNA replication (Suppl. Fig. 2). We confirmed the role of the PP4C-R2-R3β complex by doing the same set of experiments in PP4R3β-silenced (Fig. 3B) and PP4R2-silenced cells (data not shown). The increase in γ-H2AX foci in PP4R2- and PP4R3β-deficient cells that were not subjected to exogenous DNA damage suggested that the PP4 complex as a whole, and not just the catalytic subunit, was responsible for regulating basal γ-H2AX foci.
Fig. 3. Knocking down PP4C or R3β induces γ–H2AX foci in the absence of exogenous DNA damage.
PP4C (A) or PP4R3β (B) or control siRNA-transfected HeLa cells, treated or not with CPT (2 µM, 1 h), were fixed and stained with anti-γH2AX and DAPI and visualized using laser-scanning confocal microscopy. γ-H2AX foci form in untreated cells deficient in PP4C or PP4R3β.
The PP4C-R2-R3β complex is associated with chromatin and does not relocalize in response to exogenous DNA damage
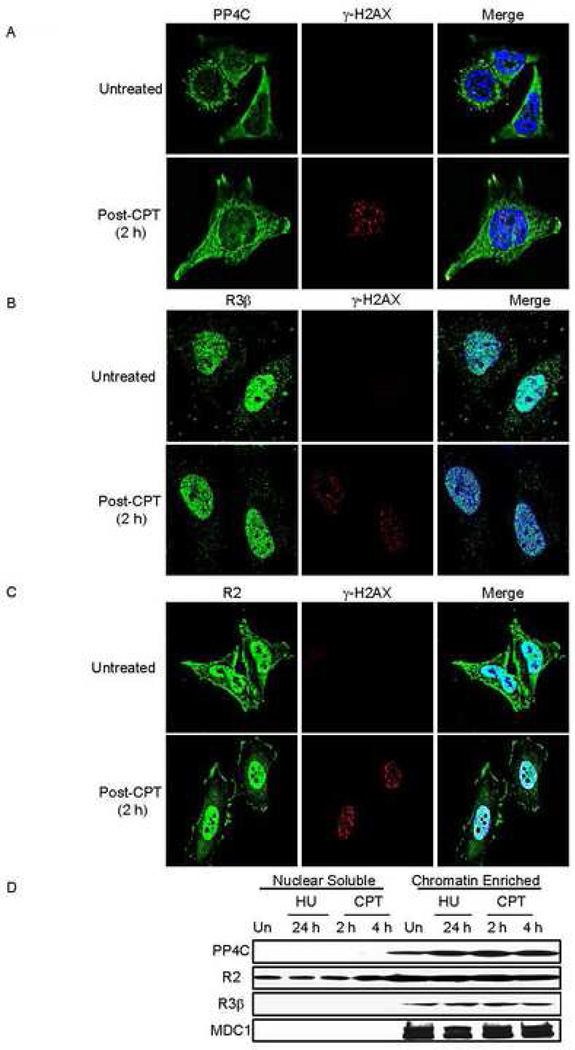
The subcellular location of PP4C and its associated subunits is not known. To study PP4 localization in the context of DNA damage, we co-stained untreated and CPT-treated HeLa cells with antibodies to γ–H2AX and the PP4 complex proteins, PP4C (Fig.4A), PP4R3β (Fig. 4B) and PP4R2 (Fig. 4C). Total cellular PP4C, PP4R2 or PP4R3β by immunoblot did not change following treatment with CPT or the dNTP synthesis-inhibitor hydroxyurea (HU) (data not shown). In untreated cells, PP4C was primarily perinuclear, with some nuclear staining (Fig. 4A). This cellular pattern did not change within the first few hours after CPT treatment. Moreover, as noted in Fig. 3A, PP4C did not colocalize with CPT-induced γ–H2AX foci. The majority of PP4R3β staining was nuclear prior to CPT treatment and this pattern remained unchanged after DNA damage (Fig. 4B). PP4R2 was visible both in the cytoplasm and nucleus before CPT treatment and in the first few hours after treatment (Fig. 4C). Although PP4C appears to be mostly extranuclear, these data suggest that the PP4C-R2-R3β complex is primarily nuclear, independently of exogenous DNA damage, and does not relocalize after exogenous DNA damage. Most cellular PP4C, which is primarily cytosolic, is likely associated with other complexes. By biochemical fractionation, the PP4C-R2-R3β nuclear complex was enriched in chromatin and not detected in the nucleoplasm (Fig. 4D). Knocking down PP4R2 (and possibly PP4R3β) reduced chromatin-associated PP4C, suggesting that these subunits may direct PP4C to chromatin (Suppl. Fig.3).
Fig. 4. Subcellular localization of PP4C, PP4R3β and PP4R2 does not substantially change in response to exogenous DNA damage.
Untreated or CPT-treated (5 µM, 1 h, 37°C) HeLa cells were washed to remove CPT and incubated for 2 h, before fixing and staining for γ–H2AX and PP4C (A), PP4R3β (B) or PP4R2 (C). Images were visualized using laser-scanning confocal microscopy. PP4C is primarily perinuclear with some nuclear staining, but PP4R3β and PP4R2 are predominantly nuclear, suggesting that the PP4C-R2-R3β complex mostly resides in the nucleus. None of these components substantially changed their localization following CPT treatment.
D) The PP4C-R2-R3β complex is associated with chromatin in fractionated nuclei from HeLa cells, treated or not with 2 mM HU for 24 h or with 10 uM CPT for 2 or 4 h. Although PP4R2 was also found in the nuclear soluble fraction, PP4C and PPR3β were exclusively detected in the chromatin fraction. MDC1 served as a control for the chromatin fraction.
