Abstract
We report the crystal structures of the ligand-binding domain (LBD) of a rat inositol 1,4,5-trisphosphate (InsP3) receptor (InsP3R) in its apo and InsP3-bound conformations. Comparison of these two conformations reveals that LBD's first β-trefoil fold (β-TF1) and armadillo repeat fold (ARF) move together as a unit relative to its second β-trefoil fold (β-TF2). Whereas apo-LBD may spontaneously transition between gating conformations, InsP3 binding shifts this equilibrium towards the active state.
Binding of InsP3 to InsP3R in the endoplasmic reticulum (ER) membrane opens the Ca2+-permeant pore in the receptor protein1-4. The resulting Ca2+ efflux from the ER elevates the cytoplasmic free Ca2+ concentration, a signal that triggers numerous important cellular processes1, 3. The LBD of InsP3R, which comprises the ∼600 amino-terminal residues5, 6, is coupled to and thereby exerts allosteric control over the trans-membrane pore domain. Even when produced as an isolated construct, LBD binds InsP3 with affinity and selectivity comparable to those of the whole InsP3R protein7-9. The LBD sequence encodes the two β-trefoil folds, β-TF1 and β-TF2, followed by an ARF10, 11. While the structural significance of the existence of two β-TF lobes in LBD has remained unknown, a construct comprising only β-TF2 and ARF (termed InsP3-binding core) binds InsP3 with even higher affinity than the entire LBD or the whole protein12. Hence, β-TF1 is viewed as a suppressor of InsP3 binding. Additional studies suggest that β-TF1 not only helps stabilize LBD but also couples its conformational changes to the gate of the ion pore9, 13, 14. The crystal structures of β-TF1 alone and of β-TF2 plus ARF bound with InsP3 have been solved separately10, 11. The latter structure reveals how InsP3 is coordinated by various side-chains in the binding sites at the ARF - β-TF2 interface, and mutation of these side-chains weakens InsP3 binding. Several low-resolution (24 – 40 Å) structures of InsP3R have also been obtained by cryo-electron microscopy15-18. Despite this remarkable progress, the fundamental question of how InsP3 binding induces the gating conformational changes of LBD remains.
To help address this question, we solved two LBD crystal structures of rat type 1 InsP3R (3.8 Å resolution; Fig. 1). Although the LBD crystal we used was grown in the presence of InsP3, the two molecules in each asymmetric unit are in distinct conformations: one with and one without substantial InsP3 occupancy. Diffraction data and structure refinement statistics are summarized in Supplementary Table 1, whereas LBD sequence and secondary structure assignments are shown in Supplementary Fig. 1. To illustrate the quality of the electron density map, we show a section of the 2Fo-Fc map and the corresponding structure in Fig. 2a.
Figure 1.
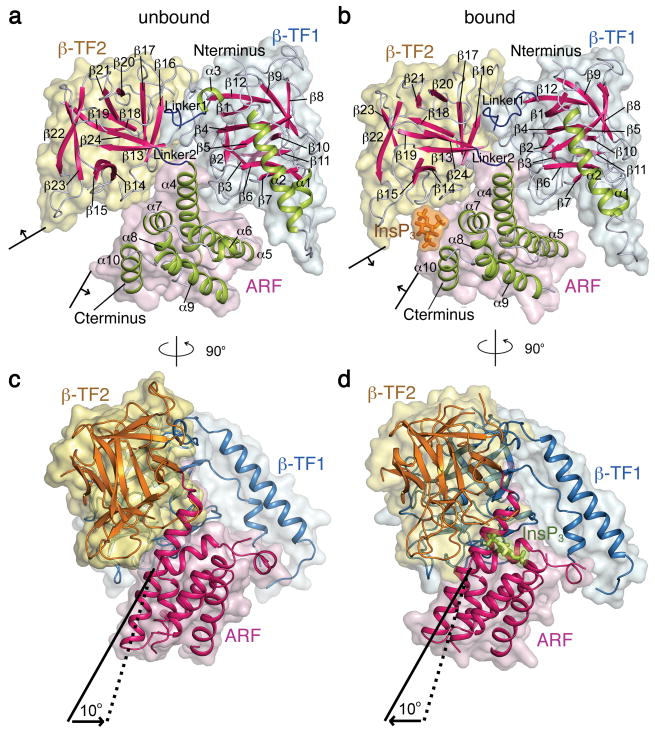
Structures of LBD. (a, b) Surface and ribbon representations of LBD structures without (a) and with (b) InsP3 bound, with β-TF2 shown in the same orientation. Surfaces of β-TF1, β-TF2, ARF, and InsP3 are colored light blue, yellow, pink, and orange, respectively, whereas α helices, β strands, linkers between lobes, and the InsP3 molecule are colored lime, magenta, blue and orange, respectively. (c, d) View of LBD structures rotated 90° from a and b where surfaces of the three lobes are colored as in a and b, and ribbon representations of β-TF1, β-TF2, and ARF are colored blue, orange and magenta, respectively. InsP3 is shown in lime sticks. Solid and dotted lines indicate the axes of helix α4 in the bound and unbound states, respectively.
