The elucidation of protein–protein interactions is fundamental to understanding biological processes. Many of these interactions are regulated by post-translational modifications (PTMs), such as phosphorylation, that are both transient and substoichiometric in nature, characteristics that makes PTMs difficult to detect by standard proteomic strategies.1 Cross-linking has long been touted as a solution to this problem since the interacting partners are covalently captured.2 Standard strategies involve the addition of exogenous cross-linkers to protein mixtures,3,4 an intrinsically unbiased approach that does not require any guiding knowledge of the structure of the system under investigation. However, such cross-linking reactions typically afford complex mixtures, the characterization of which is far from trivial.4,5 One solution to this problem is to directly anchor a cross-linker to a protein of interest, thereby reducing the complexity of the cross-linked mixture. Several approaches have been taken, including conjugation of hetero-bifunctional cross-linkers to engineered cysteines6,7 and co-option of the protein biosynthesis machinery to incorporate unnatural amino acid cross-linkers either nonspecifically8,9 or site-specifically10 into expressed proteins. Although useful, these approaches do not provide direct control over any PTMs that may regulate the bait protein, and so conceivably, not all interaction partners will be identified. In this study, we set out to address this issue by exploring whether an amino acid cross-linker and a PTM could be site-specifically introduced into the same protein, allowing the former to covalently capture an interaction dependent upon the latter.
As a test example, we chose the MH2 domain of the Smad2 signaling protein, which is known to form stable homo-trimers in response to receptor-mediated phosphorylation of serine residues 465 and 467.11 Previously, we have shown that mono-phosphorylation at Ser465 affords a destabilized homo-trimer that can be disrupted by addition of the Smad binding domain of the Smad anchor for receptor activation protein (SARA-SBD, residues 665–721) that binds exclusively to the monomeric state.12 We reasoned that this switchable oligomerization system would provide a good test of whether cross-linking could selectively capture a PTM-dependent protein–protein interaction. As shown in Figure 1, our design called for the preparation of a MH2 analogue containing both a phospho-serine (pSer) at position 465 and a photoactive cross-linker in place of the normal activating pSer at position 467. It was hypothesized that placing a cross-linker at this site would lead to productive inter-subunit cross-links based on the position of pSer467 in the native homo-trimer interface.11 This design does, however, place steric constraints on the cross-linker; standard aromatic probes (benzophenone, phenyl azides, etc.) are unlikely to fit in the binding pocket. Consequently, we turned our attention to a recently introduced diazirine-containing probe (photo-Met, 5),9 which due to its small size is less likely to disrupt the protein–protein interface.
Figure 1.
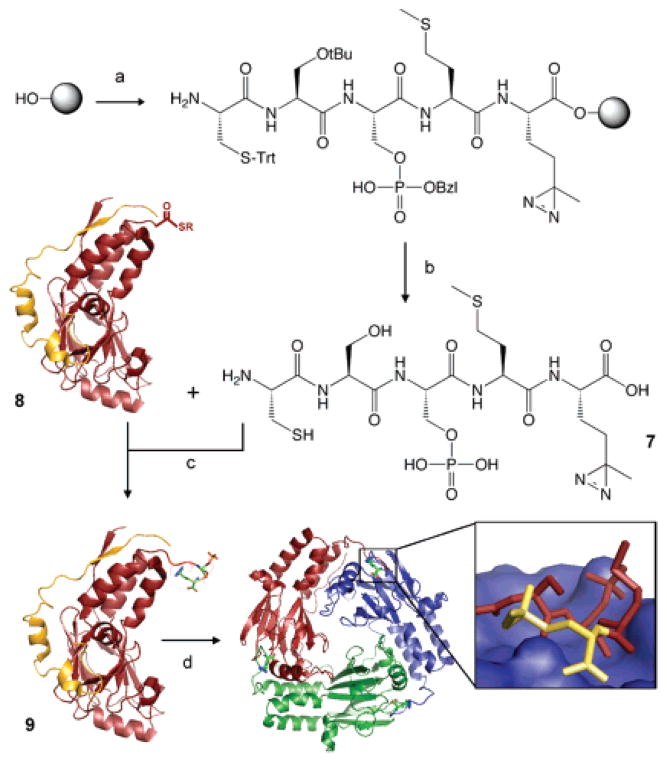
Semi-synthesis of Smad2-MH2-CSpSM-photo-Met (9). (a) Fmoc-SPPS. (b) Cleavage. (c) EPL reaction with peptide 7 and HA-MH2 thioester (red, PDB code 1DEV) bound to SARA-SBD (in yellow) 8. (d) Trimerization of ligation product 9 and release of SARA-SBD. Structural model is based on the crystal structure of the MH2 homo-trimer (PDB code 1KHX). Inset: hypothetical protein–protein interface with the tail of one MH2 subunit shown in sticks (with the diazirine moiety in yellow) and the surface of the adjacent subunit rendered in blue.