The basal increase in γ-H2AX in PP4C-silenced cells is mediated by ATR
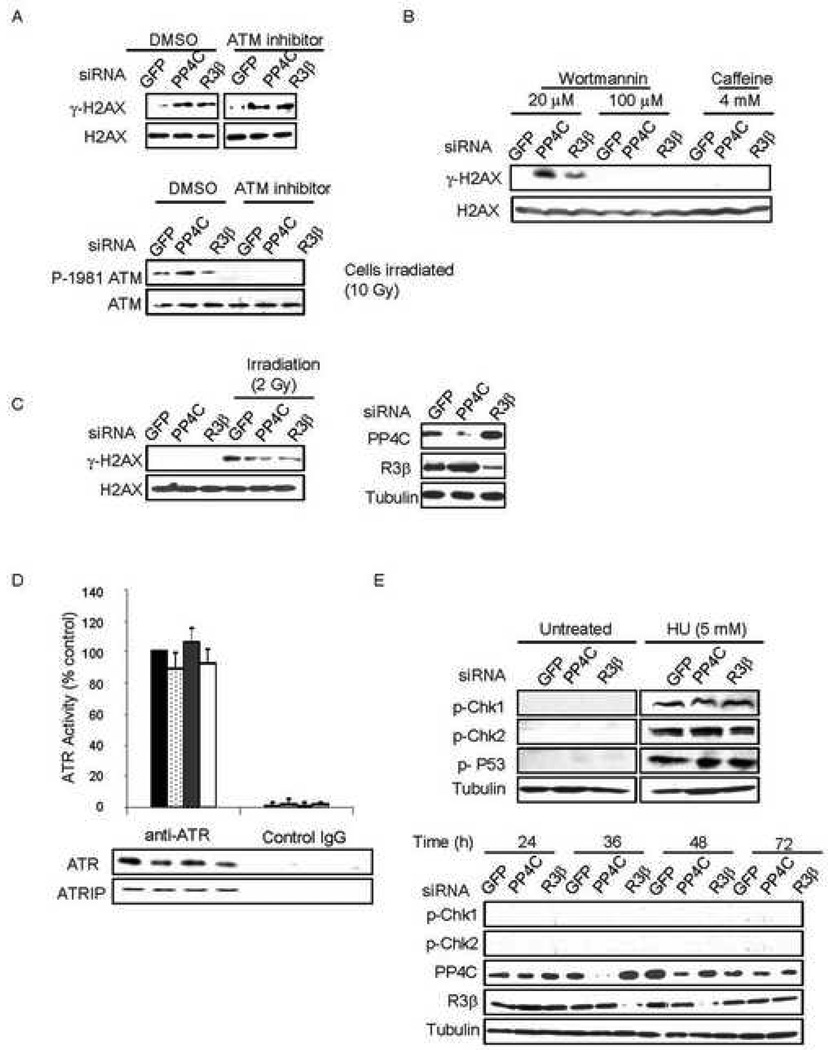
One of the earliest events in the DSB response is H2AX phosphorylation at Ser139 by one of the three PIKK, ATM, ATR or DNA-PK. However, it is not clear if γ-H2AX forms in the course of a normal cell division cycle and if so, which kinase is responsible. Recent reports show ATM-dependent H2AX phosphorylation occurs in mitosis (Ichijima et al., 2005; McManus and Hendzel, 2005). ATR phosphorylates H2AX in response to any DNA replication-mediated DNA damage (Furuta et al., 2003). To investigate which kinase was responsible for the basal increase in γ-H2AX in PP4-deficient cells, we assessed γ-H2AX levels in HeLa cells knocked down for PP4C or PP4R3β and treated with the ATM inhibitor KU55933. Lack of ATM autophosphorylation following γ-irradiation verified that ATM was completely inhibited (Fig. 5A, lower panel). In PP4-deficient cells, ATM inhibition had no effect on γ-H2AX. Wortmannin at a lower concentration (20 µM) inhibits ATM and DNA-PK, but at higher concentrations (<100 µM) inhibits all three PIKK (Sarkaria et al., 1998). Caffeine (4 mM) inhibits ATM and ATR, but does not affect DNA-PK activity (Sarkaria et al., 1999). Inhibition of ATM and DNA-PK had no effect on γ-H2AX in PP4-silenced cells, but inhibiting all three with a high dose of wortmannin completely blocked basal γ-H2AX formation. Impeding ATM and ATR with caffeine inhibited basal H2AX phosphorylation in PP4-knocked down cells (Fig. 5B). This result suggested that the basal increase in γ-H2AX in PP4-deficient cells was ATR-mediated. This was further confirmed when PP4C and PP4R3β were knocked down in ATR-deficient Seckel fibroblasts and γ-H2AX levels were not altered, either basally or after irradiation (Fig. 5C).
Fig. 5. ATR is responsible for γ-H2AX formation in PP4-deficient cells and ATR activity is not regulated by PP4.
A) ATM inhibition does not affect basal γ-H2AX levels in PP4-silenced cells. PP4C, R3β or control siRNA-transfected HeLa cells were incubated with DMSO or 5 µM KU55933 (ATM inhibitor) and whole cell extracts were analyzed by immunoblot probed for γ–H2AX or H2AX (upper panel). To verify ATM inhibition, cells were irradiated (10 Gy) and immunoblotted for phospho1981-ATM or ATM.
B) Inhibition of ATR, but not DNA-PK or ATM, decreases γ-H2AX in PP4-silenced cells. PP4C, R3β or control siRNA-transfected HeLa cells were incubated with 20 µM or 100 µM Wortmannin or with 4 mM caffeine and whole cell extracts analyzed by immunoblot for γ–H2AX or H2AX.
