SUMMARY
Complex genomic rearrangements (CGR) consisting of two or more breakpoint junctions have been observed in genomic disorders. Recently, a chromosome catastrophe phenomenon termed chromothripsis, in which numerous genomic rearrangements are apparently acquired in one single catastrophic event, was described in multiple cancers. Here we show that constitutionally acquired CGRs share similarities with cancer chromothripsis. In the 17 CGR cases investigated we observed localization and multiple copy number changes including deletions, duplications and/or triplications, as well as extensive translocations and inversions. Genomic rearrangements involved varied in size and complexities; in one case, array comparative genomic hybridization revealed 18 copy number changes. Breakpoint sequencing identified characteristic features, including small templated insertions at breakpoints and microhomology at breakpoint junctions, which have been attributed to replicative processes. The resemblance between CGR and chromothripsis suggests similar mechanistic underpinnings. Such chromosome catastrophic events appear to reflect basic DNA metabolism operative throughout an organism’s life cycle.
INTRODUCTION
Human genomic rearrangements with two or more breakpoint junctions are referred to as complex genomic rearrangements (CGRs) (Zhang et al., 2009a). CGRs have been identified frequently during characterization of nonrecurrent microduplications associated with genomic disorders. Based on the microhomologies identified at breakpoint junctions, the apparent template driven insertional complexities at breakpoints, and the fusions of distantly distributed sequences in complex genomic rearrangements, a replication based mechanism, the fork stalling and template switching (FoSTeS) model, has been proposed to explain the formation of such rearrangement complexities in the human genome (Lee et al., 2007; Slack et al., 2006). Other similar replication based models such as microhomology mediated break-induced replication (MMBIR) (Hastings et al., 2009a; Hastings et al., 2009b), microhomology/microsatellite induced replication (MMIR) (Payen et al., 2008) and microhomology-mediated replication-dependent recombination (MMRDR) (Chen et al., 2010) have also been proposed. Recent studies on genomic disorder associated nonrecurrent rearrangements identified complex genomic rearrangements on chromosome X (Carvalho et al., 2009; Liu et al., 2011) and at multiple genomic loci on autosomes such as 17p13.3 (Bi et al., 2009), 17p12 (Zhang et al., 2009b), 17p11.2 (Zhang et al., 2010), 9q34.3 (Yatsenko et al., 2009), and 1p36 (Gajecka et al., 2010).
Chromosome rearrangements are also frequently observed in cancers. At an organismal level, the rearrangements acquired in cancers differ from the ones in genomic disorders in the time they arise during the life cycle (Lupski, 2010). Genomic disorders frequently result from “constitutional” germline rearrangements that occur during gametogenesis or early postzygotic development, whereas rearrangements acquired in cancers involve “somatic” differentiated cells. Thus, genomic rearrangement may be less complex in genomic disorders than in cancers reflecting selective forces because an organism cannot endure/survive excessive toxicities from massive genomic changes early in development. However, on a cellular level, the mechanisms underlying these DNA rearrangements occurring in cells at different stages of the human life cycle (i.e. germline, postzygotic development, somatic differentiated cells) are likely the same.
Recently, the phenomenon of chromothripsis (Stephens et al., 2011), an apparent chromosome catastrophe with several copy number changes and multiple breakpoints concentrated on a single chromosome, was described in cancer cells. Remarkably, 2–3% of all cancers, and up to 25% of bone cancers, demonstrated chromothripsis. In contrast to the generally accepted concept for cancer biogenesis in which a mutational accumulation model appears operative, the profound level and complexity of rearrangements observed in chromothripsis are generated on a much shorter time scale, probably in a single mutational event. The mechanisms behind these cataclysmic genome disruptions are unknown.
We identified apparent CGR in subjects referred with developmental delay and cognitive anomalies. Using diverse high resolution genome analysis techniques, we show that such constitutional CGRs share many structural and breakpoint features with cancer chromothripsis. Constitutional CGR can involve multiple copy number changes, translocations, and inversions in a genomic region-focused manner; frequently, two or more genomic segments from separate loci are assembled into one larger rearranged piece. These characteristic features are consistent with formation of the highly complex pattern of chromosome catastrophe by a replicative mechanism in a single event. We propose that both the constitutional CGR and the chromothripsis processes reflect basic DNA metabolism and share a cellular DNA replication/repair mechanism.
RESULTS
Complex genomic rearrangements identified by clinical CMA
We investigated 17 cases – ten males and seven females, referred to the Medical Genetics Laboratories (MGL; http://www.bcm.edu/geneticlabs) for various developmental problems. Clinical chromosomal microarray analysis (CMA) by array comparative genomic hybridization (aCGH) in each subject showed apparent multiple copy number changes involving a single chromosome potentially representing a CGR (Table 1; Figure 1A). A subset of copy number changes, with a genomic size large enough to be resolved, were further confirmed by fluorescence in situ hybridization (FISH) or G-banded chromosome analyses (Figure 1B–E). Multiple rearrangement patterns were observed: a duplication (dup) followed by a normal copy (nml) sequence and then by a duplication (dup-nml-dup), a duplication followed by a deletion (del) (i.e. dup-del), a duplication followed by a normal copy sequence followed by a deletion (dup-nml-del), and a duplication followed by a triplication (trp) (i.e. dup-trp). Triplications were identified in six cases. Rearrangements with complex patterns including three or more apparent copy number changes were also identified. Additional chromosomal structural aberrations were found in case BAB2778, which has a terminal dup-nml-dup rearrangement within a ring chromosome 6 (Figure 1B).
Table 1.