Figure 2.
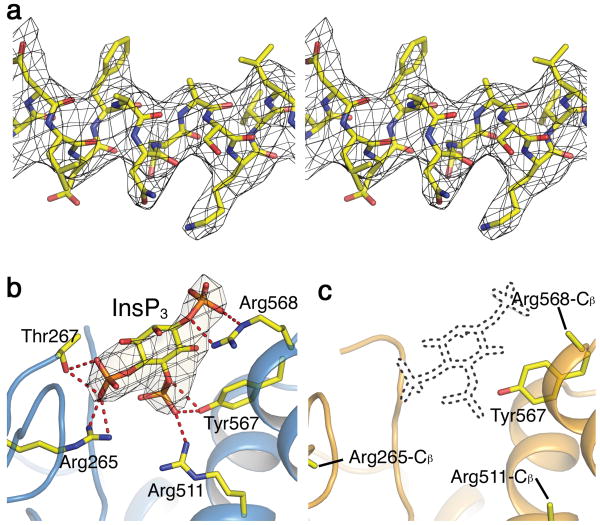
Structures and electron density maps of regions within LDB. (a) Stereo view of a section (Asp442-Leu453) of helix α4 in the InsP3-bound structure, superimposed on the corresponding 2Fo-Fc map contoured at 1 σ. (b, c) Structures of the InsP3-binding site in LBD bound (b) or unbound (c) with InsP3, superimposed on the respective Fo-Fc InsP3-omit maps contoured at 4 σ. The InsP3 molecule in panel b is shown as a stick model; the corresponding site in c is delineated by dots. The InsP3-interacting side-chains, shown as sticks in b, are mostly disordered in c. The red dotted lines in b indicate potential hydrogen bonds.
The backbone structure of a given lobe of LBD in one state is largely superimposable upon that in the other state, or upon the previously determined structures of β-TF1 (PDB 1XZZ)11 and of β-TF2 plus ARF (PDB 1N4K)10 (Supplementary Fig. 2). In both LBD structures the C-terminal region 581-602 of ARF is disordered. The three lobes in LBD: β-TF1, β-TF2 and ARF, are arranged in a triangle (Fig. 1a, b). A similar triangular architecture is adopted by its counterpart in ryanodine receptors (RyR), the other type of intracellular Ca2+ release channel19. Given that, unlike its RyR counterpart, InsP3R-LBD evolved to bind InsP3, it is unsurprising that their structures differ in the lobes' relative orientation and within individual lobes (Supplementary Fig. 3).
As expected, InsP3 binds between ARF and β-TF210. To illustrate the difference in InsP3 occupancy between the two InsP3R1-LBD conformations, we show the relevant regions of an (Fo-Fc) InsP3-omit map contoured at 4 σ and superimposed on the corresponding structures. The map was calculated using a model where neither molecule contains InsP3 (Fig. 2b, c). For one molecule in the asymmetric unit (Fig. 2b), there is clear density at the site where InsP3 is expected to bind on the basis of the previously solved structure of ARF and β-TF2 (PDB 1N4K) whereas for the other there is no visible density (Fig. 2c). Thus, the former molecule is primarily in an InsP3-bound conformation and the latter in an unbound conformation. As expected, InsP3-coordinating side-chains are disordered in the unbound structure.