C) Knocking down PP4 in ATR-deficient cells has no effect on basal γ-H2AX levels. PP4C and PP4R3β were knocked down in Seckel fibroblasts (right), and γ-H2AX levels analyzed by immunoblot in untreated cells or after irradiation (2 Gy). There was no effect of PP4-knock down on γ-H2AX in these cells (left).
D) PP4 knockdown does not affect ATR activity. ATR was immunopurified from PP4 subunit-specific or control siRNA-transfected HeLa cells. The kinase activity of the ATR immunocomplex from PP4-deficient cells was assayed relative to the activity in control cells. The lower panel shows comparable amounts of immunoprecipitated ATR and ATRIP. siRNAs used were black, control; light dots, PP4C; dark dots, PP4R2; white, PP4R3β. Mean ±SD are shown.
E) PP4 deficiency does not activate the checkpoint proteins or p53. PP4C, PP4R3β or control siRNA-transfected HeLa cells were incubated with DMSO or 5 mM HU 48 h after transfection and immunoblots were performed on whole cell extracts with the indicated antibodies (upper panel). HeLa cells transfected with PP4C, PP4R3β or control siRNAs were harvested at indicated times after transfection and immunoblots of whole cell extracts probed with the indicated antibodies (lower panel).
ATR activity and phosphorylation of checkpoint proteins is not altered in PP4 deficient cells
Free phosphatase catalytic subunits are promiscuous, with specificity imparted by their regulatory subunits. Although PP2A dephosphorylates γ-H2AX, it also enhances the kinase activity of DNA-PK (Douglas et al., 2001) and inhibits ATM (Dozier et al., 2004; Goodarzi et al., 2004). However, PP2A has no effect on ATR activity (Chowdhury et al., 2005). Since ATR is responsible for γ-H2AX formation in PP4-deficient cells, we wanted to exclude the possibility that the increase in γ–H2AX with PP4 silencing was mediated by hyperactive ATR. We therefore immunopurified ATR from PP4-silenced and control cells and assayed its kinase activity as previously described (Chiang and Abraham, 2004; Chowdhury et al., 2005). Importantly, the immunoprecipitation was done under conditions that preserved the ATR-ATRIP complex but possibly other necessary factors were missing (Fig. 5D). Knocking down PP4C, PP4R2 or PP4R3β did not alter ATR kinase activity (Fig. 5D). Moreover, ATR localization was not altered in PP4-deficient cells and ATR was not retained at γ-H2AX foci generated when PP4C was knocked down (Suppl. Fig. 2). Together these results suggest that PP4 does not affect γ-H2AX indirectly via the kinases. However, we cannot exclude transient activation of ATR in PP4C knockdown cells.
We also assessed the activation status of other bona fide ATM and ATR substrates that might indirectly regulate γ-H2AX. In PP4-knocked down cells, phosphorylation of the checkpoint proteins Chk1 (ATR substrate), Chk2 (ATM substrate), and the ATM/ATR-substrate, p53, also was unaltered (Fig. 5D, upper panel). Phospho-Chk1 or phospho-Chk2 were not detected at any time after transfecting HeLa cells with PP4C or R3β siRNAs (Fig.5D, lower panel). This suggests that PP4C does not directly regulate the activation of these factors, and that PP4-deficiency does not constitutively activate ATM or ATR. These results, taken together with the efficient γ-H2AX dephosphorylation by PP4C on mononucleosomes and the subcellular localization of the PP4C-R2-R3β complex on chromatin, suggest that PP4C likely dephosphorylates basal γ-H2AX as a direct substrate on chromatin. However, we cannot exclude the possibility that dephosphorylation of other unknown substrates by PP4C might also indirectly alter the status of γ-H2AX.
PP4C regulation of γ-H2AX only occurs post-DNA replication
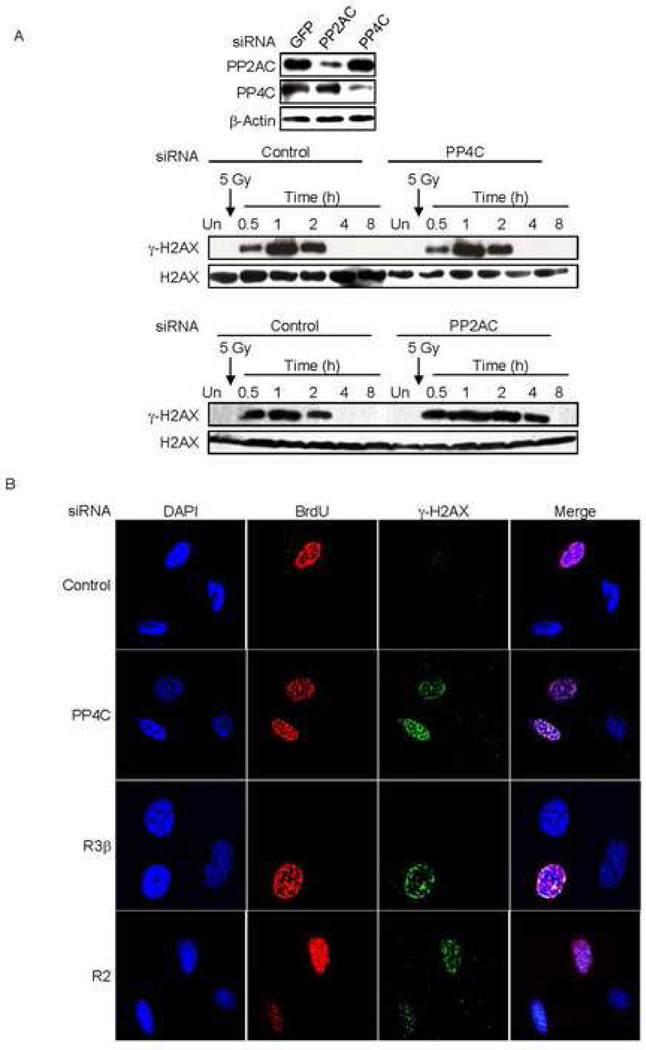