Complex genomic rearrangements assessed by gains and losses of specific human genome intervals. See also Table S1
| Subjects (BAB#) |
Sex | Location | array CGH-inferred rearrangement pattern |
Sizes of copy number changes | Clinical indication | Parental study |
Size (Mb) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | CNVs | Gain | Loss | |||||||
| Patient 1 | F | 1q32.3-q43 | del-nml-dup-nml-dup-nml-del | del 4.7 Mb-nml 11.5 Mb-dup 3.1 Mb-nml 8.1 Mb–dup 4.4 Mb-nml 0.3 Mb-del 1.8 Mb | Severe DD/ID | de novo | 34.0 | 14.0 | 7.5 | 6.5 |
| 2920 | M | 1q43-q44 | dup-del | dup 0.7 Mb-del 4 Mb | Microcephaly, epilepsy | mother nml | 4.7 | 4.7 | 0.7 | 4.0 |
| 3047 | M | 4q33-q34.1 | trp | trp 1.2 Mb | DD | mat | 1.2 | 1.2 | 1.2 | 0 |
| 2760 | F | 4q35.1-q35.2 | dup-nml-dup | dup 115 kb-nml 2.6 Mb-dup 298 kb | Autistic spectrum, DD/ID, prenatal teratogen exposure | NA | 3.0 | 0.4 | 0.4 | 0 |
| 3012 | M | 5q31.1-q31.2 | dup-trp | dup 28 kb-trp 1.9 Mb | Mild DD, DF | mat (47%) | 1.9 | 1.9 | 1.9 | 0 |
| 2778 | M | 6q27 | dup-nml-terminal dup, r(6) | dup 2.2 Mb-nml 666 kb-dup 1.0 Mb | Chromosomal abnormality, microcephaly, speech delay | father nml | 3.9 | 3.2 | 3.2 | 0 |
| 3103 | F | 7q33-ter | dup-nml-del | dup 25.6 Mb-nml 7.9 kb-del 269 kb | DF, VSD | de novo | 25.9 | 25.9 | 25.6 | 0.3 |
| 3105 | M | 9q31.1-q33.1 | dup-nml-dup-nml-dup-nml-dup-nml-dup-nml-dup-nml-dup-nml-dup-nml-dup-trp-dup-nml-dup-nml-trp-dup-nml-dup-nml-dup-nml-dup-nml-dup | dup 2.1 Mb-nml 0.8 Mb-dup 37 kb-nml 4.1 Mb-dup 0.1 Mb-nml 1.1 Mb-dup 55 kb-nml 11.2 Mb-dup 36 kb-nml 0.2 Mb-dup 0.2 Mb- nml 1.1 Mb-dup 60 kb-nml 11.7 Mb-dup 0.5 Mb-nml 8.1 Mb-dup 0.1 Mb-trp 0.1 Mb–dup 44 kb-nml 0.2 Mb-dup 0.1 Mb-nml 33 kb–trp 5.5 Mb-dup 0.4 Mb-nml 0.2 Mb-dup 20 kb–nml 35 kb-dup 0.8 Mb-nml 0.2 Mb-dup 1.4 Mb–nml 0.4 Mb-dup 29 kb | DF | de novo | 51.0 | 11.6 | 11.6 | 0 |
| 3015 | F | 12q21.1 | dup-trp | dup 9.4 kb-trp 2.2 Mb | DD, DF | mat | 2.3 | 2.2 | 2.2 | 0 |
| 2780 | M | 12q24 | dup-nml-dup | dup 250 kb-nml 162 kb-dup 10.7 Mb | DD | de novo | 11.1 | 10.9 | 10.9 | 0 |
| 2785 | F | 13q34 | dup-nml-dup | dup 508 kb-nml 127 kb-dup 500 kb | Moderate DD/ID, DF | mat | 1.1 | 1.0 | 1.0 | 0 |
| 3011 | M | 14q12-q21.3 | dup-nml-del | dup 2.8 Mb-nml 1.7 Mb-del 15.4 Mb | DD, DF | de novo | 19.9 | 18.2 | 2.8 | 15.4 |
| 3050 | M | 15q26.3 | dup-trp-dup-trp-dup | dup 1.3 Mb-trp 0.9 Mb-dup 40 kb-trp 0.3 Mb–dup 41 kb | DD, DF, MCA, immunodeficiency | de novo | 2.5 | 2.5 | 2.5 | 0 |
| 2783 | M | 18p11.32 | dup-nml-dup | dup 590 kb-nml 1.4 Mb-dup 443 kb | ID | mother nml | 2.4 | 1.0 | 1.0 | 0.0 |
| Patient 2 | F | 22q11.1-q13.33 | del-nml-dup-nml-dup-nml-del-nml-dup | del 2.0 Mb-nml 5.1 Mb-dup 2.6 Mb-nml 0.3 Mb–dup 1.7 Mb- nml 0.8 Mb-dup 4.0 Mb-nml 2.5 Mb–dup 2.2 Mb-nml 8.8 Mb-del 0.4 Mb-nml 3.1 Mb–dup 1.6 Mb | Chromosomal abnormality, epilepsy | NA | 35.1 | 14.5 | 12.1 | 2.4 |
| 3104 | M | 22q12.2-q13.31 | del-nml-dup-nml-dup-nml-dup-nml-del-nml-del-nml-del | del 0.4 Mb-nml 295 kb-dup 4.3 Mb-nml 175 kb-dup 0.5 Mb-nml 1.1 Mb-dup 0.3 Mb-nml 10 Mb–del 76 kb-nml 23 kb-del 2.8 Mb-nml 738 kb-del 0.9 Mb | Hypotonia, Seizure | NA | 21.8 | 9.4 | 5.1 | 4.3 |
| 3032 | F | Xq27.1 | dup-trp-dup-trp-dup | dup 194 kb-trp 191 kb-dup 18 kb-trp 28 kb-dup 200 kb | Moderate DD/ID | mother nml | 0.7 | 0.7 | 0.7 | 0.0 |
Legends: dup- duplication, trp- triplication, del- deletion, nml- normal, DD- Developmental delay, ID- intellectual disability, DF- dysmorphic features, VSD-ventricular septal defect, MCA- multiple congenital anomalies, NA- not available, mat- maternal.
Figure 1.
Cases with complex rearrangements identified by clinical CMA. (A) Array CGH data for subjects on the left with the interpreted rearrangement patterns depicted on the right. The piled rectangles depict copy number status for the region (one for deletion, two for normal diploid copy, three for duplication, and four for triplication). The V-shaped lines connecting different segments indicate regions of unknown copy number state due to lack of interrogating oligonucleotide coverage in the array. (B)–(E) Representative FISH and chromosome analyses independently confirm the CMA findings and provide positional information. The chromosomes with rearrangements are indicated by arrows. (B) Metaphase FISH analysis using two probes located within the duplications show that the rearrangement was within a ring chromosome 6 in BAB2778. (C)–(D) G-banded chromosome (C) and FISH (D) analyses using two probes within the two duplications show that neither of the two individual duplicated segments appear to be simple tandem events; subsequent sequence analysis demonstrated that one duplication in BAB2780 is inserted in between the other duplication. (E) Interphase FISH analysis revealed three red signals, which confirmed the triplication in BAB3050. The BAC probes used were indicated. See Figure S1 for additional rearrangements with CMA results indicating a duplication next to a terminal deletion.
All CGRs show localization to a single chromosome. The copy number changes in most cases (15/17) were confined within the distal half of the involved chromosomal arms. Three cases (BAB2778, 3103 and patient 2) contained a terminal rearrangement. BAB3103 had a terminal deletion whereas BAB2778 had a terminal duplication. Patient 2 had a terminal duplication that was part of its very complex rearrangement. Additional CMA cases with complex rearrangements involving terminal changes frequently consist of a terminal deletion followed by an inverted duplication (Figure S1). The rearrangements were apparently distributed on all chromosomes.
Of the 17 patients, parental studies when available showed that five patients had a de novo rearrangement whereas four patients had a maternally inherited rearrangement, demonstrating transmission of the CGR. The mother of individual BAB3012 with a triplication was mosaic for the triplication in 47% of the cells examined by FISH analysis and the mosaicism was independently confirmed by aCGH (data not shown). These data suggest a postzygotic, mitotic origin for this CGR.