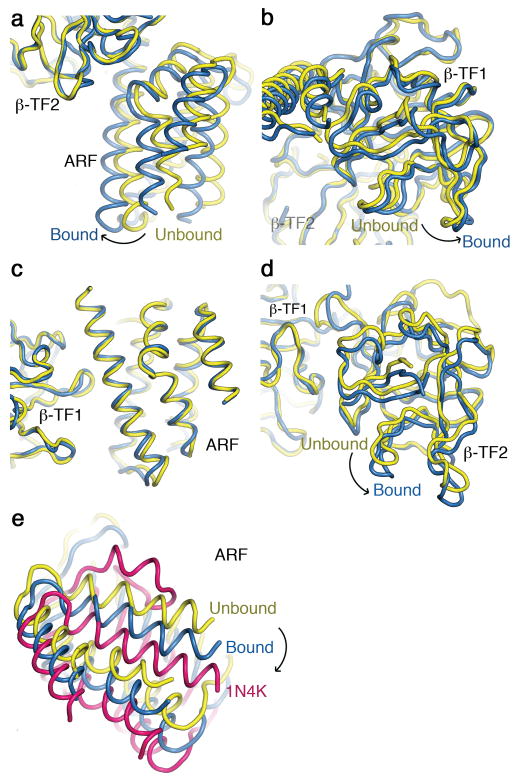
The main global difference between LBD structures with and without InsP3 bound is the relative orientation of the lobes (Fig. 1). To better illustrate this, we superimposed the two structures using the β-TF2 backbone as reference. In this alignment, ARF and β-TF1 undergo about a 10° rigid-body rotation (and slight translation) between the two states, such that ARF moves closer to β-TF2 to bind InsP3 (Fig. 1c and 1d; Fig. 3a, b). This motion is more evident in a movie that alternates between the bound and unbound structures of LBD (Supplementary Movie 1). We also aligned bound and unbound structures of LBD, using the other trefoil fold (β-TF1) as reference. This second alignment reveals that the entire ARF plus β-TF1 portion of the two structures is largely superimposable between the two states (Fig. 3c) whereas the orientation of β-TF2 with respect to β-TF1 differs (Fig. 3d). This comparison reveals that the interface between the two β-TFs is dynamic, allowing them to undergo modest relative motion. The interface between ARF and β-TF2 is not only dynamic but also InsP3 mediated. Given that the relative orientation of ARF and β-TF1 undergoes little change between bound and unbound states, the interface between them must be relatively static during that transition. The two linkers connecting the three lobes are in close proximity near one end of pseudo-three-fold axis (Fig. 1a, b). These linkers have previously been suggested to be flexibile14. In the LBD structures, the linker joining the β-TFs consists of about ten residues and is sufficiently ordered to reveal the main-chain density (Supplementary Fig. 4). It appears to differ somewhat between bound and unbound states, where flexible Gly236 and Gly237 may facilitate the linker's motion (Fig. 1a, b; Supplementary Fig. 4). On the other hand, the linker between ARF and β-TF2, formed by only four residues (PVSP), differs little between the two states (Fig. 1a, b).
Figure 3.
Comparison of InsP3-bound and -unbound LBD structures. (a, b) ARF (a) and β-TF1 (b) structures of bound (blue) and unbound (yellow) LBD are aligned using β-TF2 as a reference. (c, d) ARF (c) and β-TF2 (d) structures of bound (blue) and unbound (yellow) LBD are aligned using β-TF1 as a reference. (e) Shown are ARF structures of InsP3-bound (blue) and -unbound (yellow) LBD as well as that of the InsP3-bound partial LBD (magenta) (PDB 1N4K), all aligned using β-TF2 as reference. For clarity β-TF1 or β-TF2 or both are removed and, for easy comparison, the C-terminal 581-602 region of the partial LBD (PDB 1N4K) is not shown, as it is disordered in both LBD structures.