Because PP4C depletion affected basal DNA damage-initiated γ-H2AX, we wanted to determine what kind of endogenous DNA damage might be linked to elevated basal γ-H2AX in PP4-silenced cells. Endogenous DNA damage in a cell may occur during DNA replication or in response to metabolic byproducts and reactive oxygen species (ROS) (Tanaka et al., 2007). In order to determine whether cell division is necessary for basal γ-H2AX formation in PP4-deficient cells we isolated human peripheral blood monocytes and differentiated them into macrophages. These are terminally differentiated cells that are unable to divide. We partially silenced PP4C and PP2AC (Fig. 6A, top panel) and then irradiated the cells to induce DNA damage. PP4C knockdown did not alter γ-H2AX in macrophages either basally or after irradiation (Fig. 6A, middle panel, Suppl. Fig. 4). However, PP2AC silencing led to γ-H2AX persistence in irradiated macrophages (Fig. 6A, lower panel). This result suggested that γ-H2AX formation is driven by cell division and not by spontaneous DNA lesions in PP4-silenced cells. To confirm this and directly establish a link between DNA replication and γ-H2AX in PP4-silenced cells, we silenced PP4C complex proteins in serum-starved 293T cells. Serum starvation synchronizes cells in the G0/G1 phase. We then released the G0/G1 block by adding serum and added BrdU to label cells undergoing DNA replication. We allowed the cells to proceed through the S-phase and then analyzed them for BrdU incorporation and γ-H2AX foci. In control siRNA treated cells, no γ-H2AX foci (other than a few isolated foci occasionally seen in normally dividing cells) formed in the replicating BrdU+ cells. However, upon partially depleting any of the PP4 complex proteins, only cells that had incorporated BrdU had increased γ-H2AX foci (Fig. 6B). These results in nondividing and synchronized cells strongly suggest that γ-H2AX induction in PP4-deficient cells is due to DNA replication-mediated DNA damage.
Fig. 6. γ-H2AX formation in PP4-knocked down cells is dependent on cell division.
A) Knocking down PP4C in non-dividing cells (macrophages) has no effect on γ-H2AX. Human macrophages were transfected with control siRNAs or siRNAs targeting PP2AC or PP4. Immunoblots showed significant knockdown of PP2AC and PP4C (upper panel). Macrophages in which PP4C (middle panel) or PP2AC (lower panel) were knocked down were irradiated (5 Gy) and immunoblots of whole cell extracts at indicated times probed for γ–H2AX or H2AX. Macrophages with lower amounts of PP4C showed no change in γ-H2AX relative to control cells, but γ-H2AX persisted longer in cells with reduced PP2AC.
B) γ-H2AX foci formation in PP4-deficient cells is induced by DNA replication. Serum starved 293T cells were transfected with siRNAs targeting PP4C, PP4R2 or PP4R3β. Transfected cells were released from the G0/G1 block by adding serum and BrdU was added to visualize cells that had undergone DNA replication. After 12 h, cells were washed before fixing and staining for BrdU, γ–H2AX and DAPI. Images were visualized using laser-scanning confocal microscopy. After partial depletion of PP4 complex proteins, only cells that were BrdU positive (had undergone DNA replication) showed γ-H2AX foci.
PP4 dephosphorylation of γ-H2AX is required for efficient DNA damage repair during replication
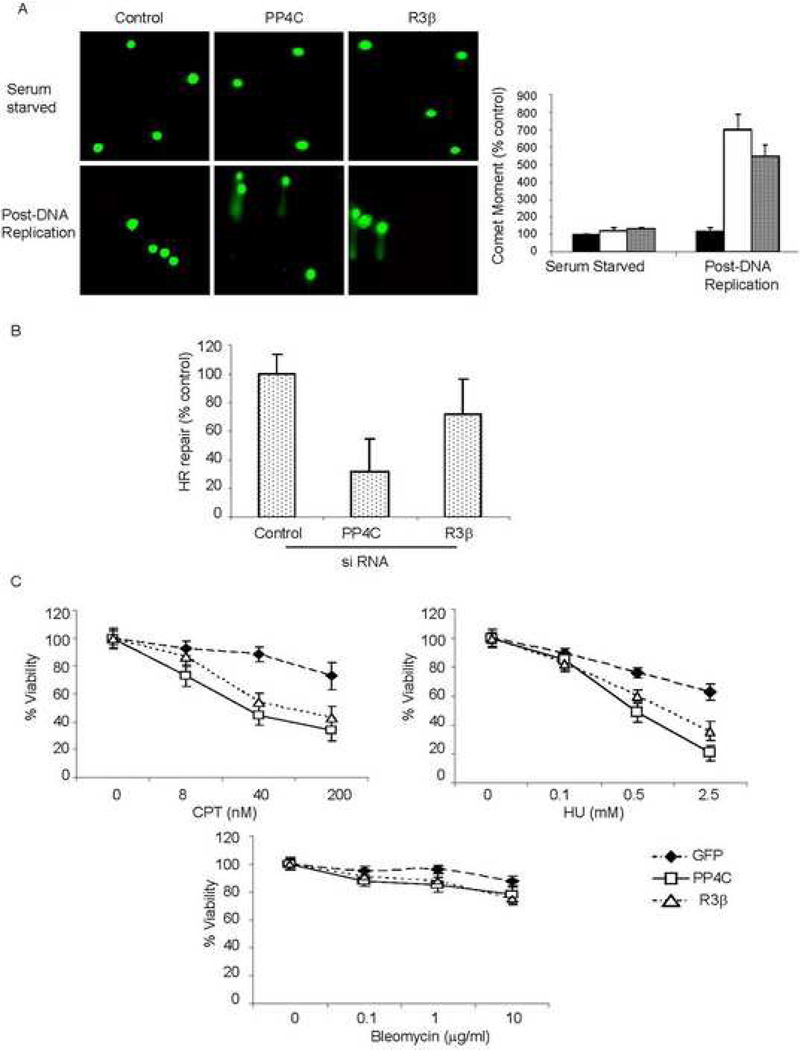
To determine whether PP4 expression and basal γ–H2AX formation affects DNA repair, we measured DSB persistence in HeLa cells, transfected with PP4C, PP4R3β or control siRNAs, using single cell gel electrophoresis (comet assay). The comet moment quantifies the extent of unrepaired DNA damage. In asynchronously dividing cells not subjected to exogenous DNA damage the basal comet moments were significantly increased when the PP4 complex proteins, PP4C and PP4R3β, were knocked down (Suppl. Fig. 5). However, CPT treated control and PP4-silenced cells had comparable amounts of DNA damage (Suppl. Fig. 5). To focus on DNA replication-mediated damage, we repeated these experiments in synchronized cells. We serum starved 293T cells and measured DNA damage by comet assay before and after releasing the G0/G1 block. As expected, DNA damage in PP4-knocked down serum-starved cells was not increased (Fig.7A, upper panel). However, when these cells were released from serum-deprivation and the cells allowed to undergo DNA replication, unresolved DNA damage increased 5–6 fold in cells partially depleted of either PP4C or PP4R3β (Fig. 7A, lower panel). This effect of PP4 on DNA repair post-DNA replication suggests that unresolved γ-H2AX foci impede repair of DNA replication-mediated DSB.