Characterization of complex genomic rearrangements by high density arrays
More extensive genome resolution of these complex rearrangements was achieved by aCGH using custom designed high density Agilent 60K or 180K arrays, or commercially available Nimblegen or Agilent arrays (Figure 2). These studies confirmed the CMA findings, refined the genomic intervals involved, and mapped most breakpoint junctions to a genomic region of a few kb or less in size (Table S2). The genomic sizes of the intervals involved in these CGR ranged from 0.7 Mb to 51 Mb, whereas the combined individual intervals with copy number changes within the CGR ranged from 0.4 Mb to 25.9 Mb in size; the latter reduced amounts reflecting normal copy intervals within the CGRs. For a single CNV event, the sizes ranged between 9.4 kb and 25.6 Mb in size. Two copy number changes were either adjacent to each other or separated by a normal copy sequence as small as 7.9 kb or as large as 11.7 Mb in size.
Figure 2.
Characterization of the rearrangements using high density arrays. (A) Array CGH using customized Agilent arrays. Copy number changes (indicated by black circles) in addition to the ones detected by clinical CMA were seen for patients BAB3012, 3015, 3103, and 3032. The boxed regions were enlarged in the left bottom panels. For both BAB3012 and 3015, a small duplication was detected proximal and next to the triplicated regions. For BAB3103, a 7.9 kb normal copy sequence was detected between the terminal deletion and the duplicated segment. BAB3032 had a trp-dup rearrangement revealed by CMA. High-density array showed a duplication proximal to the triplication and a 18 kb duplication within the triplicated segment. (B) Array CGH using Nimblegen 4.2 M arrays. BAB3050 had a dup-trp rearrangement identified by CMA. High density array showed a 40 kb duplication within the triplicated segment which was highlighted in a blue circle. This duplication is within a CNV in the database of genomic variants, and most likely is not part of the complex event, but rather reflects relative signal intensities from a benign CNV. See also Table S2.
Higher resolution aCGH analysis revealed additional complexity in eight cases, BAB3012, 3015, 3103, 3032, 3104, 3105, patient 1 and patient 2. Duplications were identified proximal and adjacent to triplications in two of the three cases with a single triplication detected initially by clinical CMA. The sizes of the duplications in these two cases, BAB3012 and BAB3015, were 28 kb and 9 kb, respectively. BAB3103 had a duplication of 25.6 Mb in size apparently juxtaposed to a terminal deletion of 269 kb identified by CMA. High density aCGH revealed that the duplication and deletion are separated by a 7.9 kb normal copy sequence. BAB3032 had a trp-dup in Xq revealed by CMA. High density array detected a 194 kb duplication adjacent and proximal to the triplication and an 18 kb duplication within the triplicated segment.
Some genomic rearrangements are very complex
Four cases, BAB3104 and 3105, patient 1 and patient 2, had highly complex rearrangements. Using a multitude of techniques, we found that these cases all had a combination of interspersed multiple copy number changes, including duplications, deletions or triplications together with additional structural changes, such as translocations or inversions in one single chromosome.
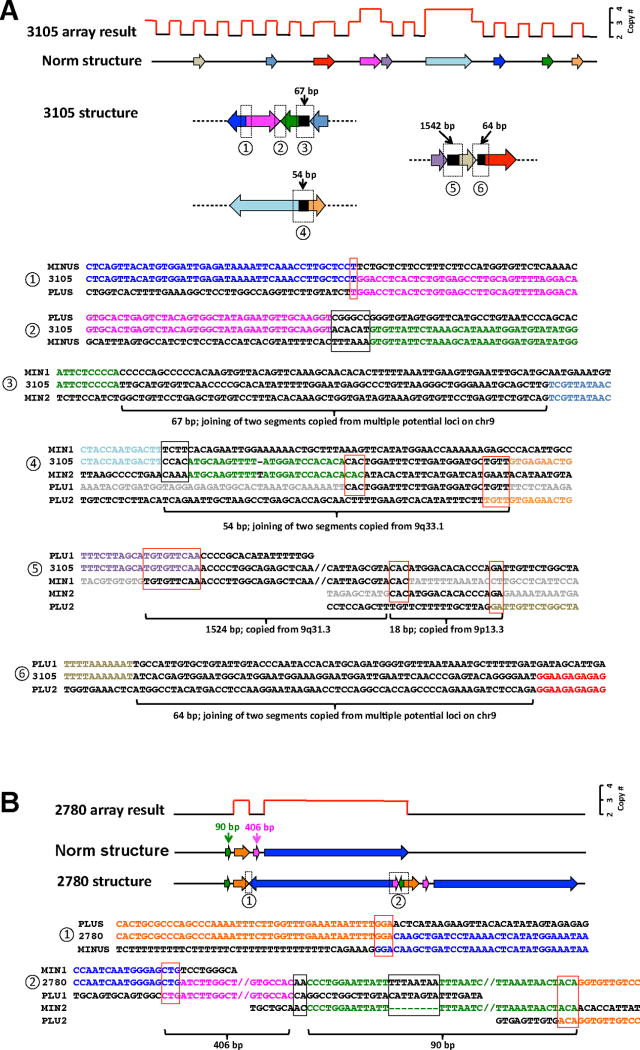
Patient BAB3105, a one and a half-year-old boy with dysmorphic features, had 18 copy number change events, with the rearrangement pattern nml-dup-nml-dup-nml-dup-nml-dup-nml-dup-nml-dup-nml-dup-nml-dup-nml-dup-trp-dup-nml-dup-nml-trp-dup-nml-dup-nml-dup-nml-dup-nml-dup-nml (Figures 3A–B and S2A–F). Partial chromosome analysis showed that this highly complex rearrangement was constrained within one chromosome with the abnormal chromosome 9 having additional material inserted proximal to 9q21; the chromosome homologue was cytogenetically normal (Figure S2H). Multiple FISH analyses were performed to determine whether the gained material in the duplications and triplications were inserted in tandem with their original copies or translocated into different loci. Interestingly, for all the intervals tested (four duplications and one triplication), FISH results demonstrated breakpoint clustering with all the additional copies translocated to a proximal region at 9q21, adjacent to the pericentric heterochromatin (Figures 3A and S2I-L). In addition, FISH also revealed an inversion event with one copy of the 5.5 Mb triplicated segment inversely oriented in comparison to the other two copies (Figure 3A). Breakpoint sequencing analyses successfully resolved six breakpoint junctions (Figures 3A and 4A). A striking breakpoint-clustering pattern was evident, whereby all junctions presented fusion of dispersed duplicated or triplicated segments. The FISH and breakpoint junction data are consistent with a general impression that most, if not all, of the additional copies of duplications and triplications in BAB3105 are randomly conjoined and assembled into a large “breakpoint junction cluster”.
Figure 3.