From a symmetry viewpoint, the following description offers a simple synopsis of LBD's overall architecture and how it transitions between the two observed states (Fig. 1; Supplementary Movie 1). The two β-TF lobes are similar (Fig. 1), except that β-TF1 has a prominent helix-turn-helix, arm-like motif (colored lime in a and b or blue in c and d). These two lobes form a pseudo-duplex and share a dynamic interface that allows modest relative rotational motion. Each β-TF lobe also interfaces with ARF. A significant breakdown of this pseudo-symmetry occurs at the interfaces between ARF and each of the two β-TFs. The interface between ARF and β-TF1 is relatively static in the two states and InsP3 independent, whereas the interface between ARF and β-TF2 is dynamic and InsP3 dependent. The former, static, interface ensures a largely concerted motion of ARF and β-TF1 whereas the latter confers InsP3-regulated conformational changes upon LBD. The movement of ARF relative to β-TF2 renders the InsP3-binding site between them either suited or unsuited for capturing InsP3. Successful capture of an InsP3 molecule then “locks” LBD in a bound state that favors opening of the ion pore. If β-TF1 indeed couples InsP3-dependent conformational states of LBD to the gate9, 13, it would cause the gate to open while the other lobe (β-TF2) of the duplex and ARF move closer together to capture an InsP3 molecule. It then follows that the relatively static interface between β-TF1 and ARF would in turn couple two remote and functionally coordinated regions, the InsP3-interacting region in ARF and the gate-coupling region in β-TF1, and thereby confer efficient allosteric regulation upon InsP3R.
Regarding β-TF1's interaction with ARF, mutations of β-TF1 such as L30K, L32K and D34K in the β2-β3 loop and others in the β5-β6 loop are known to enhance InsP3 binding to LBD11 (Supplementary Fig. 5). However, neither the structure of β-TF1 nor that of ARF plus β-TF2 has yielded clues as to which regions of ARF or β-TF2 interact with the β2-β3 and β5-β6 loops of β-TF110, 11. In our LBD structures, the β2-β3 and β5-β6 loops of β-TF1 extend toward helix α4 of ARF (Supplementary Fig. 5). The electron density map exhibits continuous densities extending from the two β-TF1 loops to the ARF helix. In light of a previous report that the D448N mutant does not express20, we point out that the density between helix α4 of ARF and the β2-β3 loop of β-TF1 appears to correspond to the Asp448 side chain in the helix, which could be within hydrogen bond distance from the backbone amide group of Leu32 at the tip of the β2-β3 loop.
Interactions between ARF and β-TF1 evidently impact InsP3 binding, as perturbing their interface or removing β-TF1 enhances InsP3 binding11, 12. To gain further insight, using β-TF2 as reference, we aligned our two LBD structures with the InsP3-bound (ARF - β-TF2) structure of Bosanac et al.10 (PDB 1N4K). Compared to our InsP3-bound structure, the ARF in their structure is further rotated with respect to our unbound structure (Fig. 3e; for clarity β-TF1 and β-TF2 are removed). Given that their structure lacks the suppressor β-TF1and thus has a higher affinity for InsP3, it most likely represents a more tightly bound state.
In the structure of ARF plus β-TF2 previously solved by Bosanac et al.10 (PDB 1N4K), the Gly487-Pro502 region forms a hairpin motif that lies parallel to its neighboring helices α5 and α9. It may be noteworthy that in our LBD structures this hairpin motif in ARF, although not fully ordered, appears to point towards helix α4 that contains Asp97, Glu99, and Glu104 (supplementary Fig. 6). This structural feature and the previous finding10 that mutation of these residues affects InsP3 binding raise the issue of whether the ARF hairpin motif interacts with the arm motif of β-TF1.
In summary, the present crystal structures of LBD in both InsP3-bound and -unbound conformations – two snapshots of the LBD dynamic process – provide the structural information necessary for uncovering the mechanism underlying allosteric regulation of InsP3R. The two β-TFs of LBD form a pseudo-duplex with a dynamic interface that permits relative motion. The two lobes of this pseudo-duplex offer two separate interfaces to the ARF lobe as well. The interface between β-TF1 and ARF is relatively static so that these two lobes move largely together with respect to β-TF2. Such relative motion must in turn be coupled, presumably via β-TF19,13, to the ion-pore gate so that LBD exerts allosteric control over the gate. In contrast, the interface between β-TF2 and ARF is more dynamic and regulated by InsP3. Given these characteristics, LBD, when not bound by InsP3, would spontaneously transition between gating conformations but, when bound by InsP3, would be locked in a state that favors opening of the ion pore.
