Abstract
Many aminoacyl-tRNA synthetases (AARSs) prevent mistranslation by relying upon proofreading activities at multiple stages of the aminoacylation reaction. In leucyl-tRNA synthetase (LeuRS), editing activities that precede or are subsequent to tRNA charging have been identified. Although both are operational, either the pre-or post-transfer editing activity can predominate. Yeast cytoplasmic LeuRS (ycLeuRS) misactivates structurally similar noncognate amino acids including isoleucine and methionine. We show that ycLeuRS has a robust post-transfer editing activity that efficiently clears tRNALeu mischarged with isoleucine. In comparison, the enzyme's post-transfer hydrolytic activity against tRNALeu mischarged with methionine is weak. Rather, methionyl-adenylate is cleared robustly via an enzyme-mediated pre-transfer editing activity. We hypothesize that similar to E. coli LeuRS, ycLeuRS has coexisting functional pre and post-transfer editing activities. In the case of ycLeuRS, a shift between the two editing pathways is triggered by the identity of the noncognate amino acid.
The translational machinery relies on aminoacyl-tRNA synthetases (AARSs) to attach correct amino acids to cognate tRNAs1. To ensure fidelity in protein synthesis, about half of the AARSs, including leucyl-tRNA synthetase (LeuRS) have evolved editing mechanisms against structurally similar noncognate amino acids to clear mistakes that originate in the synthetic active site2. The absence of these editing mechanisms results in statistical mutations in the proteome3.
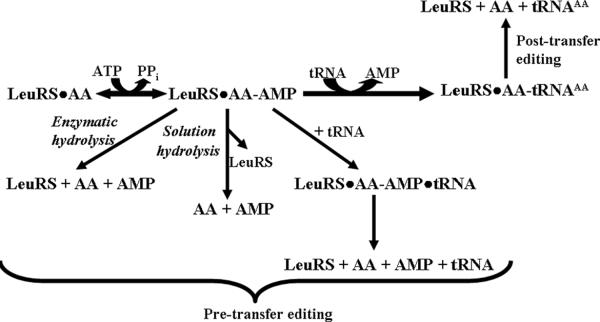
Amino acid editing may occur either before (pre-transfer)4,5 or after (post-transfer)6 the activated amino acid is transferred to the tRNA7. Thus, pre-transfer editing targets the misactivated aminoacyl-adenylate intermediate (AA-AMP) for hydrolysis, while post-transfer editing cleaves misaminoacylated tRNA (AA-tRNAAA; Scheme 1) to clear the AARSs mistakes2,8. For LeuRS, the post-transfer editing active site is housed in a discrete insertion domain, called connective poly-peptide 1 (CP1) that is separated from the aminoacylation active site by approximately 35 Å9.
Scheme 1.
Pre and post-transfer amino acid editing pathways.
Yeast cytoplasmic LeuRS (ycLeuRS) has been proposed to have relatively poor initial substrate discrimination10 and misactivates structurally similar or isosteric standard and nonstandard amino acids such as isoleucine, methionine, norvaline, and homocysteine10. An early report by Englisch et al. also suggested that ycLeuRS edited predominantly by a pre-transfer editing mechanism, while E. coli LeuRS clears mistakes exclusively by post-transfer editing mechanism10. In E. coli LeuRS, we have shown that the CP1 editing domain that is responsible for post-transfer editing masks a pre-transfer editing activity that is associated with the canonical amino-cylation core11,12. In ycLeuRS, we hypothesize that both pre and post-transfer editing mechanisms coexist and that one predominates similar to E. coli LeuRS. Here, we determined that a shift between the two AARS fidelity mechanisms can be dependent on the identity of the standard amino acids that threaten LeuRS fidelity. A partition of fidelity mechanisms has also been reported for the human cytoplasmic LeuRS (hcLeuRS) against nonstandard biosynthetic intermediates norvaline and α-aminobutyrate13. This multi-step strategy for quality control could be akin to the evolution of auxiliary editing domains such as ybak to clear mischarged CystRNAPro in trans, in addition to Ala-tRNAPro that is produced and then cleared by cis editing activity of prolyl-tRNA synthetase14,15.
