Abstract
Meningococcal disease is a widely distributed complex disease affecting all age categories. It can cause severe meningitis and septicemia, especially in unvaccinated infants and young children. The causative agent, Neisseria meningitidis (Nm), can be phenotypically and genetically differentiated into serogroups and sequence types (STs) and has a highly dynamic population structure. To obtain a deeper understanding of the epidemiology of Nm, we sequenced seven Nm genomes. Large-scale genomic analysis was conducted with these 7 Nm genomes, 27 additional Nm genomes from GenBank, and 4 other Neisseria genomes. We observed extensive homologous recombination in all gene functional categories among different Nm genomes. Homologous recombination is so frequent that it has resulted in numerous chimeric open reading frames, including genes in the capsule biosynthesis cluster and loci targeted by commercial vaccines. Our results reveal that, despite widespread use, evolutionary relationships inferred from the standard seven-gene multilocus sequence typing (MLST) method could not predict virulence gene content or strain phenotype. In fact, up to 28% of the virulence-associated genes could differ between strains of identical STs. Consistent with previous studies, we found that allelic recombination is also associated with alterations in antibiotic susceptibility. Overall, these findings emphasize the extensive genomic plasticity of Nm and the limitations of standard molecular methods to quantify this genotypic and phenotypic diversity.
Keywords: homologous recombination, horizontal gene transfer, genome evolution, multilocus sequence typing, virulence gene
Introduction
Neisseria meningitidis (Nm) is an encapsulated Gram-negative bacterium. Although it is a common commensal with a carriage prevalence of about 20% in human populations (Claus et al. 2005), Nm can cause severe meningitis and septicemia (Stephens et al. 2007). The genetic basis for the remarkable differences in meningococcal virulence is still a matter of investigation (Schoen et al. 2008). Some efforts have been made to identify virulence factors that are associated with the invasive disease (Dunning Hotopp et al. 2006). However, many designated virulence genes in Nm were also found to be present in nonpathogenic species such as Neisseria lactamica (Snyder and Saunders 2006; Bennett et al. 2010). Similarly, commensal and nonpathogenic species serve as reservoirs of virulence alleles and they engage extensively in the exchange of genes and gene pieces, largely via homologous recombination (Nakamura et al. 2004; Marri et al. 2010).
The high frequency of recombination results in a highly dynamic population structure (Holmes et al. 1999). Multilocus sequence typing (MLST) groups sequence types (STs) into clonal complexes (CCs) by their similarity to a central allelic profile (genotype) (Maiden et al. 1998). Since its initial introduction (Maiden et al. 1998), MLST has been embraced for characterizing bacterial variation and facilitating epidemiological investigations (Maiden 2006). Despite the increasing popularity of MLST, it has been recognized that MLST cannot be used to infer phenotypic characteristics. For instance, MLST cannot be used to predict vaccine coverage due to the poor correlation between MLST and antigenic variability (Bambini et al. 2009) and the isolates of identical ST, but distinct serogroup have been described (Yazdankhah et al. 2004). Evolutionary relationships of Nm may be better resolved by using more loci (Didelot et al. 2009) or whole genome approaches. A recent analysis using genomic data has suggested that the Nm species population has evolved into distinct phylogenetic clades larger than the lineages identified by MLST (Budroni et al. 2011).
Despite the wide recognition of homologous recombination in the Nm species, the extent of recombination still demands a systematic investigation. In this study, we sequenced 7 Nm isolates, 6 of which form 3 pairs with identical serotypes and serosubtypes. We then conducted a large-scale comparative analysis on these seven genomes together with additional 31 Neisseria genomes. We attempted to assess the extent of homologous recombination, especially among strains with identical STs and CCs, and address potential functional implications of these recombination events through analysis of genes involved in antibiotic resistance, capsule biosynthesis, and vaccine efficacy.