Supplementary Material
Acknowledgments
We thank T.C. Südhof (Stanford University) for sharing the InsP3R1-cDNA, J.K. Foskett (University of Pennsylvania) for providing the InsP3R1-cDNA-containing plasmid and for discussion, K. Schmitz (University of Pennsylvania) for assistance in crystal diffraction, G. Van Duyne (University of Pennsylvania) for both the modified pET21 plasmid containing a TEV site and the TEV-cDNA-containing plasmid, staffs of the synchrotron beam lines at the Advanced Photon Source (GM/CA-CAT 23-ID-B and 23-ID-D) and the Advanced Light Source (8.2.1 and 8.2.2) for technical assistance, Y. Xu (University of Pennsylvania) for technical assistance, and P. De Weer (University of Pennsylvania) for critical review and discussion of our manuscript. This study was supported by the Howard Hughes Medical Institute.
Footnotes
Accession code. Protein Data Bank: The atomic coordinates and structure factors for InsP3 ligand-binding domain structures have been deposited with accession code 3T8S.
Author Contributions: C-C. L., K.B. and Z.L. performed the experiments, analyzed the data, and prepared the manuscript.
References
- 1.Berridge MJ, Lipp P, Bootman MD. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 2.Taylor CW, da Fonseca PC, Morris EP. Trends Biochem Sci. 2004;29:210–219. doi: 10.1016/j.tibs.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 3.Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 4.Foskett JK, White C, Cheung KH, Mak DO. Physiol Rev. 2007;87:593–658. doi: 10.1152/physrev.00035.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mignery GA, Sudhof TC. EMBO J. 1990;9:3893–3898. doi: 10.1002/j.1460-2075.1990.tb07609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyawaki A, et al. Proc Natl Acad Sci U S A. 1991;88:4911–4915. doi: 10.1073/pnas.88.11.4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newton CL, Mignery GA, Sudhof TC. J Biol Chem. 1994;269:28613–28619. [PubMed] [Google Scholar]
- 8.Yoshikawa F, et al. Biochem Biophys Res Commun. 1999;257:792–797. doi: 10.1006/bbrc.1999.0498. [DOI] [PubMed] [Google Scholar]
- 9.Rossi AM, et al. Nat Chem Biol. 2009;5:631–639. doi: 10.1038/nchembio.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosanac I, et al. Nature. 2002;420:696–700. doi: 10.1038/nature01268. [DOI] [PubMed] [Google Scholar]
- 11.Bosanac I, et al. Mol Cell. 2005;17:193–203. doi: 10.1016/j.molcel.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 12.Yoshikawa F, et al. J Biol Chem. 1996;271:18277–18284. doi: 10.1074/jbc.271.30.18277. [DOI] [PubMed] [Google Scholar]
- 13.Uchida K, Miyauchi H, Furuichi T, Michikawa T, Mikoshiba K. J Biol Chem. 2003;278:16551–16560. doi: 10.1074/jbc.M300646200. [DOI] [PubMed] [Google Scholar]
- 14.Chan J, et al. J Mol Biol. 2007;373:1269–1280. doi: 10.1016/j.jmb.2007.08.057. [DOI] [PubMed] [Google Scholar]
- 15.Jiang QX, Thrower EC, Chester DW, Ehrlich BE, Sigworth FJ. EMBO J. 2002;21:3575–3581. doi: 10.1093/emboj/cdf380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamada K, Terauchi A, Mikoshiba K. J Biol Chem. 2003;278:52881–52889. doi: 10.1074/jbc.M309743200. [DOI] [PubMed] [Google Scholar]
- 17.Serysheva II, et al. J Biol Chem. 2003;278:21319–21322. doi: 10.1074/jbc.C300148200. [DOI] [PubMed] [Google Scholar]
- 18.Wolfram F, Morris E, Taylor CW. Biochem J. 2010;428:483–489. doi: 10.1042/BJ20100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tung CC, Lobo PA, Kimlicka L, Van Petegem F. Nature. 2010;468:585–588. doi: 10.1038/nature09471. [DOI] [PubMed] [Google Scholar]
- 20.Joseph SK, Brownell S, Khan MT. Cell Calcium. 2005;38:539–546. doi: 10.1016/j.ceca.2005.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.