Highly complex rearrangements in BAB3105. (A) Schematic drawing of copy number and rearrangement structure profiles on chromosome 9q summarized from aCGH, FISH and G-banded chromosome analysis results. Bars (red for duplication and blue for triplication) above the chromosome ideogram and piled rectangular boxes below the ideogram represent the copy number states for each genomic segment. The sizes of the rearrangements and normal copy number intervals are listed in megabases. The arrows indicate translocations (upward facing) and insertions (downward facing) as indicated by FISH analysis using locus-specific BAC clones. All these additional segments of the 5.1 Mb duplication, 2.1 Mb duplication, 0.78 Mb duplication, 1.4 Mb duplication, and 5.5 Mb triplication were translocated close to the pericentric region, proximal to the original location of the 2.1 Mb duplication region. The orders of these additional segments are deduced from FISH results. It is unknown whether the additional 1.4 Mb duplicated segment is located proximal to or between the two copies of the 5.5 Mb triplicate segments. Two representative FISH images are shown (Figure 3A). The FISH image on the left shows that the duplicated (RP11-35N6) and triplicated (RP11-18B16) segments in 9q31-q33 were translocated close to the 9q21 region. The FISH image on the right shows that the two additional copies of the 5.5 Mb triplication marked by an arrow are in an inverted orientation with each other. RP11-203L12 is mapped at distal end and RP11-104M22 is mapped at the proximal end of the triplicated segment. Note that the additional complexities of the rearranged chromosome are represented as curvilinear connections of breakpoint regions. The purple curves indicate junctions between two segments with copy number gain revealed by breakpoint sequencing. (B) Nimblegen 4.2M aCGH plots for the trio. The displayed regions correspond to the regions in (A). Parental aCGH analyses demonstrated that all the copy number changes were de novo. Triplication was indicated by blue and duplication was indicated by red. The pedigrees are on the bottom. (C) Representative SNP transmission patterns that suggest a paternal interchromosome origin of the rearrangements. (D) Zoomed-in view of the SNP array data for the 5.5 Mb triplication and the 0.78 Mb duplication in BAB3105. Five different genotypes are observed across the entire triplicated region. See also Figure S2
Figure 4.
Representative breakpoint sequences. In each panel, the top graph shows schematic representation of the aCGH result. Regions of copy number gains are highlighted in red. The sizes of different segments are not in proportion to the actual rearrangement size. Below the array result is the schematic diagram illustrating the structure of a reference genomic region corresponding to the region in the array result. Below the reference structure is the proposed rearranged structure in the patient, which is surmised from breakpoint sequencing data. Specific breakpoint sequences and alignments to the reference sequences are shown below. Reference sequences are named as PLUS, MINUS, PLU1 (plus1), and MIN1 (minus1), etc. in order to indicate the orientation of DNA strand. A transition between the plus and minus strands within a breakpoint indicates that such a rearrangement causes an inversion. Microhomologies found at the breakpoints are boxed in red, whereas short sequences insertions at the breakpoints with no microhomology are boxed in black. (A) BAB3105. Six breakpoint junctions were obtained, providing information about their relative positions in the rearranged chromosome. In breakpoints #3–6, novel sequence insertions, ranging from 54–1542 bp, were identified at the fusion point. (B) BAB2870. In breakpoint junction #2, a 406 bp and a 90 bp segments copied from nearby genomic sequences are inserted. There is one 8 bp novel sequence inserted within the 90 bp segment (green). See also Figure S3.
Breakpoint junction sequence also revealed that novel sequence insertions were frequently observed at junctions (5/6), with four of them being relatively long sequences (54–1542 bp) (Figure 4A). These insertions are all non-random sequences because their sequences match to either a single interval or a joining of two intervals from chromosome 9 in the reference genome. Microhomology and inversion are also frequently observed in this subject. For example, the breakpoint #5 in Figure 4A was a head-to-tail junction of gained genomic sequences corresponding to the purple and the grey segments (Figure 4A), whose mapping in the reference genome were located ~34 Mb apart. A 1542 bp sequence was inserted at the junction. The 1542 bp insertion consists of two segments: one 1524 bp segment that matches to a region on the opposite DNA strand ~ 1500 bp distal to the reference purple arrow sequence and one 18 bp segment that matches to a region on 9p13.3. Both of the two segments were inserted into the junction with flanking microhomology. The constellation of the above features strongly suggests a replicative mechanism for formation of the highly complex chromosome catastrophe event in BAB3105. A potential replication fork collapse at 9q21 could explain the breakpoint clustering therein.
The parental aCGH analysis (Figure 3B) and chromosome analyses (data not shown) independently confirmed that all the highly complicated rearrangements on chromosome 9 were de novo events in the patient. Further experiments were performed to narrow the time frame when these catastrophic rearrangements occurred. High density informative SNP array analysis in the trio suggested that the additional copies of the duplicated and triplicated genomic segments were all (8/8) derived from the paternal allele that was not transmitted (Figures 3C–D and S2G), indicating that the rearrangements arose in the father, either in early development as a postzygotic event, or in germline during spermatogenesis. PCR analysis using patient-breakpoint-junction-specific primers and paternal lymphoblast DNA as template failed to yield the junction product, suggesting that the father, at least in the blood tissue, was unlikely to be somatically mosaic for the rearrangements (data not shown). In aggregate, these data suggest that the CGR in BAB3105 likely occurred during spermatogenesis in the father.
Patient BAB3104 was referred for CMA due to hypotonia and epilepsy. In total, there were seven copy number changes and 14 breakpoints over a 21.8 Mb interval (Figures 5A–B and S3A). Multiple FISH experiments demonstrated extensive structural changes occurring in chromosome 22 (Figures S3B–G). The proposed structure and derivation resulting in the CGR are illustrated in Figure 5C.
Figure 5.
Highly complex rearrangements in BAB3104. (A) Copy number profiles in BAB3104 revealed by aCGH. Deletions are shown in green and duplications in red. (B) Nimblegen 2.1M aCGH plot in BAB3104. Note that a 23 kb copy number normal region is indicated by an arrow. (C) The rearrangement structure in BAB3104 proposed based on multiple FISH results. To the left is an ideogram of a normal chromosome 22. Shown in middle is the proposed replicative process that produced the genomic imbalances. The segments involving rearrangement are indicated by colored brackets. The figure to the right shows the derivative chromosome 22 with the segments involved in the final order based on FISH analyses. The mapping positions of FISH BAC RP11- probes used are indicated. The orientation of these segments is not certain. See also Figure S4.
The rearrangements studied from two other patients also presented localized but extensive chromosomal copy number and structural changes. Patient 1, an 11-year-old girl with severe developmental delay and intellectual disability, had a de novo complex rearrangement including four copy number changes – two deletions and two duplications between bands 1q32.2 and 1q43, spanning the distal half of the long arm of chromosome 1 (Figures 6A and S4A–B). All four copy number changes were separated by normal copy number segments in their original positions. Data from partial chromosome and metaphase FISH analyses suggested additional inversion and revealed that the two duplicated segments were brought into proximity after rearrangement (Figures 6A and S4C–G). Patient 2, a 3-year-old girl with an epilepsy disorder who was referred due to abnormal chromosome findings, had two losses and five gains interspersed along the entire long arm of chromosome 22 (Figures 6B and S4H). Retrospective chromosome analysis (Figure S4I) and FISH results (data not shown) showed that the rearranged chromosome 22 was nearly metacentric in appearance without stalk and satellites and that the duplicated segments on the long arm were translocated into the short arm.