Fig. 7. PP4 is required to repair DNA replication-mediated DNA damage.
A) Knocking down the PP4 complex impairs DNA repair in dividing cells. PP4C, PP4R3β or control siRNA-transfected 293T cells were serum starved for 2 d. Cells were either mainta ined in low serum or released from G0/G1 block by adding serum and analyzed by single cell gel electrophoresis (comet assay) 12 h later. Representative images are shown above. The comet tail moment of 75 cells for each condition was quantified using NIH Image software. The comet moment was normalized to that of control cells (black bar) and expressed as a percentage. DNA damage in the PP4C (white bar, p<0.02) and R3β (dotted bar, p<0.02) deficient cells was significantly increased only after DNA replication resumed.
B) Measurement of HR-mediated repair of an I-SceI-induced DSB. U2OS cells carrying the recombination substrate (DR-GFP) were transfected with PP4C, PP4R3β or control siRNAs. I-SceI expression plasmid was transfected after 24 h and GFP+ cells measured 48 h later. HR repair was significantly impaired in cells transfected with PP4C (p<0.01) or R3β (p< 0.01) siRNA.
C) PP4-silenced cells are hypersensitive to DNA replication inhibitors. Cell viability was analyzed by MTT assay relative to untreated cells. Curves were generated from 3 independent experiments. PP4C or PP4R3β deficient cells were hypersensitive to CPT (p<0.001, left) and HU (p<0.002, right), but not to bleomycin (lower) relative to control cells.
In panels (A–C), the mean ±S.D. of representative experiments are shown.
DSB induced during DNA replication are typically repaired by homologous recombination (HR) (Ben-Yehoyada et al., 2007). To test whether PP4 complex-deficient cells displayed differences in HR repair, we assayed for HR-mediated repair of an I-SceI-induced DSB using a recombination substrate DR-GFP (Weinstock et al., 2006). With this reporter assay, if the DSB is repaired by HR, GFP is expressed (typically in <5% of cells), which is quantifiable by flow cytometry. HR was significantly reduced in cells treated with PP4C and PP4R3β siRNAs (Fig. 7B). Since DNA replication-mediated DSB are repaired by HR, the effect of the PP4-complex on HR is consistent with its role in resolving DNA lesions during replication. Interestingly, knocking down PP4C affects HR more than PP4R3β, suggesting that other PP4C complexes (R3β-independent) may also participate in regulating HR.
To investigate the biological significance of the increased DNA damage in PP4-deficient cells, we examined their sensitivity to different DNA damaging agents (Fig. 7C). PP4 complex deficient cells were hypersensitive to both CPT and HU, but were not particularly sensitive to bleomycin. HU and CPT induce DNA breaks by inhibiting DNA replication, whereas bleomycin induces DNA damage independently of replication by direct Fe-mediated oxidation of DNA sugars. These results suggest that the PP4 complex may be specifically involved in the removal of γ-H2AX caused during DNA replication and impeding this step sensitizes cells to further replication-mediated DNA damage. It should be noted that PP4 depletion reduces cell viability even in undamaged cells (data not shown).
Discussion
The cellular response to DNA damage inflicted by external sources has been well studied for many decades, but it is unclear whether all DSB induced during DNA replication elicit a cellular response comparable to exogenous damaging agents. γ-H2AX clearly is important for repairing replication-induced DNA damage (Furuta et al., 2003; Pommier et al., 2003), but its precise role is not certain. Recent studies suggest that it may not be involved in recruiting repair factors (Bartek and Lukas, 2007), but rather play an important role in stabilizing the repair complex and the broken ends (Celeste et al., 2003b). In this study we identify a trimeric phosphatase complex (PP4-PP4R2-R3β) that specifically eliminates γ-H2AX formed without exogenous DNA damage. We show that PP4 dephosphorylates γ–H2AX by demonstrating in vitro phosphatase activity on nucleosomal γ-H2AX and increased γ–H2AX and persistence of γ–H2AX foci when any of the PP4 complex components is silenced. γ-H2AX foci detected in the absence of PP4 is induced by ATR specifically in the course of DNA replication and associates with DNA repair factors like MRE11 and PCNA. In PP4-deficient cells the activation status of ATR and the ATM/ATR target checkpoint proteins Chk1, Chk2, and p53, were not altered. Although we cannot formally exclude the possibility that PP4 regulates γ-H2AX foci indirectly via some other factor, our data suggest that PP4 directly dephosphorylates γ-H2AX generated during DNA replication. Moreover, unresolved γ-H2AX impedes the repair of DNA damage that occurs during DNA replication, as we previously also found for repair of exogenous DNA damage.