Materials and Methods
Genomes Sequenced and Analyzed
The seven clinical Nm strains were all blood isolates from cases of invasive disease. Six of them represent three matched pairs (table 1). Paired strains are of identical serotype and serosubtype and were isolated from elderly patients from Ontario, Canada. However, they are temporally distinct and were obtained 4–6 years apart. The seventh isolate, Nm1140, is an unusual invasive strain that could not be serogrouped by serogroup-specific capsular antigen antisera (National Microbiology Laboratory, Public Health Agency of Canada) or real time PCR (Public Health Laboratories, Public Health Ontario; Taha 2000; Corless et al. 2001; Mothershed et al. 2004). Genome sequences for the seven Nm strains were generated via GS-FLX (Roche/454 Life Sciences, Brantford, CT) pyrosequencing. Draft genomes were assembled using the gsAssembler (Roche) and CAP3 (Huang and Madan 1999), and further edited and visualized using the Phred/Phrap/Consed software package (Gordon et al. 2001). The seven draft genomes have been submitted to GenBank and the accession numbers are AGBN00000000 (Nm8187), AGBO00000000 (Nm3127), AGBP00000000 (Nm2732), AGBQ00000000 (Nm6938), AGBR00000000 (Nm6756), AGBS00000000 (Nm8663), and AGBT00000000 (Nm1140). Genome sequences for additional 27 Nm, 1 N. lactamica (Nlact), and 3 Neisseria gonorrhoea (Ng) strains were obtained from GenBank (supplementary table S1, Supplementary Material online).
Table 1.
Nm Isolates Sequenced in the Study
| Isolates | Serogroup | Serosubtype | ST | CC | Year | Patient Age/Gender | Penicillin MIC (μg/ml) |
| Nm8187 | Y | NT/P1.5 | 3,923 | 167 | 2002 | 77/M | 0.12 |
| Nm3127 | Y | NT/P1.5 | 3,980 | 167 | 2006 | 73/F | 0.50 |
| Nm6938 | W135 | NT/P1.6 | 22 | 22 | 2001 | 72/M | ≤0.06 |
| Nm2732 | W135 | NT/P1.6 | 22 | 22 | 2007 | 96/F | 0.12 |
| Nm8663 | Y | 14/P1.- | 23 | 23 | 2002 | 80/M | ≤0.06 |
| Nm6756 | Y | 14/P1.- | 23 | 23 | 2008 | 73/M | ≤0.06 |
| Nm1140 | NG-cnla | — | 1,136 | 1,136 | 2009 | 10/M | ≤0.06 |
NG, not groupable; cnl, capsule null locus.
Virulence Genes and Homolog Identification
A set of 177 putative virulence genes in the Neisseria genus was originally identified in Marri et al. (2010) by searching published literature and available databases. Out of these 177 genes, 164 genes are present in the Nm species, 153 of them are present in the MC58 genome (Tettelin et al. 2000), and the remaining 11 are absent from MC58 but present in the Z2491 genome (Parkhill et al. 2000). Sequences of these 153 MC58 genes and 11 Z2491 genes were extracted from the reannotated genomes in the NeMeSys database (Rusniok et al. 2009) and used as the query sequences to identify virulence genes present in other genomes (for a complete list of these virulence genes, see supplementary table S2, Supplementary Material online). The nucleotide sequences of the examined genes were used as queries in a BlastN search (Camacho et al. 2009). Homologues of these genes were identified using BlastN with an E-value <10−20 and match length >85%. To identify orthologues, we required genes present as single-copy per genome.
Multilocus Sequence Typing
A standard seven-locus scheme (Maiden et al. 1998) was used to determine multilocus ST. The determination of STs was conducted on the Neisseria MLST website (http://pubmlst.org/neisseria/) developed by Jolley and Maiden (2010) and based at the University of Oxford, UK.