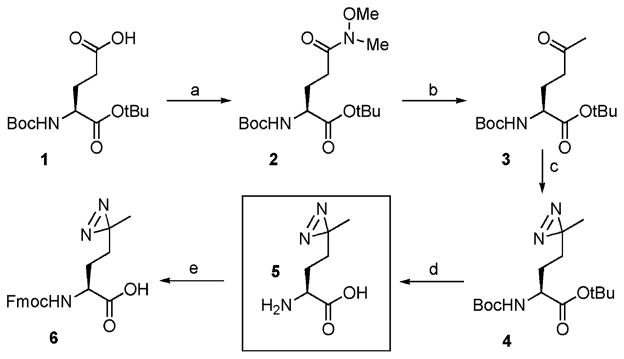
The previously described synthesis of photo-Met required an enzymatic resolution step and so proceeded with low overall yield.9 To avoid this problem, we developed a four-step stereoselective route based on the use of L-glutamic acid as a starting material (Scheme 1). Briefly, Boc-L-glutamic acid α-tert-butyl ester (1) was converted into the corresponding γ-Weinreb amide 2, which was then transformed into ketone 3 by treatment with methyl Grignard.13 The photoactive diazirine moiety was then installed by reaction of 3 with liquid ammonia followed by hydroxylamine-O-sulfonic acid to give the diazo cyclic intermediate, with subsequent oxidation to give 4. Deprotection and ensuing reprotection with Fmoc-OSu gave compounds 5 and 6, respectively. The overall yield of compound 5 was 32%, approximately six times higher than the previously reported synthesis.9 Protected amino acid 6 was then used in the assembly of phospho-peptide 7 using Fmoc-based SPPS chain assembly protocols.13 Following cleavage from the support, we purified the desired peptide 7 by RP-HPLC and confirmed its identity by MS. Note, a minor side product (~7% of the crude) was identified that, based on MS and UV analysis, was consistent with loss of N2 from the diazirine and addition of a benzyl group.
Scheme 1.
a Conditions: (a) N-methylmorpholine, isobutyl chloroformate, N,O-dimethylhydroxylamine, CH2Cl2, 2 h, 99%; (b) Me–MgBr, toluene, 3 h, 67%; (c) (i) NH3, hydroxylamine-O-sulfonic acid, (ii) TEA, I2, MeOH, 49%; (d) 4 M HCl, THF, 20 h, 99%; (e) NaHCO3, Fmoc-OSu, 1,4-dioxane, 20 h, 97%.
Expressed protein ligation (EPL) was then used to synthesize the desired semi-synthetic protein 9 through the reaction of peptide 7 and protein thioester 8. The latter corresponds to a 1:1 complex between the Smad2 MH2 domain (residues 241–462 fused to a N-terminal HA tag) and SARA-SBD and was prepared using an established intein-fusion strategy.14 The ligation reaction was performed in the dark and was monitored by SDS-PAGE,13 which indicated almost complete conversion of 8 into 9 after 40 h, with no detectable decomposition of the diazirine moiety as indicated by MS or UV. Size exclusion chromatography was used to purify the ligation product from residual starting materials.13 Notably, ligation product 9 eluted much earlier than residual unreacted 8 and had an elution volume similar to that of the bis-phosphorylated MH2 domain, which is known to run as a homo-trimer under the conditions used.12 Moreover, RP-HPLC analysis of purified 9 indicated that it had dissociated from SARA-SBD, which is again diagnostic of Smad homo-oligomerization.15
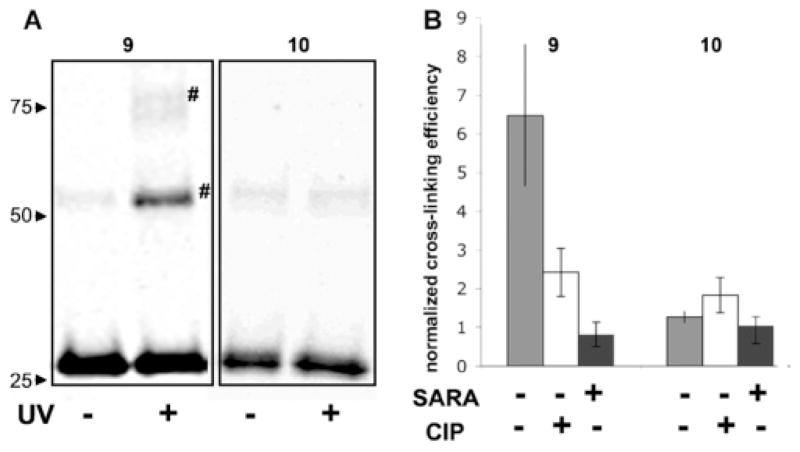
Next, we tested the ability of the diazirine cross-linker to capture Smad oligomerization. In the first experiment, we compared diazirine-containing protein 9 with the control protein, HA-MH2-CSpSMpS (10), which lacks the cross-linker. The proteins were irradiated with a He–Cd laser at 325 nm and incubated on ice for 20 min. The samples were then resolved by SDS-PAGE and analyzed by western blotting using an anti-HA antibody (Figure 2A). In the case of protein 9, irradiation led to the generation of two major cross-linked species whose apparent molecular weight was consistent with dimeric and trimeric versions of Smad2-MH2. These high molecular weight species were barely detectable in the non-irradiated sample of 9 and in the control protein 10, either before or after UV irradiation.16
Figure 2.