In yeast, mutation of the genes encoding for a phosphatase complex composed of PP4C-R2-R3β homologues (Pph3-Yb1046w-Psy2) also led to a basal increase in γ-H2AX, but the cause and consequence of this increase were not clear. Here we address those key issues in the context of PP4 deficiency in mammalian cells. We demonstrate that the cause of the basal increase in γ-H2AX in PP4-deficient cells is DNA replication-mediated damage. PP4-deficient cells are specifically hypersensitive to DNA replication inhibitors. This further supports the idea that PP4 is playing a role in the repair of DNA lesions induced by replication. The only phenotypic effect of Pph3 deficiency in yeast was the phospho-Rad53 mediated prolongation of the DNA damage-induced cell cycle checkpoint (Keogh et al., 2006). In mammalian cells, there was no effect of PP4 deficiency on the Rad53 homologue, Chk2. The two mammalian homologs of Psy2 are R3β and R3α (Gingras et al., 2005). We failed to detect any interaction between Chk2 and R3β (data not shown). It is possible that some other PP4 complex, perhaps containing R3α, might interact with Chk2. However, a PP2A complex has been shown to dephosphorylate Chk2 in mammalian cells (Dozier et al., 2004). The yeast study with Pph3 deletion mutants and our work with PP4 silenced mammalian cells share many important features; in both systems a highly homologous complex is required to resolve an increase in basal γ-H2AX levels. Whether or not the major role of yeast HTP-C is to resolve DNA replication-mediated damage will require further study.
In PP4-deficient cells we found a basal increase in DNA damage even in the absence of an exogenous damaging agent. However, despite unresolved replication-mediated DNA damage, PP4-silenced cells did not arrest during S phase or at the G2/M checkpoint (data not shown) and checkpoint proteins were not activated. Why then does this DNA damage not activate cell cycle checkpoint proteins, like Chk1 and Chk2? One possibility is that checkpoint activation might require a higher degree of DNA damage than what occurs during ordinary replication. Another explanation could be via ‘checkpoint adaptation’, originally defined in S. cerevisiae as the ability to divide despite the presence of damaged DNA (Toczyski et al., 1997). The molecular mechanism or consequence of checkpoint adaptation in human cells remains unknown. The effect of PP4-deficiency on checkpoint adaptation needs to be further investigated.
How does persistence of γ-H2AX foci in PP4-deficient cells lead to an increase in basal DNA damage? There are many possible explanations. γ-H2AX recruits and/or stabilizes a DNA repair complex at the site of DSB. The orchestration of DNA repair might involve a series of regulated steps, one of which is blocked by γ-H2AX. Some critical repair factor(s) might either not be recruited or activated until γ-H2AX is resolved. Another possibility is that some repair factors present in limiting amounts and recruited by γ-H2AX might be unnecessarily sequestered by ectopic γ-H2AX in PP4-deficient cells, consequently preventing it from functioning at other DSBs. The association of MRE11 with γ–H2AX foci in PP4C-silenced cells supports this idea. MRE11 is necessary to recognize a DSB, and sequestering MRE11 and keeping it from other DSB sites may affect the efficacy of DSB repair (Assenmacher and Hopfner, 2004). Alternatively, some of the factors recruited include nucleases like RAD50, which have a limited and transient role at a DSB site (Assenmacher and Hopfner, 2004); sequestering such factors at the DSB could increase DNA damage.
We previously showed that another phosphatase PP2A directly dephosphorylates γ-H2AX formed by exogenous DNA damaging agents (Chowdhury et al., 2005). Why does the cell need multiple phosphatases to eliminate γ-H2AX? Our data suggest that distinct phosphatases differentially regulate γ-H2AX that originates from different stressors and/or from different degrees of DNA damage. For relatively low levels of DNA damage that occur during DNA replication, the PP4 complex is both necessary and sufficient to remove γ-H2AX, but higher levels of damage during DNA replication induced by CPT or hydroxyurea (HU), might require both PP2A and PP4 to eliminate γ-H2AX. The effect of PP4-deficiency on the HR-mediated repair of the I-SceI break (Fig. 7B) suggests that PP4 may have a role in the repair of any DSB, induced exogenously or endogenously, that is repaired by HR. Although the kinases, ATM and ATR have distinct roles in γ-H2AX formation, there is also considerable functional overlap in their activity (Abraham, 2004). We speculate that the phosphatases PP2A and PP4 might function in an analogous manner. Future studies with PP4 and PP2A will further elucidate their potentially overlapping roles in eliminating γ-H2AX and enable us to understand the need for multiple phosphatases in regulating γ-H2AX and DNA repair.
Experimental Procedures (additional experimental details in Supplemental Online Material)
Immunofluorescence, Chromatin Fractionation and Co-Immunoprecipitation
Immunofluorescence, chromatin fractionation and co-immunoprecipitation were performed essentially as described (Chowdhury et al.,2005; (Xu and Stern, 2003) (see Supplemental Data for details).