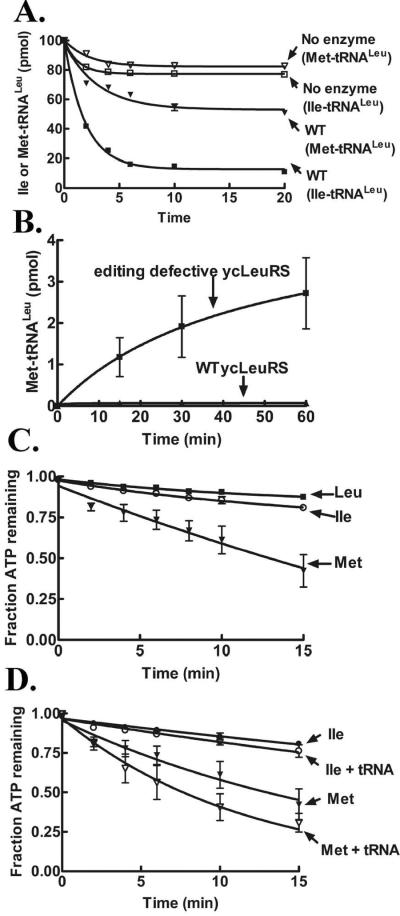
We cloned the gene for yeast cytoplasmic tRNALeu (yctRNALeuCAA) into the pTrc-99 vector16 for overexpression in E. coli. The yctRNALeu was extracted with phenol/tris saturated solution, followed by denaturing gel purification. ycLeuRS with a six histidine tag, was purified via affinity purification17. Using in vitro deacylation assays that incorporated purified yctRNALeu mischarged with [3H]-isoleucine, we determined that ycLeuRS has a robust post–transfer editing mechanism that clears mischarged Ile-tRNALeu (Figure 1A). In comparison, this yeast enzyme exhibited significantly reduced hydrolytic activity against [3H]-Met-tRNALeu (Figure 1A). This amino acid-dependent difference in post-transfer editing was surprising in that it contrasts with other LeuRSs that effectively deacylate tRNALeu mischarged with either isoleucine or methionine13,18–21.
Figure 1.
In vitro enzymatic activities of ycLeuRS. Deacylation of [3H]-Ile-tRNALeu and [3H]-Met-tRNALeu by 1 μM ycLeuRS (A). Misaminoacylation of yctRNALeu with 25 μM [3H]-methionine (422 μCi/mL) by 1 μM LeuRS (B). ATP hydrolysis in absence (C) or presence (D) of tRNA. The reactions contained 250 μM ATP (600 μCi/mL), amino acids (2.5 mM leucine or 100 mM isoleucine or methionine) and 1 μM ycLeuRS (C). Assays are described in detail in the Supporting Information.
Despite its weak post-transfer editing activity, we did not observe accumulation of mischarged Met-tRNALeu for wild type ycLeuRS (Figure 1B), as compared to an editing defective mutant (D419A) that we previously characterized17. In ycLeuRS, mutation of the universal Asp419 to alanine disrupts overall editing (pre- and post-transfer)17. Thus, the mutant enzyme mischarges yctRNALeu with noncognate amino acids. It remains unclear how this single site impacts both pre- and post-transfer editing. Because of this striking difference in deacylation activities, we hypothesized that methionine that is misactivated by ycLeuRS is more efficiently cleared by an alternate fidelity pathway to post-transfer editing.
Consistent with previous activation measurements of other noncognate amino acids by ycLeuRS10, we measured a poor discrimination factor of 1/72 for methionine (data not shown). Since ycLeuRS lacks a robust post-transfer editing activity to clear Met-tRNALeu, we predicted that the methionyl adenylate intermediate might be cleared by pre-transfer editing. Pre-transfer editing is typically characterized by increased consumption of ATP in the presence of noncognate amino acids13,22,23. We analyzed both ATP hydrolysis and AMP formation using TLC-based assays24 that utilize [α-32P]-ATP to visualize separated ATP, AMP and AA-AMP. In absence of tRNA, ATP hydrolysis for methionine was stimulated relative to cognate leucine (Figure 1C) yielding an accumulation of 25 μM of AMP with a kobs of 2.0 ± 0.3 min−1 for AMP formation (Table 1). By comparison, isoleucine-dependent AMP formation was only slightly elevated with a kobs of 0.7 ± 0.1 min−1 in the absence of tRNA.