Cross-linking experiments. (A) Anti-HA western blot of semi-synthetic proteins 9 and 10 with and without UV irradiation, # indicates cross-links. (B) Effect of CIP treatment or addition of excess of SARA-SBD on the cross-linking efficiency of proteins 9 and 10. Shown is the normalized cross-linking efficiency (change of the ratio between oligomer and monomer bands before and after UV irradiation, ±SD, n = 3).
In order to test that a native interaction had been covalently captured by the reaction, we probed the dependence of the cross-linked species on phosphorylation and trimer formation. Accordingly, cross-linking reactions were performed in samples pretreated with calf intestinal phosphatase (CIP) or in the presence of a ~20-fold molar excess of SARA-SBD, a concentration sufficient to disrupt homo-trimers of singly phosphorylated MH2.12 As shown in Figure 2B, incubation of 9 with CIP or SARA-SBD substantially reduced the amounts of cross-linked proteins. Thus, the high molecular weight species generated upon irradiation of 9 are dependent upon phosphorylation and protein oligomerization, and by extension, the cross-links are located at the native protein–protein interface.
In conclusion, we have developed an efficient synthesis of the photo-Met cross-linker and demonstrated that this amino acid is compatible with SPPS and EPL. To our knowledge, this represents the first time this or, indeed, any cross-linker has been introduced into a protein by EPL. In the present example, we exploited the unique power of semi-synthesis to incorporate multiple different probes into the same protein by preparing a version of Smad2-MH2 containing both photo-Met and an activating phospho-serine. Through judicious placement of the cross-linker, we have been able to covalently capture a transient MH2–MH2 interaction that is dependent upon the PTM in the protein. This work has important implications for the area of comparative proteomics where the role of PTMs on regulating protein associations is often a key focus.1 Given the generality of the EPL approach to protein semi-synthesis,17 it will be possible to use differentially modified bait proteins in comparative cross-linking studies.
Supplementary Material
Acknowledgments
We thank Michael Hahn for helpful suggestions. This work was supported by NIH grants GM55843 and EB001991 (to T.W.M.) and by fellowships from the Spanish Ministry of Science and Education (M.V.P.) and American Cancer Society (M.R.P.).
Footnotes
Supporting Information Available: Full experimental procedures and spectroscopic data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Kluger R, Alagic A. Bioorg Chem. 2004;32:451–72. doi: 10.1016/j.bioorg.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Trester-Zedlitz M, Kamada K, Burley SK, Fenyo D, Chait BT, Muir TW. J Am Chem Soc. 2003;125:2416–25. doi: 10.1021/ja026917a. [DOI] [PubMed] [Google Scholar]
- 4.Liu B, Burdine L, Kodadek T. J Am Chem Soc. 2006;128:15228–35. doi: 10.1021/ja065794h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinz A. Mass Spectrom Rev. 2006;25:663–82. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 6.Giron-Monzon L, Manelyte L, Ahrends R, Kirsch D, Spengler B, Friedhoff P. J Biol Chem. 2004;279:49338–45. doi: 10.1074/jbc.M409307200. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Ebright YW, Ebright RH. Science. 1994;265:90–2. doi: 10.1126/science.8016656. [DOI] [PubMed] [Google Scholar]
- 8.Kurzchalia TV, Wiedmann M, Girshovich AS, Bochkareva ES, Bielka H, Rapoport TA. Nature. 1986;320:634–6. doi: 10.1038/320634a0. [DOI] [PubMed] [Google Scholar]
- 9.Suchanek M, Radzikowska A, Thiele C. Nat Methods. 2005;2:261–7. doi: 10.1038/nmeth752. [DOI] [PubMed] [Google Scholar]
- 10.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Proc Natl Acad Sci USA. 2002;99:11020–4. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu JW, Hu M, Chai J, Seoane J, Huse M, Li C, Rigotti DJ, Kyin S, Muir TW, Fairman R, Massague J, Shi Y. Mol Cell. 2001;8:1277–89. doi: 10.1016/s1097-2765(01)00421-x. [DOI] [PubMed] [Google Scholar]
- 12.Ottesen JJ, Huse M, Sekedat MD, Muir TW. Biochemistry. 2004;43:5698–706. doi: 10.1021/bi0498407. [DOI] [PubMed] [Google Scholar]
- 13.See Supporting Information.
- 14.Hahn ME, Muir TW. Angew Chem. 2004;43:5800–3. doi: 10.1002/anie.200461141. [DOI] [PubMed] [Google Scholar]
- 15.Pellois JP, Hahn ME, Muir TW. J Am Chem Soc. 2004;126:7170–1. doi: 10.1021/ja0499142. [DOI] [PubMed] [Google Scholar]
- 16.The trace amounts of high MW species in the control lanes are attributed to non-covalent oligomers.
- 17.Muralidharan V, Muir TW. Nat Methods. 2006;3:429–38. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.