Determination of Nonvertically Acquired Genes between Identical-ST Genomes
We assumed that, if the strains from the same ST are truly clonal, all nucleotide differences within the same ST should be solely due to point mutations. Given the identical seven MLST loci, we can estimate an upper bound of nucleotide divergence for these genome pairs. If these mutations are independent random events, under a binomial distribution, the P value to observe no change in a 3288-nucleotide sequence (the total concatenated length of the seven MLST sequences) is 0.001 when the divergence is set to be 0.0021 (and is 0.05 when the divergence is 0.00091). We assumed that the divergence is 0.0021 between any two genomes from the same ST, and genes that had more than the expected number of nucleotide changes at a significance level of 0.001 were deemed as nonvertically acquired genes. All statistical tests were conducted using the R package (R Development Core Team 2009). It is important to note that the significance level of 0.001 was chosen to obtain a more conservative estimate for the identification of nonvertical genes, and the use of a relaxed significance level of 0.05 would have resulted in the identification of more nonvertical genes.
Simulations were performed to assess the robustness of the nonvertical genes determination. In brief, the MC58 whole genome sequence was used as a reference genome and substitutions were randomly introduced as in the Jukes–Cantor model (Jukes and Cantor 1969) to generate “derived” genomes and, as done in Hao and Golding (2008), no indels were allowed. Nucleotide sequences of the MC58 single-copy genes (1822 in total) were used as query sequences in a BlastN search (Camacho et al. 2009) against the derived genome sequences, and the best Blast hits were collected as orthologues for further analysis.
Phylogenetic Analyses
Homologous sequences were aligned using MUSCLE (Edgar 2004). DNA distance among sequences was measured using dnadist of the PHYLIP package (Felsenstein 1989) version 3.6. The taxon with the shortest phylogenetic distance was identified as the nearest neighbor (Koski and Golding 2001). Nucleotide diversity (Tajima's π), which is the average number of nucleotide differences per site between any two DNA sequences from the sample population (Tajima 1983), was calculated using the BioPerl Perlymorphism module (Stajich and Hahn 2005). Phylogenetic trees were constructed using a maximum likelihood method via the RAxML program (Stamatakis 2006) under a GTR + Γ + I substitution model.
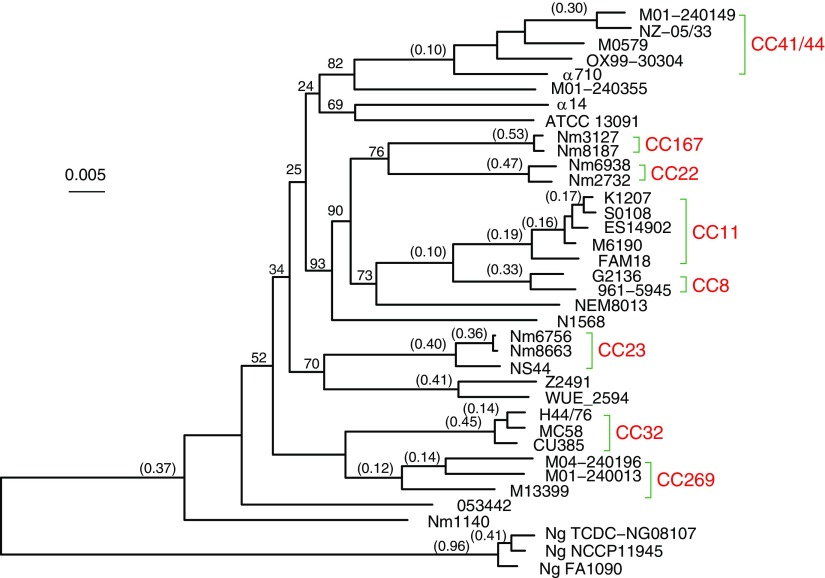
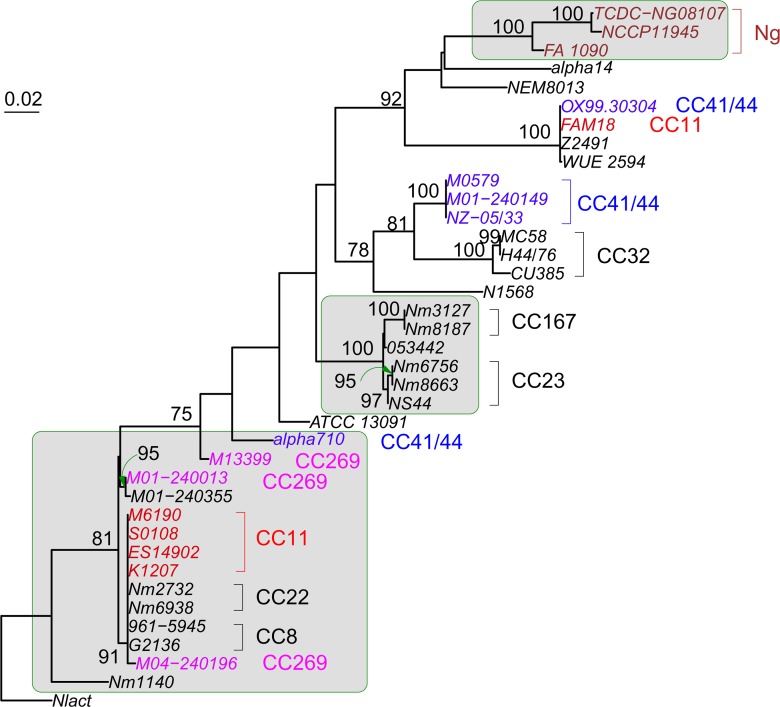
There were 1072 single-copy genes universally present in all 38 examined Neisseria genomes (for a complete list of the 1,072 genes, see supplementary table S3, Supplementary Material online). The phylogeny of the concatenated sequences of the 1,072 genes (992,796 characters) was constructed using the RAxML program (fig. 1). Five hundred bootstrap replicates were generated. Furthermore, a maximum likelihood tree was constructed for each individual gene using the RAxML program, and the proportion of individual gene trees that support each branch of the concatenated 1072-gene tree was calculated and shown in figure 1.