Figure 6.
Highly complex rearrangements in patient 1 and patient 2. Copy number profiles and schematic drawings of the genomic rearrangements are shown on chromosome 1q for patient 1 (A) and chromosome 22q for patient 2 (B). Structural changes are revealed by FISH or G-banded chromosome analyses. Insertional translocations are indicated by arrows. Inversion in patient 1 is indicated by a curly bracket. See Figure S5 for aCGH, FISH, and chromosome analyses data.
The common gestalt of a region-focused complex rearrangement and breakpoint clustering, with multiple CNVs and other structural changes that is observed in these subjects bears a striking resemblance with the phenomenon of chromosome shattering (chromothripsis) recently reported in 2–3% of all cancers (Stephens et al., 2011).
Low-copy repeats are enriched in the breakpoints with highly complicated rearrangements
Genome architecture incites genomic instability. Low-copy repeats (LCR) represent paralogous segments of the human genome usually greater than 10 kb in length with > 97% sequence identity (Stankiewicz and Lupski, 2002). LCR have been shown to mediate recurrent rearrangements by NAHR and stimulate nonrecurrent rearrangements (Carvalho et al., 2009; Stankiewicz et al., 2003). To examine whether the LCRs might potentially incite instability of a region during CGR formation, we analyzed genomic DNA sequences of breakpoint intervals, as delineated by high resolution arrays, for the presence of segmental duplications (repeat sequences of > 1 kb and > 90% identity; Bailey et al., 2002) or other short repeats. Of the 116 segments encompassing 658,119 bp of reference genome sequence that were analyzed, six segmental duplications (91,782 bp) and seven short repeats from the self-chain tracks in the UCSC genome browser, i.e. representing both direct and inverted repeats that can be shorter than the 1 kb, were identified in 13 breakpoints of seven cases (Figure S5 and Table S2). The percentage of segmental duplication in these breakpoint regions (13.95%) is significantly higher than the genome-wide average (5.53%) (Chi square test, P<0.0001). Most of the repeat-associated breakpoints (8/13) were found in the highly complicated rearrangements with four or more copy number changes. Noteworthy, paralogous LCR pairs, i.e. homologous sequences that were probably derived from a common ancestral segment (Bailey et al., 2002), were identified flanking the duplication proximal and adjacent to a triplication in BAB3032 and were inversely orientated. In addition, two inversely orientated LCRs were found flanking a normal copy sequence between two duplicated regions in BAB3105.
Breakpoint junction sequencing analyses in eight CGRs
Long range PCR and DNA sequencing of the breakpoint junctions were attempted for all novel joints with the hypothesis that examination of rearrangement products could allow inferences regarding potential mechanisms for their formation. Besides the six junctions sequenced from BAB3105, fourteen breakpoint sequences were obtained from eight other patients with CGRs, many of which revealed the presence of microhomologies, inversions (the fusion of two reference sequences belonging to opposite DNA strands), short DNA sequence inserts, or additional small copy number gains at the breakpoint junctions.
CGRs in four individuals (BAB2760, 2780, 2783 and 2785) had an interstitial nml-dup-nml-dup-nml pattern. Breakpoint sequencing revealed that the CGRs in these four patients share the same configuration in their final structures: one copy of the duplicated segment is inserted in an inverted orientation in between the two copies of the other duplicated segments (Figure 4B and S6A–B) (Carvalho et al., 2011). For the CGRs in individuals BAB2760 and 2783, sequences of one breakpoint showed that the rearrangement occurred in two paralogous Alu repeats in an inverted orientation (Figure S6A). The lengths of perfect homology at the breakpoints in these two Alu elements are below the length of minimum efficient processing segment required for homologous recombination, making NAHR unlikely to be the possible mechanism. The sequence homology between Alu elements can serve as microhomology and may facilitate template switch during a replicative process. Intriguingly, additional complexities were identified in the breakpoints in two CGRs. In BAB2780, two segments flanking the proximal duplicated interval, 90 bp and 406 bp in size, were found adjacent to each other in one breakpoint. In addition, an 8 bp novel sequence was inserted within the 406 bp segment (Figure 4B). In BAB2785, an 89 bp segment is inserted within one of the breakpoints, resulting in triplication of this segment (Figure S6B). Next and proximal to this segment is a 13 bp fragment, which could be a sequence synthesized during the repair of this breakpoint, or a result of complex joining of two shorter sequences that could be copied from multiple genomic intervals located within the rearranged chromosome region. These additional small-scale complexities may reflect that a low-processivity polymerase, perhaps allowing iterative template switches, could have been used to initiate the replicative repair when the replication fork collapsed (Hastings et al., 2009a).
Individual BAB2778 had a duplication followed by a normal copy sequence, then a terminal duplication in a ring chromosome 6. DNA sequence was obtained for one breakpoint showing that one copy of the two duplicated segments was adjacent to another consistent with an inverted segment (Figure S6C).
Two types of structures were revealed in three patients with triplications. In BAB3032 and 3015, the triplicated segments were inserted in an inverted orientation between the two copies of the duplicated segments: i.e. dup-trp/inv-dup (Figure S6D) (Carvalho et al., in press); whereas in BAB3050, the triplicated segment was apparently in the same orientation as the duplicated segments (Figure S6E). One pair of inverted LCRs flanks the first duplicated segment in the CGR in BAB3032.
Although the cases with breakpoint sequences presented above do not have as many copy number changes as the four chromothripsis-like CGR cases, most of them present evidence of breakpoint clustering, i.e. dispersed genomic fragments involving breakpoints are jumbled together forming a junction that apparently contains a medley of multiple genomic fragments, which is also a key feature of chromothripsis (Stephens et al., 2011). This may potentially be a vestige of replicative repair. It also suggests that the complex joining is a result of one single catastrophic event, as opposed to progressive rearrangement, in which the expectation would be that the individual duplications or deletions arise independently at their original loci as simple deletions or tandem duplications.
Clinical phenotypes of patients with complex rearrangements
The complex rearrangements were identified among patients referred by clinical geneticists for genomic studies using chromosomal microarray analysis, i.e. CMA. Most were referred because of developmental delay (DD) in attaining milestones in motor and language development (10/12). Clinical information from 14 patients is summarized in Table S1. The combination of intellectual disability, failure to thrive, behavioral problems, dysmorphic features and congenital anomalies was present in > 4/12 subjects. However, DD was observed in 12/12 patients (Table S1).