Table 1.
Rate constants for tRNA-independent AARS pre-transfer editing activities of standard amino acidsa
| Enzyme-dependent AMP formation (min−1) | Non-enzymatic hydrolysis of AA-AMP (min−1) | Fold difference | Ref | |
|---|---|---|---|---|
| LeuRS Met | 2.0 | 0.100 | 20 | This work |
| IleRS Val | 3.2 | 0.120 | 30 | 23 |
| GlnRS-bGln | 1.5 | 0.042 | 35 | 24 |
| ProRS Ala | 27.2 | 0.113 | 240 | 22,25 |
| ValRS Thr | 22.2 | 0.070 | 300 | 23 |
For enzymatic AMP formation, kobs was reported for all systems, except IleRS and ValRS activation of valine and threonine respectively, for which kcat was reported. For non-enzymatic solution hydrolysis, kobs was reported.
The GlnRS-Gln system requires presence of tRNAGln.
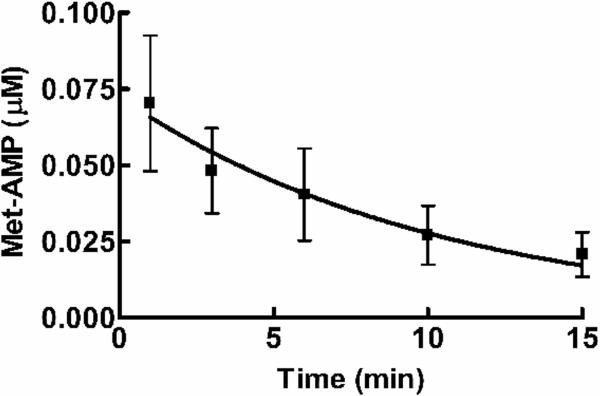
Similar increases in AMP formation have been measured for other AARSs in the presence of noncognate amino acids as an indicator for tRNA independent pre-transfer editing activity (Table 1). The addition of tRNA enhanced ATP hydrolysis for methionine, but failed to significantly stimulate isoleucine-dependent ATP hydrolysis (Figure 1D), suggesting an increase in overall editing against methionine. This is consistent with early reports10 where tRNA addition was reported to weakly stimulate pre-transfer editing of noncognate amino acids by ycLeuRS10. Hydrolysis of AA-AMP can be catalyzed by the enzyme or depend on a selective release mechanism22,25. The latter relies on enzyme ejection of adenylate intermediate into the aqueous environment for hydrolysis of noncognate AA-AMP (Scheme 1)2,8,22. To distinguish between these two possibilities for Met-AMP hydrolysis, we performed a chase assay23 using reaction conditions that were identical to the AMP formation assays described above. Enzyme-synthesized AA-AMP accumulated for 10 min, followed by addition of a large molar excess of non-radioactive ATP (25 mM) to displace AA-AMP from the synthetic active site. Enzyme-independent hydrolysis of methionyl-adenylate in solution occurred at a rate of 0.1 ± 0.001 min−1 (Figure 2), which is 20-fold slower than the rate of methionine-dependent AMP formation by ycLeuRS (2.0 min−1) (Table 1). This is consistent with other AARSs that have been proposed to edit by tRNA-independent pre-transfer editing mechanisms (Table 1). We estimate that enzyme-associated hydrolysis of the methionyladenylate would account then for approximately 95% of the tRNA-independent pre-transfer editing activity of ycLeuRS against methionine.
Figure 2.
Non-enzymatic hydrolysis of methionyladenylate. TLC-based ATP chase assays include 251 μM ATP (1800 μCi/mL), 100 mM methionine and 5 μM ycLeuRS. AA-AMP was chased from the enzyme active site into solution by adding 100-fold excess of non-radioactive 25 mM ATP. Assays are described in Supporting Information.