FIG. 1.—
Maximum likelihood tree constructed using the concatenated sequences of 1,072 universally present genes. The tree was rooted using N. lactamica as the outgroup (not shown). Five hundred bootstrap replicates were generated, and only the bootstrap values <99% are shown. A maximum likelihood tree was also constructed for each individual gene and the proportion of genes (out of 1,072) that support each branch was calculated and shown in parentheses only for the relatively well-supported branches (with a support ≥0.1). CCs are red.
Phylogenetic Incongruence and Intragenic Recombination
Phylogenetic incongruence between different genes was examined by the approximately unbiased (AU) test (Shimodaira 2002). The AU test was first performed on virulence genes. Because the AU test requires the same set of taxa present for both gene sequences, to obtain a more comparable interpretation, the AU test was restricted to only the virulence genes that are universally present in the 38 studied genomes (61 in total). The AU test was also performed on the 1,072 individual universal genes against the concatenated 1072-gene tree. To reduce any concern about phylogenetic uncertainty due to low sequence diversity, the AU test was performed on the same number of “super-genes” (1,072 for universal genes and 61 for universally present virulence genes) generated by randomly concatenating 3, 6, or 9 corresponding genes. The site-by-site likelihoods for the trees were calculated with PUZZLE (Strimmer and von Haeseler 1996), the AU test was then implemented using CONSEL (Shimodaira and Hasegawa 2001) to assign the tree probabilities.
The network-like relationship was constructed using the SplitsTree program with Neighbor-Net distance transformation (Huson and Bryant 2006). ClonalFrame 1.1 (Didelot and Falush 2007) was employed to estimate the ratio of probabilities that a given site is altered through recombination and mutation (the r/m ratio). Intragenic recombination was detected using Phi (Bruen et al. 2006), RDP (Martin and Rybicki 2000), and OnePop (Hao 2010). Three tests (NSS, Max χ2, and Phi) were performed using the Phi package. The other two programs OnePop and RDP make use of essentially the same methodology, but OnePop removes the examined region from the calculation of sequence divergence (for more details, see Hao 2010) to improve recombination detection power among closely related sequences. To be conservative, when using the Phi package, significance was determined only when the P values in all three tests are <0.05. Significant P values in RDP and OnePop are required to be <0.05 after stringent Bonferroni correction for multiple tests.