We hypothesized that the rearrangements with large combined copy number changes may be de novo and associated with more severe phenotypes (Table 1). Indeed, of the ten cases with combined copy number changes larger than 2.5 Mb, six rearrangements were de novo events and the other four had either no or incomplete parental studies. These de novo cases were associated with severe DD/intellectual disability, epilepsy, and dysmorphic features. In contrast, in the seven cases with relatively smaller copy number changes (0.4 Mb – 2.2 Mb), four rearrangements were inherited and the other three had incomplete parental information. Interestingly, in each of these CGRs, only copy number gains were present, and most of these patients showed a comparatively milder phenotype such as moderate DD. It is unclear at the present time whether these relatively smaller complex rearrangements are clinically significant. The fact that four rearrangements were maternally inherited, but none were paternally inherited is consistent with the previous observations in complex chromosomal rearrangements that familial transmission is mainly observed through female carriers (Batista et al., 1994).
As noted for cancer chromothripsis, such complex genomic rearrangement can have “multigenic” consequences wherein many genes have an altered dosage or copy number, others are potentially disrupted, and there may be novel gene fusion formed at the multiple breakpoints. Perhaps the observed common clinical phenotype of neurodevelopmental delay reflects multiple potential gene disruptions or dosage alterations by CGR and the large number of genes in the human genome that contribute to neurological function.
DISCUSSION
Do somatic chromothripsis events occur in a mechanistically similar manner to constitutional CGR?
Recently, the phenomenon of chromosome shattering termed chromothripsis, i.e. a massively complex genomic rearrangement which occurs in a single catastrophic event involving local apparent shattering of a chromosome and subsequent reassembly proposed to be potentially mediated by non-homologous end joining, was described in 2–3% of all cancers analyzed (Stephens et al., 2011). This phenomenon was also reported in the germline (Borg et al., 2005; Kloosterman et al., 2011). The attractively simple idea that chromothripsis stems from the shattering of chromosomes or regions of chromosomes, and their re-ligation with scant regard for their site of origin, is based on data obtained by next generation paired-end sequencing. This technique provides excellent data on the relative positions of sequences, but cannot directly reveal copy number dosages. Whereas next generation sequencing can infer copy number dosage based on “depth of read coverage”, in the analysis the raw read length data are first processed through matching to a reference haploid human genome build; a filtering process notoriously challenged by low copy repeats, repetitive sequences, and other sequence complexities.
By use of a combination of molecular techniques, we have obtained descriptions of chromosomes that have experienced a chromothripsis-like process that includes full detail of copy number, position and orientation in non-cancerous patients. This has revealed that, in addition to inversion and translocation, there is extensive duplication and triplication of sequence. We also show that some novel junctions have microhomology and small-scale complexity in the form of apparent templated insertion of fragments of nearby sequence at the junctions. These features are characteristic of events that we have previously attributed to postulated replicative processes of chromosomal structural changes, long-distance template-switching by FoSTeS/MMBIR; the latter apparent insertional complexity near the junctions can be attributed to a synthesis product resultant from a new low-processivity fork as proposed for the MMBIR model (Hastings et al., 2009a). Interestingly, DNA polymerase(s) involved in break-induced replication (BIR) may also have poor fidelity (Deem et al., 2011; Hicks et al., 2010).
Although a duplication might potentially result from “chromosome shattering” and re-ligation, either because the chromosomes have replicated, or by involvement of both homologues, explanation of triplication in this way would become complicated. Furthermore, such a “shattering and re-ligation” mechanism predicts the presence of a deletion reciprocal to any duplication, and this is not observed. A more parsimonious explanation for the origin of the extra copies is that they were formed by replication. Although re-replication, the inappropriate firing of replication origins, might explain some cases of over-replication (Doksani et al., 2009; Green et al., 2010), BIR could also lead to over-replication, either by use of ectopic homology or non-homologous processes using microhomology to anneal single-strands that act as primers for DNA replication. Thus, MMBIR provides an explanation for duplication and triplication as well as deletion, inversion, and translocation. In addition, MMBIR can explain both the observed microhomology at selected breakpoints and the insertion of short segments flanked by microhomology around the junctions. Can MMBIR also account for the multiple breaks over extended regions described in chromothripsis?
We have previously pointed out that replication forks formed by BIR are believed to differ from those formed at an origin in that they might involve a Holliday junction (Hastings et al., 2009a). The Holliday junction might be resolved at the time of formation of the replication fork, or it might follow the fork. If a replication fork stalls, a following Holliday junction might process through the fork leading to fork collapse. This was offered as the origin of discontinuities on the scale of hundreds of kilobases or megabases seen in many duplications. Another way in which BIR differs from origin-dependent replication is that it appears, at least in some cases, to replicate to the telomere, thereby ignoring intervening replicon signals. This was shown in yeast in the “break copy” duplication model, wherein a foreign chromosome fragment can invade into an endogenous target chromosome and initiate a unidirectional replication fork that proceeds to duplicate all sequences distal to the site of initiation (Morrow et al., 1997). Smith and Symington later showed that BIR can go all the way from near the centromere to the end of the chromosome arm in yeast (Smith et al., 2007). Multiple template-switches using homologies or microhomologies is also a signature of BIR (Hicks et al., 2010; Smith et al., 2007). From these considerations, one may argue that, once a replication fork has collapsed, and been restarted by BIR, chromosomal structural changes of all kinds can form over distances of tens of megabases of sequence on the same chromosome. In this context, it is also of interest to note both the observed long arm and the distal chromosome “preference” of the constitutional CGR events we described here. Viewed from this mechanistic perspective, chromothripsis is less likely to represent a “blowing apart” of a chromosome and putting the pieces of the puzzle together that is peculiar to a cancer cell (Stephens et al., 2011), but rather may reflect an inherent cellular DNA replicative/repair process for maintaining genome stability. Perhaps the phenomenon termed chromothripsis might be better referred to as “chromoanasynthesis” (chromosome reconstitution or chromosome reassortment).
Time frame of formation of the chromosome catastrophe CGR event
As indicated by Stephens et al., chromothripsis occurs in a single catastrophic event, rather than in a step-wise cumulative way (Stephens et al., 2011). Furthermore, recent studies of multiple myeloma identified large numbers of genomic rearrangements with the hallmarks of chromothripsis in 1.3% of samples (Magrangeas et al., in press). This catastrophic event confers a poor outcome as indicated by shortened time to relapse and death within two years. Several lines of evidence support the contention that in the constitutional CGR cases in our cohort, in particular the four chromothripsis-like cases, the majority of rearranged genomic intervals also arose as one singular event. First, the multiple rearrangements are localized to a focused region on one homologue of a chromosome, most frequently in the long arms. In a mutation accumulation model, the rearrangements would be expected to disperse randomly over the genome. Second, there is no evidence of differential level of mosaicism among individual rearrangements, which would be the expectation when new rearrangements accumulate as cells proliferate. Third, clustering and juxtaposition of breakpoints indicate that these breakpoints were interrelated when they were formed. Notably, inversion of genomic segments gives proximity of what appears to be distant breakpoints (Branzei and Foiani, 2010; Carvalho et al., in press; Futcher, 1986).