Adenylate hydrolysis by the homologous IleRS has been proposed to involve its translocation from the synthetic site to the CP1 domain editing site in a tRNA-dependent manner26. In contrast for E. coli LeuRS, a robust pre-transfer editing activity was only stimulated when the CP1 domain was deleted from the enzyme11 or when a mutation at CP1 domain-based Ala293 was introduced12. Similarly, pre-transfer editing activity is activated in ProRS25 and ThrRS27 mutants under circumstances where post-transfer activity was selectively inactivated. Pre-transfer editing activities that are tRNA independent have also been reported for SerRS28, which lacks a specialized editing domain, as well as in GlnRS24 which apparently does not require editing activity. In addition, MetRS clears homocysteine in a pre-transfer editing cyclization mechanism to produce thiolactone in the synthetic active site29.
Segregation of amino acids to different editing pathways has been reported for hcLeuRS, which edits biosynthetic intermediates (α-amino butyrate and norvaline) under in vitro conditions via pre- and post-transfer hydrolysis, respectively13. A tRNA-independent pre-transfer editing for Aquifex aeolicus LeuRS has also been identified to clear norvalyl-adenylate30. Our results demonstrate that pre- and post-transfer editing co-exist in ycLeuRS, as also found previously for E. coli LeuRS11,12. Methionine-stimulated ATP hydrolysis and AMP formation in the absence of tRNA as well as ATP chase experiments support that quality control for ycLeuRS is highly dependent on pre-transfer editing under in vitro conditions. It is possible that the inclusion of tRNA shifts the editing mechanism preference and increases overall global editing. The same conditions that incorporate isoleucine rather than methionine suggest that the ycLeuRS relies on post-transfer editing for isoleucine clearance. Despite that most in vitro enzyme experiments fail to recapitulate the dynamic cellular environment, direct in vitro comparison for ycLeuRS suggests that methionine and isoleucine partition for clearance between different editing pathways. It remains possible that ycLeuRS relies on a combination of editing mechanisms, but our results support that the preference of editing pathways can be dependent on substrate identity.
We hypothesize that these two fidelity mechanisms in ycLeuRS are not redundant, but adapted to accommodate diverse specificities of the noncognate amino acids that challenge LeuRS fidelity in the cell3,20. Under this scenario, ycLeuRS pre- and post-transfer editing sites would have evolved in a complementary way, similar to AARSs that are dependent on independent auxiliary editing domains. Thus, in the case of ycLeuRS, we propose that pre- and post-transfer editing pathways partition in a way that is dependent on the chemical structure of the noncognate amino acid to ensure that all errors are efficiently targeted for clearance by ycLeuRS. This is critical to the cell since disruptions or absence of editing activities in LeuRS can cause mistranslation and cell death3,20.
Supplementary Material
Acknowledgement
This work was supported by National Institutes of Health Grant GM063789.
Footnotes
Supporting Information Available: Details on all materials and methods.
References
- (1).Ibba M, Soll D. Annu. Rev. Biochem. 2000;69:617. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- (2).Mascarenhas AP, Martinis SA, An S, Rosen AE, Musier-Forsyth K. In: Protein Engineering. RajBhandary UL, Koehrer C, editors. Springer-Verlag; 2008. p. 153. [Google Scholar]
- (3).Li L, Boniecki M, Jaffe J, Imai B, Yau PM, Luthey-Schulten ZA, Martinis SA. Proc. Natl. Acad. Sci. U S A. 2011;108:9378. doi: 10.1073/pnas.1016460108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Baldwin AN, Berg P. J. Biol. Chem. 1966;241:839. [PubMed] [Google Scholar]