Antimicrobial Susceptibility Testing
Antimicrobial Susceptibility Testing (AST) using the agar dilution method was performed according to the CLSI M7-A7 method standard (CLSI 2006). Test inocula were prepared from a direct colony suspension obtained after 24 h growth on sheep blood agar (incubated at 35 °C in 5% CO2). Colonies were suspended in brain heart infusion broth to a 0.5 McFarland standard (108 CFU/ml). A final cell density of approximately 104 CFU/spot was delivered onto Mueller–Hinton agar supplemented with 5% sheep blood by using a replicator and was incubated at 35 °C in 5% CO2 for 24 h prior to determination of minimum inhibitory concentrations (MICs). Isolates were tested for susceptibility to penicillin (0.004 to 8.0 μg/ml) at the concentrations noted (table 1), and interpretive criteria were used according to the CLSI document M100-S21, 2011 (CLSI 2011). Testing was repeated on a different day and yielded identical results.
Results and Discussion
Population Structure and Phylogenetic Incongruence
On the concatenated 1072-gene tree, branches leading to the individual CCs are all supported by a 100% bootstrap value, whereas most branches representing deeper relationship among the CCs are poorly resolved (fig. 1). Phylogenetic networks based on the concatenated sequences revealed a similar picture such that the strains from the same CC are closely related and form monophyletic groups, yet the overall topological structure is star-like, suggesting poorly resolved deep phylogenetic relationships among the CCs (supplementary fig. S1, Supplementary Material online). This supports the view that Nm population has experienced selective sweeps, rapid population growth, and very high levels of recombination, all of which remove phylogenetic signals and result in a star-like phylogeny (Smith et al. 1993). Unlike the bootstrap support, the support by individual gene trees was universally low except for a value of 0.96 on the branch leading to the Ng species (fig. 1). This suggests that most individual gene trees are topologically different from the concatenated gene tree. For instance, the branch leading to the Nm species is supported by only 37% of individual gene trees. In other words, about 63% of genes support a closer relationship between a fraction of the Nm strains and either Ng or Nlact rather than a monophyletic origin of Nm. The support observed for individual CCs ranges from 0.12 (CC269) to 0.53 (CC167). The AU test was applied on each universal gene against the concatenated 1072-gene tree (supplementary table S4, Supplementary Material online). One thousand forty-one genes (97.11%) showed significant incongruence (P < 0.01) with the concatenated 1072-gene tree. Furthermore, a significant amount of universally present genes were involved in intragenic recombination. The detected proportion of genes involved in intragenic recombination is 48.51% by RDP, 66.51% by Phi, and 79.48% by OnePop (table 2). All together, it suggests extensive homologous recombination at both the gene and intragenic levels among the Nm genomes.
Table 2.
Genes Involved in Intragenic Recombination Detected by Various Programs
| Recombinants |
|||||
| Genesa |
Total | RDPb | Phic | OnePopb | |
| Universal | Total | 1,072 | 520(48.51%) | 713(66.51%) | 852(79.48%) |
| Information storage and processing | 206 | 103(50.00%) | 131(63.59%) | 152(73.79%) | |
| Cellular processes and signaling | 194 | 100(51.55%) | 140(72.16%) | 160(82.47%) | |
| Metabolism | 382 | 215(56.28%) | 288(75.39%) | 338(88.48%) | |
| Poorly characterized | 194 | 79(40.72%) | 123(63.40%) | 150(77.32%) | |
| Not in COG | 123 | 35(28.46%) | 56(45.53%) | 78(63.41%) | |
| Virulence | Total | 164 | 82(50.00%) | 95(57.93%) | 117(71.34%) |
| Universally present | 61 | 32(52.46%) | 37(60.66%) | 52(85.25%) | |
Universal genes were grouped according to their Clusters of Orthologous Groups of proteins (COG) functional categories. Information storage and processing includes COG categories J, A, K, L, and B; cellular processes and signaling includes COG categories D, Y, V, T, M, N, Z, W, U, and O; Metabolism includes C, G, E, F, H, I, P, and Q; Poorly characterized includes R and S. Because some genes have been annotated into multiple COG categories, the sum of individual groups is slightly larger than the total number of single-copy genes.