In vitro enzymatic analyses
Mononucleosomes containing human γ-H2AX were prepared as described previously (Chowdhury et al., 2005). PP4 and PP2A enzymes were expressed in vitro using PROTEINscript II T7 Kit (Ambion), and immunopurified using anti-HA beads (Sigma). Phosphatase reactions with γ-H2AX mononucleosomes were performed as described (Zabrocki et al., 2002) in 20 mM Tris-HCl, pH 7.4, 50 mM NaCl, 0.2 mM EDTA, 0.2% β-mercaptoethanol for 30 minutes at 30°C. Reaction mixtures were resolved by 12% SDS-PAGE, and relative phosphatase activity determined by loss of γ–H2AX immunoreactivity.
Silencing of PP4 subunits and PP2A
PP4 subunits and PP2A were silenced using standard transfection procedures, the siRNA sequences and details are provided in the Supplemental Data.
ATR activity assay
This assay was done as described (Chiang and Abraham, 2004). For details see Supplemental Data.
Single Cell Gel Electrophoresis (comet) assay
Single cell comet assays were performed as per manufacturer’s instructions (Trevigen). For details see the Supplemental Data.
HR Assay
HR assay was as described (Weinstock et al., 2006). Briefly, 2×105 U2OS cells with a single, stably integrated copy of the transgenic reporter DR-GFP plated overnight in 24-well plates were first transfected with PP4C, PP4R3β or control siRNAs and 24 h later transfected with 0.8 µg of I-SceI expression plasmid (pCBA Sce) using Lipofectamine 2000. Two days later, GFP positive cells were assayed by FACScan.
Supplementary Material
Acknowledgment
This work was supported by NIH grants AI45587 (J.L.) and CA88873 (G.P.P.), a Leukemia & Lymphoma Society Fellowship (D.C.), startup funds from CNU, NSFC (30570371 and 90608014 and 30711120570), the Program for New Century Excellent Talents in University (NCET-06-0187), Beijing Natural Science Foundation Program and Scientific Research Key Program of Beijing Municipal Commission of Education (KZ200810028014), and Funding Project for Academic Human Resources Development in Institutions of Higher Learning Under the Jurisdiction of Beijing Municipality (PHR(IHLB), 075313607) (X.X.). We thank Eric W. McIntush (Bethyl Laboratories) for protein phosphatase antibodies and Rohit Panchakshari for help with Figure 1A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham RT. PI 3-kinase related kinases: 'big' players in stress-induced signaling pathways. DNA Repair (Amst) 2004;3:883–887. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Assenmacher N, Hopfner KP. MRE11/RAD50/NBS1: complex activities. Chromosoma. 2004;113:157–166. doi: 10.1007/s00412-004-0306-4. [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- Ben-Yehoyada M, Gautier J, Dupre A. The DNA damage response during an unperturbed S-phase. DNA Repair (Amst) 2007;6:914–922. doi: 10.1016/j.dnarep.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Carnegie GK, Sleeman JE, Morrice N, Hastie CJ, Peggie MW, Philp A, Lamond AI, Cohen PT. Protein phosphatase 4 interacts with the Survival of Motor Neurons complex and enhances the temporal localisation of snRNPs. J Cell Sci. 2003;116:1905–1913. doi: 10.1242/jcs.00409. [DOI] [PubMed] [Google Scholar]
- Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003a;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003b;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- Chiang GG, Abraham RT. Determination of the catalytic activities of mTOR and other members of the phosphoinositide-3-kinase-related kinase family. Methods Mol Biol. 2004;281:125–141. doi: 10.1385/1-59259-811-0:125. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Keogh MC, Ishii H, Peterson CL, Buratowski S, Lieberman J. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Cohen PT, Philp A, Vazquez-Martin C. Protein phosphatase 4--from obscurity to vital functions. FEBS Lett. 2005;579:3278–3286. doi: 10.1016/j.febslet.2005.04.070. [DOI] [PubMed] [Google Scholar]
- Douglas P, Moorhead GB, Ye R, Lees-Miller SP. Protein phosphatases regulate DNA-dependent protein kinase activity. J Biol Chem. 2001;276:18992–18998. doi: 10.1074/jbc.M011703200. [DOI] [PubMed] [Google Scholar]
- Dozier C, Bonyadi M, Baricault L, Tonasso L, Darbon JM. Regulation of Chk2 phosphorylation by interaction with protein phosphatase 2A via its B' regulatory subunit. Biol Cell. 2004;96:509–517. doi: 10.1016/j.biolcel.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair (Amst) 2004;3:959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Franco S, Gostissa M, Zha S, Lombard DB, Murphy MM, Zarrin AA, Yan C, Tepsuporn S, Morales JC, Adams MM, et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol Cell. 2006;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Furuta T, Takemura H, Liao ZY, Aune GJ, Redon C, Sedelnikova OA, Pilch DR, Rogakou EP, Celeste A, Chen HT, et al. Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J Biol Chem. 2003;278:20303–20312. doi: 10.1074/jbc.M300198200. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Caballero M, Zarske M, Sanchez A, Hazbun TR, Fields S, Sonenberg N, Hafen E, Raught B, Aebersold R. A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Mol Cell Proteomics. 2005;4:1725–1740. doi: 10.1074/mcp.M500231-MCP200. [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Jonnalagadda JC, Douglas P, Young D, Ye R, Moorhead GB, Lees-Miller SP, Khanna KK. Autophosphorylation of ataxia-telangiectasia mutated is regulated by protein phosphatase 2A. Embo J. 2004;23:4451–4461. doi: 10.1038/sj.emboj.7600455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- Hastie CJ, Vazquez-Martin C, Philp A, Stark MJ, Cohen PT. The Saccharomyces cerevisiae orthologue of the human protein phosphatase 4 core regulatory subunit R2 confers resistance to the anticancer drug cisplatin. Febs J. 2006;273:3322–3334. doi: 10.1111/j.1742-4658.2006.05336.x. [DOI] [PubMed] [Google Scholar]