- (5).Fersht AR. Biochemistry. 1977;16:1025. doi: 10.1021/bi00624a034. [DOI] [PubMed] [Google Scholar]
- (6).Eldred EW, Schimmel PR. J. Biol. Chem. 1972;247:2961. [PubMed] [Google Scholar]
- (7).Schimmel P, Schmidt E. Trends Biochem. Sci. 1995;20:1. doi: 10.1016/s0968-0004(00)88937-9. [DOI] [PubMed] [Google Scholar]
- (8).Ling J, Reynolds N, Ibba M. Annu. Rev. Microbiol. 2009;63:61. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- (9).Cusack S, Yaremchuk A, Tukalo M. EMBO J. 2000;19:2351. doi: 10.1093/emboj/19.10.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Englisch S, Englisch U, von der Haar F, Cramer F. Nucleic Acids Res. 1986;14:7529. doi: 10.1093/nar/14.19.7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Boniecki MT, Vu MT, Betha AK, Martinis SA. Proc. Natl. Acad. Sci. U S A. 2008;105:19223. doi: 10.1073/pnas.0809336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Williams AM, Martinis SA. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3586. doi: 10.1073/pnas.0507362103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Chen X, Ma JJ, Tan M, Yao P, Hu QH, Eriani G, Wang ED. Nucleic Acids Res. 2011;39:235. doi: 10.1093/nar/gkq763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).An S, Musier-Forsyth K. J. Biol. Chem. 2005;280:34465. doi: 10.1074/jbc.M507550200. [DOI] [PubMed] [Google Scholar]
- (15).Ruan B, S□ll D. J. Biol. Chem. 2005;280:25887. doi: 10.1074/jbc.M502174200. [DOI] [PubMed] [Google Scholar]
- (16).Martin F, Eriani G, Eiler S, Moras D, Dirheimer G, Gangloff J. J. Mol. Biol. 1993;234:965. doi: 10.1006/jmbi.1993.1651. [DOI] [PubMed] [Google Scholar]
- (17).Lincecum TL, Jr., Tukalo M, Yaremchuk A, Mursinna RS, Williams AM, Sproat BS, Van Den Eynde W, Link A, Van Calenbergh S, Gr□tli M, Martinis SA, Cusack S. Mol. Cell. 2003;11:951. doi: 10.1016/s1097-2765(03)00098-4. [DOI] [PubMed] [Google Scholar]
- (18).Zhai Y, Martinis SA. Biochemistry. 2005;44:15437. doi: 10.1021/bi0514461. [DOI] [PubMed] [Google Scholar]
- (19).Mursinna RS, Martinis SA. J. Am. Chem. Soc. 2002;124:7286. doi: 10.1021/ja025879s. [DOI] [PubMed] [Google Scholar]
- (20).Karkhanis VA, Mascarenhas AP, Martinis SA. J Bacteriol. 2007;189:8765. doi: 10.1128/JB.01215-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Chen JF, Guo NN, Li T, Wang ED, Wang YL. Biochemistry. 2000;39:6726. doi: 10.1021/bi000108r. [DOI] [PubMed] [Google Scholar]
- (22).Hati S, Ziervogel B, Sternjohn J, Wong FC, Nagan MC, Rosen AE, Siliciano PG, Chihade JW, Musier-Forsyth K. J. Biol. Chem. 2006;281:27862. doi: 10.1074/jbc.M605856200. [DOI] [PubMed] [Google Scholar]
- (23).Dulic M, Cvetesic N, Perona JJ, Gruic-Sovulj I. J. Biol. Chem. 2010;285:23799. doi: 10.1074/jbc.M110.133553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Gruic-Sovulj I, Uter N, Bullock T, Perona JJ. J. Biol. Chem. 2005;280:23978. doi: 10.1074/jbc.M414260200. [DOI] [PubMed] [Google Scholar]
- (25).Splan KE, Ignatov ME, Musier-Forsyth K. J. Biol. Chem. 2008;283:7128. doi: 10.1074/jbc.M709902200. [DOI] [PubMed] [Google Scholar]
- (26).Nomanbhoy T, Hendrickson T, Schimmel P. Mol Cell. 1999;4:519. doi: 10.1016/s1097-2765(00)80203-8. [DOI] [PubMed] [Google Scholar]
- (27).Minajigi A, Francklyn CS. J. Biol. Chem. 2010;285:23810. doi: 10.1074/jbc.M110.105320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Gruic-Sovulj I, Rokov-Plavec J, Weygand-Durasevic I. FEBS Lett. 2007;581:5110. doi: 10.1016/j.febslet.2007.09.058. [DOI] [PubMed] [Google Scholar]
- (29).Kim HY, Ghosh G, Schulman LH, Brunie S, Jakubowski H. Proc. Natl. Acad. Sci. U. S. A. 1993;90:11553. doi: 10.1073/pnas.90.24.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Zhu B, Yao P, Tan M, Eriani G, Wang ED. J Biol. Chem. 2009;284:3418. doi: 10.1074/jbc.M806717200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.