After Bonferroni correction for multiple tests at the 0.05 level.
Significance was determined when P < 0.05 was observed in all three tests (NSS, Max χ2, and Phi) in the Phi package.
Similarly, a high frequency of recombination was observed within the Ng species. Out of the 1,072 genes, there are 436 genes supporting the clustering pattern ([TCDC-NG08107, NCCP11945], FA1090), 150 genes supporting ([NCCP11945, FA1090], TCDC-NG08107), 140 genes supporting ([TCDC-NG08107, FA1090], NCCP11945), and 346 genes that are not phylogenetically informative. That is, among the phylogenetically informative genes, 60.06%, 20.06%, and 19.28% of genes support the three possible phylogenetic relationships, respectively. If more Ng genomes are analyzed, the portion of genes that support the same phylogenetic relationship is expected to be further reduced. In fact, Ng has been suggested to have higher frequency of intraspecies recombination than Nm (Perez-Losada et al. 2006).
Extensive Recombination in Virulence Genes
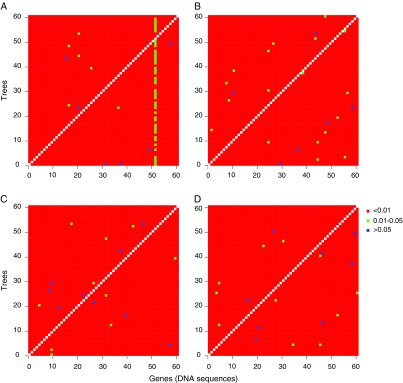
Among the 61 universally present virulence genes, 1,830 possible gene pairs were examined by the AU test (fig. 2), of which 1,773 (96.88%) pairs showed significant phylogenetic incongruence (P < 0.01) supported by both gene nucleotide sequences, and all pairs showed significant incongruence supported by at least one gene. Similar results were observed when analyzing translated protein sequences. One thousand thirteen (93.61%) protein pairs showed significant incongruence supported by both sequences and 1,828 (99.89%) protein pairs showed significant incongruence supported by at least one sequence (supplementary fig. S2, Supplementary Material online). The slightly decreased number of significant protein pairs could be explained by fewer informative sites due to synonymous substitutions as well as the reduced length of amino acid sequences relative to gene sequences. It is also notable that several genes show marginally significant or nonsignificant P values against many other gene topologies (seen as vertical blocks of marginally significant or nonsignificant P values in fig. 2 and supplementary fig. S2, Supplementary Material online). The less than significant P values are due to the lack of informative signals in these particular gene sequences (e.g., low sequence diversity), and the data illustrating these sequence features are not shown. Importantly, when the AU test was performed on the randomly concatenated genes, all pairs showed significant incongruence supported by at least one gene, and the number of pairs that showed significant incongruence supported by both genes are 1,805 (98.63%) for concatenation of 3 genes, 1,811 (98.96%) for concatenation of 6 genes, and 1,811 (98.96%) for concatenation of 9 genes. This simulation reduced the concern about topology alteration due to insufficient phylogenetic signal.
FIG. 2.—
Heatmap of AU test, P values obtained by comparing universally present virulence gene trees based on nucleotide sequences. The AU test was performed on the original 61 universally present virulence genes (A) and also performed on 61 artificially constructed “super-genes” by randomly concatenating 3 (B), 6 (C), or 9 (D) universally present virulence genes. In each panel, there are 1,830 tested gene pairs and 3,660 P values. The 52nd gene in panel A is pilN, which has the least nucleotide diversity (supplementary fig. S2, Supplementary Material online) among the 61 virulence genes.