- Honkanen RE, Golden T. Regulators of serine/threonine protein phosphatases at the dawn of a clinical era? Curr Med Chem. 2002;9:2055–2075. doi: 10.2174/0929867023368836. [DOI] [PubMed] [Google Scholar]
- Hu MC, Tang-Oxley Q, Qiu WR, Wang YP, Mihindukulasuriya KA, Afshar R, Tan TH. Protein phosphatase X interacts with c-Rel and stimulates c-Rel/nuclear factor kappaB activity. J Biol Chem. 1998;273:33561–33565. doi: 10.1074/jbc.273.50.33561. [DOI] [PubMed] [Google Scholar]
- Ichijima Y, Sakasai R, Okita N, Asahina K, Mizutani S, Teraoka H. Phosphorylation of histone H2AX at M phase in human cells without DNA damage response. Biochem Biophys Res Commun. 2005;336:807–812. doi: 10.1016/j.bbrc.2005.08.164. [DOI] [PubMed] [Google Scholar]
- Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V, Goris J, Van Hoof C. PP2A: the expected tumor suppressor. Curr Opin Genet Dev. 2005;15:34–41. doi: 10.1016/j.gde.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kim JA, Downey M, Fillingham J, Chowdhury D, Harrison JC, Onishi M, Datta N, Galicia S, Emili A, et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- McManus KJ, Hendzel MJ. ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Mol Biol Cell. 2005;16:5013–5025. doi: 10.1091/mbc.E05-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihindukulasuriya KA, Zhou G, Qin J, Tan TH. Protein phosphatase 4 interacts with and down-regulates insulin receptor substrate 4 following tumor necrosis factor-alpha stimulation. J Biol Chem. 2004;279:46588–46594. doi: 10.1074/jbc.M408067200. [DOI] [PubMed] [Google Scholar]
- Pandey AV, Mellon SH, Miller WL. Protein phosphatase 2A and phosphoprotein SET regulate androgen production by P450c17. J Biol Chem. 2003;278:2837–2844. doi: 10.1074/jbc.M209527200. [DOI] [PubMed] [Google Scholar]
- Petrini JH, Stracker TH. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol. 2003;13:458–462. doi: 10.1016/s0962-8924(03)00170-3. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Redon C, Rao VA, Seiler JA, Sordet O, Takemura H, Antony S, Meng L, Liao Z, Kohlhagen G, et al. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat Res. 2003;532:173–203. doi: 10.1016/j.mrfmmm.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, Eisenreich TR, Chen J, Dickins RA, Lowe SW, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM, Abraham RT. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- Sarkaria JN, Tibbetts RS, Busby EC, Kennedy AP, Hill DE, Abraham RT. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 1998;58:4375–4382. [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Shui JW, Hu MC, Tan TH. Conditional knockout mice reveal an essential role of protein phosphatase 4 in thymocyte development and pre-T-cell receptor signaling. Mol Cell Biol. 2007;27:79–91. doi: 10.1128/MCB.00799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson B, Brautigan DL. Protein phosphatase PP6 N terminal domain restricts G1 to S phase progression in human cancer cells. Cell Cycle. 2007;6:1386–1392. doi: 10.4161/cc.6.11.4276. [DOI] [PubMed] [Google Scholar]
- Stucki M, Jackson SP. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst) 2006;5:534–543. doi: 10.1016/j.dnarep.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Kajstura M, Halicka HD, Traganos F, Darzynkiewicz Z. Constitutive histone H2AX phosphorylation and ATM activation are strongly amplified during mitogenic stimulation of lymphocytes. Cell Prolif. 2007;40:1–13. doi: 10.1111/j.1365-2184.2007.00417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toczyski DP, Galgoczy DJ, Hartwell LH. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell. 1997;90:1097–1106. doi: 10.1016/s0092-8674(00)80375-x. [DOI] [PubMed] [Google Scholar]
- Toyo-oka K, Mori D, Yano Y, Shiota M, Iwao H, Goto H, Inagaki M, Hiraiwa N, Muramatsu M, Wynshaw-Boris A, et al. Protein phosphatase 4 catalytic subunit regulates Cdk1 activity and microtubule organization via NDEL1 dephosphorylation. J Cell Biol. 2008;180:1133–1147. doi: 10.1083/jcb.200705148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006;409:524–540. doi: 10.1016/S0076-6879(05)09031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Stern DF. NFBD1/KIAA0170 is a chromatin-associated protein involved in DNA damage signaling pathways. J Biol Chem. 2003;278:8795–8803. doi: 10.1074/jbc.M211392200. [DOI] [PubMed] [Google Scholar]
- Yeh PY, Yeh KH, Chuang SE, Song YC, Cheng AL. Suppression of MEK/ERK signaling pathway enhances cisplatin-induced NF-kappaB activation by protein phosphatase 4-mediated NF-kappaB p65 Thr dephosphorylation. J Biol Chem. 2004;279:26143–26148. doi: 10.1074/jbc.M402362200. [DOI] [PubMed] [Google Scholar]
- Zabrocki P, Van Hoof C, Goris J, Thevelein JM, Winderickx J, Wera S. Protein phosphatase 2A on track for nutrient-induced signalling in yeast. Mol Microbiol. 2002;43:835–842. doi: 10.1046/j.1365-2958.2002.02786.x. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ozawa Y, Lee H, Wen YD, Tan TH, Wadzinski BE, Seto E. Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes Dev. 2005;19:827–839. doi: 10.1101/gad.1286005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Mihindukulasuriya KA, MacCorkle-Chosnek RA, Van Hooser A, Hu MC, Brinkley BR, Tan TH. Protein phosphatase 4 is involved in tumor necrosis factor-alpha-induced activation of c-Jun N-terminal kinase. J Biol Chem. 2002;277:6391–6398. doi: 10.1074/jbc.M107014200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.