If replicative mechanisms are used, CGR can occur during DNA replication in gametogenesis and, in some cases, as postzygotic events (Zhang et al., 2009b). Here, we provide evidence for examples of both types (BAB3105 and 3012). Given the resemblance of key features between constitutional CGR and cancer chromothripsis, we propose that such chromosome catastrophe events can occur by essentially similar mechanisms in a variety of stages in an organism’s life cycle.
In summary, genomic studies of rearrangements associated with cancer and genomic disorders reveal unanticipated complexities. Such observations have important ramifications, such as inducing multigenic changes in a singular event, with implications for both cancer genesis and species evolution. Elucidating the mechanism underlying such apparent “one-off” events is essential to understanding mutational processes. Our findings in CGR suggest replicative processes may be operative in both genomic disorders and cancers.
EXPERIMENTAL PROCEDURES
Patients
Patients with complex rearrangements were identified by CMA clinical testing at Baylor College of Medicine (BCM) MGL. Patient samples chosen for further higher resolution genome analysis studies had evidence for either a triplication or at least two separate copy number changes in one chromosome, with no additional clinically significant genomic findings in other chromosomes. In addition, rearrangements involving a terminal deletion in one arm and a terminal duplication in the other arm of the same chromosome were not included because they tend to be products of unbalanced translocation. In all, 17 cases were included in this study out of over 23,000 CMA cases studied in the MGL. Clinical information was collected after procuring informed consents approved by the Institutional Review Board for Human Subject Research at BCM.
CMA using clinical BCM arrays
Clinical CMA was performed on custom designed Agilent arrays as described (Boone et al., 2010; Cheung et al., 2005; Lu et al., 2007; Ou et al., 2008). Patients were identified either by the BAC emulation oligo-based arrays with at least one BAC clone for one chromosome band at a resolution of 550 band level, or by the oligo-based arrays with one probe for every 30 kb interval.
High density aCGH
To characterize further the complex rearrangements identified by CMA, two customized Agilent arrays were designed including an 8X 60K array format for investigation of the rearrangements in seven patients, BAB2760, 2778, 2785, 3012, 3015, 3047, and 3050, and a 4X180K array format for four patients BAB2780, 2783, 3032, and 3103. The designed probe density was 2–4 oligonucleotides per kb for intervals with copy number changes and breakpoint regions as assessed by transitions between two separate copy number states. The high density arrays also interrogate the flanking genomic regions of up to 5 Mb in size with probe density of 1–3 oligos per kb.
Patients BAB2780, 2920, 3011, 3050, 3104, 3105, and patients 1 and 2 were analyzed by Nimblegen 2.1M or 4.2M arrays. Patient BAB3104 was also analyzed by a whole genome 244K array (Agilent Technologies, Inc); assays were performed according to the manufacturers’ instructions.
Fluorescence in situ hybridization
After identification of a complex rearrangement by clinical CMA, confirmatory FISH analyses were performed on Phytohemagglutinin-stimulated cultured blood cells using standard procedures (Cheung et al., 2005). For the two-color FISH analyses the test probes were directly labeled by nick-translation with rhodamine (red), while the control probes were directly labeled with FITC (green). To confirm a deletion detected by CMA, ten metaphase cells were examined using the centromere probe as a control. To confirm a copy number gain detected by CMA, 50 interphase nuclei were examined. A region was considered to be duplicated when two red signals, or triplicated when three signals were observed in more than 70% of the cells examined. Metaphase cells were also examined to determine chromosome/genome position and investigate whether the duplicated segments were translocated to a different location.
SNP array analysis
SNP array analysis was performed on Illumina Infinium HD assay platform using HumanOmni1-Quad BeadChip (Illumina Inc.) DNA whole genome amplification, fragmentation, hybridization, enzymatic single base extension, slides staining and washing were performed according to the manufacturer’s instructions (Illumina, Inc.). The Illumina iScan System was used for image registration, image extraction and data output. The GenomeStudio software (Illumina Inc.) was used for genotyping data normalization, genotype calling, clustering, data intensity analysis, calculation of Log R ratio and B-allele frequency. Illumina CNV partition statistical algorithm was used for copy number variation and copy number neutral absence of heterozygosity analyses. The reference model file provided by Illumina Inc. was used as a reference for data analysis. Parent of origin was determined by comparison of proband’s and parental genotypes and B-allele frequencies for SNPs in the rearranged regions on chromosome 9.
Breakpoint analysis
Genomic coordinates for potential breakpoints were estimated from aCGH interrogating oligonucleotides revealing gain/no gain or loss/no loss copy number transitions. Rearrangement breakpoints located in subtelomeric or pericentromeric regions were not included in this analysis. The presence of LCRs was revealed by examining the segmental_dups track in the UCSC genome browser (assembly hg19), while the presence of both direct and inverted short repeats was revealed by examining the self_chain track.
The breakpoint junctions were amplified by long-range PCR using the TAKARA LA Taq™ kit (TAKARA Bio Inc.) as previously described (Zhang et al., 2009b). PCR products were sequenced by the Sanger dideoxy method and DNA sequences were compared to the human genome reference assembly (hg19).
Supplementary Material
HIGHLIGHTS.
Constitutional CGR share features with chromothripsis observed in cancer cells
CGR can involve replication-based mechanisms
Chromosome catastrophe can occur throughout an organism’s life cycle
Acknowledgments
We thank the patients and their families for participation in this research, the Molecular Cytogenetics Laboratories in the MGL at BCM (https://www.bcm.edu/geneticlabs) for providing technical support, Dr. Katarzyna Derwinska for helpful discussion, and Roche NimbleGen, Inc. for supplying NimbleGen aCGH materials. This work was supported in part by National Institute of Neurological Disorders and Stroke, NIH grant R01 NS058529 to J.R.L., the National Institutes of General Medical Science grant R01GM064022 to P.J.H., and the BCM IDDRC P30HD024064 funded from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. A.E. is supported by DK081735-02 and SCSN is supported by a fellowship grants by the LCRC from the Osteogenesis Imperfecta Foundation and the National Urea Cycle Disorders Foundation.