Intragenic recombination was also found to play an important role during the evolution of virulence genes. In fact, more than half of the virulence genes were suggested to be involved in the intragenic recombination by RDP (50.00%), Phi (57.93%), and OnePop (71.34%). The percentages were slightly higher when the analyses were restricted to the universally present virulence genes (table 2). Furthermore, the median r/m ratio in virulence genes, as a direct measure of the relative impact of recombination on sequence diversification, was observed as 5.37 (supplementary fig. S3, Supplementary Material online). That is, recombination introduces 5.37 times more nucleotide substitutions than do point mutations during the evolution of virulence genes. This is consistent with the estimates in previous studies (e.g., 4.75 in Feil et al. 2001 and 6.71 in Vos and Didelot 2009) and is higher than the estimates in many other bacterial species (e.g., 3.7 for Haemophilus influenzae, 1.5 for Pseudomonas syringae, 0.9 for Legionella pneumophila, 0.7 for Bacillus cereus, 0.7 for Escherichia coli, and 0.1 for Staphylococcus aureus [Vos and Didelot 2009]).
Virulence genes are not the only genes affected by recombination (supplementary fig. S3, Supplementary Material online). Universally present genes also have relatively high r/m ratios, which are not significantly different from the ratios in virulence genes. It has been found that virulence genes have a slightly greater diversity, though not significant, at the sequence level than the universally present genes (supplementary fig. S3, Supplementary Material online), which is consistent with the statement that the high sequence diversity in virulence genes serves as an excellent resource for creating novel adaptive recombinant genes (Marri et al. 2010). The r/m ratios were examined in different functional categories as defined in the Clusters of Orthologous Groups of proteins database (Tatusov et al. 2000). The r/m ratios range from 4.94 for genes involved in cellular processes and signaling to 7.76 for genes involved in information storage and processing (supplementary fig. S4, Supplementary Material online). Furthermore, a significant proportion of genes from all functional categories were observed to undergo intragenic recombination (table 2). All together, this suggests that not only virulence genes but also genes in all functional categories are involved in extensive recombination.
How Clonal Is a CC in Nm?
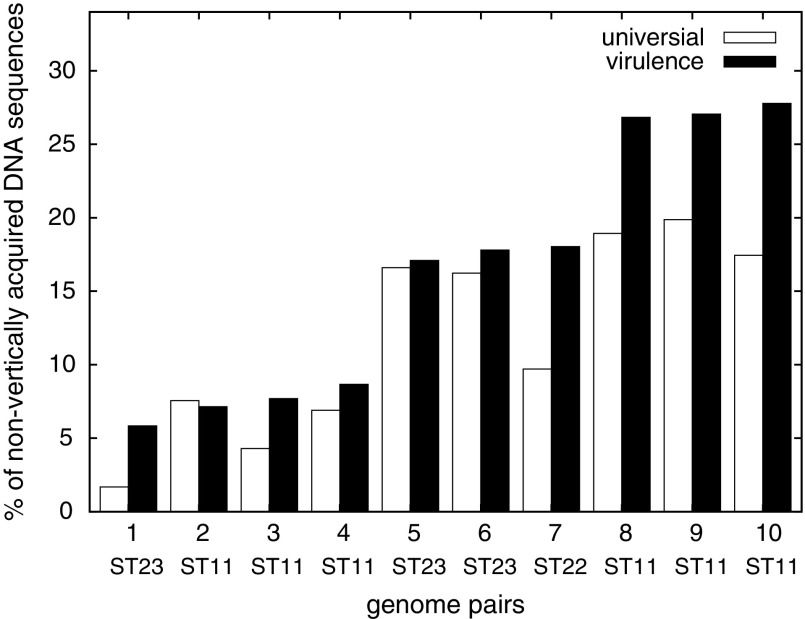
Given the high frequency of recombination at both the gene and intragenic levels, it is important to examine the extent of recombination within a CC. As such, we sought to address how many virulence genes were involved in recombination within the same ST. Unfortunately, it has been well demonstrated that existing recombination detection programs do not effectively identify recombination among low-diversity sequences (Drouin et al. 1999; Posada and Crandall 2001; Wiuf et al. 2001; Posada 2002). In this study, we took a novel approach by estimating the maximum possible nucleotide differences caused by mutation and identifying genes that exceed such estimates. A significant percentage of nonvertically acquired virulence genes were found in 10 identical-ST genome pairs, ranging from 5.8% in Nm6756-Nm8663 to 27.8% in FAM18-ES14902 (fig. 3). In other words, the strains in the same ST are not strictly clonal. Differences were observed in up to 27.8% of the virulence genes and some of the differences impact protein sequences and could result in functional consequences.