Footnotes
Disclosure J.R.L. is a consultant for Athena Diagnostics, has stock ownership in 23 and Me and Ion Torrent Systems, and is a coinventor on multiple United States and European patents for DNA diagnostics. The Department of Molecular and Human Genetics derives revenue from clinical testing by high resolution human genome analysis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Batista DA, Pai GS, Stetten G. Molecular analysis of a complex chromosomal rearrangement and a review of familial cases. Am J Med Genet. 1994;53:255–263. doi: 10.1002/ajmg.1320530311. [DOI] [PubMed] [Google Scholar]
- Bi W, Sapir T, Shchelochkov OA, Zhang F, Withers MA, Hunter JV, Levy T, Shinder V, Peiffer DA, Gunderson KL, et al. Increased LIS1 expression affects human and mouse brain development. Nat Genet. 2009;41:168–177. doi: 10.1038/ng.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone PM, Bacino CA, Shaw CA, Eng PA, Hixson PM, Pursley AN, Kang SH, Yang Y, Wiszniewska J, Nowakowska BA, et al. Detection of clinically relevant exonic copy-number changes by array CGH. Hum Mutat. 2010;31:1326–1342. doi: 10.1002/humu.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg K, Stankiewicz P, Bocian E, Kruczek A, Obersztyn E, Lupski JR, Mazurczak T. Molecular analysis of a constitutional complex genome rearrangement with 11 breakpoints involving chromosomes 3, 11, 12, and 21 and a approximately 0.5-Mb submicroscopic deletion in a patient with mild mental retardation. Hum Genet. 2005;118:267–275. doi: 10.1007/s00439-005-0021-0. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Leaping forks at inverted repeats. Genes Dev. 2010;24:5–9. doi: 10.1101/gad.1884810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho C, Bartnik M, Pehlivan D, Fang P, Shen J, Lupski J. Evidence for disease penetrance relating to CNV size: Pelizaeus-Merzbacher disease and manifesting carriers with a familial 11 Mb duplication at Xq22. Clin Genet. 2011 doi: 10.1111/j.1399-0004.2011.01716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Ramocki MB, Pehlivan D, Franco LM, Gonzaga-Jauregui C, Fang P, McCall A, Pivnick EK, Hines-Dowell S, Seaver L, et al. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet. doi: 10.1038/ng.944. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Zhang F, Liu P, Patel A, Sahoo T, Bacino CA, Shaw C, Peacock S, Pursley A, Tavyev YJ, et al. Complex rearrangements in patients with duplications of MECP2 can occur by fork stalling and template switching. Hum Mol Genet. 2009;18:2188–2203. doi: 10.1093/hmg/ddp151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JM, Cooper DN, Ferec C, Kehrer-Sawatzki H, Patrinos GP. Genomic rearrangements in inherited disease and cancer. Semin Cancer Biol. 2010;20:222–233. doi: 10.1016/j.semcancer.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Cheung SW, Shaw CA, Yu W, Li J, Ou Z, Patel A, Yatsenko SA, Cooper ML, Furman P, Stankiewicz P, et al. Development and validation of a CGH microarray for clinical cytogenetic diagnosis. Genet Med. 2005;7:422–432. doi: 10.1097/01.gim.0000170992.63691.32. [DOI] [PubMed] [Google Scholar]
- Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, Mathur R, Chabes A, Malkova A. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doksani Y, Bermejo R, Fiorani S, Haber JE, Foiani M. Replicon dynamics, dormant origin firing, and terminal fork integrity after double-strand break formation. Cell. 2009;137:247–258. doi: 10.1016/j.cell.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher AB. Copy number amplification of the 2 micron circle plasmid of Saccharomyces cerevisiae. J Theor Biol. 1986;119:197–204. doi: 10.1016/s0022-5193(86)80074-1. [DOI] [PubMed] [Google Scholar]
- Gajecka M, Saitta SC, Gentles AJ, Campbell L, Ciprero K, Geiger E, Catherwood A, Rosenfeld JA, Shaikh T, Shaffer LG. Recurrent interstitial 1p36 deletions: Evidence for germline mosaicism and complex rearrangement breakpoints. Am J Med Genet A. 2010;152A:3074–3083. doi: 10.1002/ajmg.a.33733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green BM, Finn KJ, Li JJ. Loss of DNA replication control is a potent inducer of gene amplification. Science. 2010;329:943–946. doi: 10.1126/science.1190966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009a;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009b;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Guryev V, van Roosmalen M, Duran KJ, de Bruijn E, Bakker SC, Letteboer T, van Nesselrooij B, Hochstenbach R, Poot M, et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet. 2011;20:1916–1924. doi: 10.1093/hmg/ddr073. [DOI] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Liu P, Erez A, Sreenath Nagamani SC, Bi W, Carvalho CM, Simmons AD, Wiszniewska J, Fang P, Eng PA, Cooper ML, et al. Copy number gain at Xp22.31 includes complex duplication rearrangements and recurrent triplications. Hum Mol Genet. 2011;20:1975–1988. doi: 10.1093/hmg/ddr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Shaw CA, Patel A, Li J, Cooper ML, Wells WR, Sullivan CM, Sahoo T, Yatsenko SA, Bacino CA, et al. Clinical implementation of chromosomal microarray analysis: summary of 2513 postnatal cases. PLoS One. 2007;2:e327. doi: 10.1371/journal.pone.0000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. New mutations and intellectual function. Nat Genet. 2010;42:1036–1038. doi: 10.1038/ng1210-1036. [DOI] [PubMed] [Google Scholar]
- Magrangeas F, Avet-Loiseau H, Munshi NC, Minvielle S. Chromothripsis identifies a rare and aggressive entity among newly diagnosed multiple myeloma patients. Blood. doi: 10.1182/blood-2011-03-344069. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow DM, Connelly C, Hieter P. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics. 1997;147:371–382. doi: 10.1093/genetics/147.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Z, Kang SH, Shaw CA, Carmack CE, White LD, Patel A, Beaudet AL, Cheung SW, Chinault AC. Bacterial artificial chromosome-emulation oligonucleotide arrays for targeted clinical array-comparative genomic hybridization analyses. Genet Med. 2008;10:278–289. doi: 10.1097/GIM.0b013e31816b4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen C, Koszul R, Dujon B, Fischer G. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 2008;4:e1000175. doi: 10.1371/journal.pgen.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet. 2006;2:e48. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Shaw CJ, Dapper JD, Wakui K, Shaffer LG, Withers M, Elizondo L, Park SS, Lupski JR. Genome architecture catalyzes nonrecurrent chromosomal rearrangements. Am J Hum Genet. 2003;72:1101–1116. doi: 10.1086/374385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko SA, Brundage EK, Roney EK, Cheung SW, Chinault AC, Lupski JR. Molecular mechanisms for subtelomeric rearrangements associated with the 9q34.3 microdeletion syndrome. Hum Mol Genet. 2009;18:1924–1936. doi: 10.1093/hmg/ddp114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Carvalho CM, Lupski JR. Complex human chromosomal and genomic rearrangements. Trends Genet. 2009a;25:298–307. doi: 10.1016/j.tig.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009b;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Seeman P, Liu P, Weterman MA, Gonzaga-Jauregui C, Towne CF, Batish SD, De Vriendt E, De Jonghe P, Rautenstrauss B, et al. Mechanisms for nonrecurrent genomic rearrangements associated with CMT1A or HNPP: rare CNVs as a cause for missing heritability. Am J Hum Genet. 2010;86:892–903. doi: 10.1016/j.ajhg.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.