FIG. 3.—
Percentage of nonvertical nucleotide sequences between genome pairs in the same ST. Results are shown for both virulence-associated genes (virulence) and universally present genes (universal). Among the strains with identical STs (4 ST11s, 3 ST23s, and 2 ST22s), there are 10 possible genome-pair combinations: 1) Nm6756-Nm8663, 2) S0108-ES14902, 3) K1207-S0108, 4) K1207-ES14902, 5) NS44-Nm8663, 6) NS44-Nm6756, 7) Nm2732-Nm6938, 8) K1207-FAM18, 9) S0108-FAM18, and 10) ES14902-FAM18.
To further assess the robustness of the detection of nonvertically acquired genes, we conducted a series of simulations. As shown in supplementary fig. S5, Supplementary Material online, when the genome-wide sequence divergence is 0.0021 (i.e., the upper bound divergence used in the determination of nonvertical genes), there are only 26.6% identical genes between all genomes, whereas there are 63.5% identical genes between the most diverse genome pair within an ST (FAM18-S0108). To have a similar fraction of genes identical as in FAM18-S0108, the genome-wide sequence divergence should be close to 0.0005. Therefore, the estimates of nonvertically acquired virulence genes based on a sequence divergence of 0.0021 should be considered conservative. Furthermore, DNA distance increased much faster in the five examined genome pairs than in the simulated genomes. This is consistent with the explanation that extensive recombination has introduced substantial sequence divergence in many genes. It is clear that recombination within an ST is not restricted to virulence genes, instead, recombination is common across the genome and the nonvertically acquired genes within an ST could be up to 28% or more of the total gene content.
It is important to note that the observed substantial amount of nonvertically acquired genes within an ST should not be interpreted as a contradiction to the existence of population structure shown in previous studies (Gupta et al. 1996, 1998; Buckee et al. 2008). Consistent with these studies, all major CCs were found to be supported by a 100% bootstrap value in figure 1. In other words, despite the substantial amount of nonvertically acquired genes, it is still possible to obtain a relatively well-supported relationship. However, the relationships of the strains may not represent the relationships for individual genes in the analysis. Similarly, Posada and Crandall (2002) have shown that chimeric sequences of 25% foreign and 75% native origins could still resemble native sequences in phylogenetic analysis.
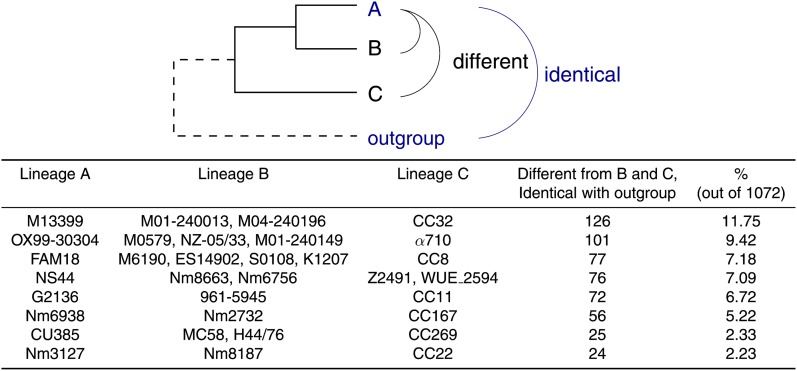
Knowing the significant number of genes that differ between strains in the same ST, we sought to address the significance of horizontal transfer/recombination between CCs. Figure 4 shows that 11.75% of genes in a CC269 strain M13399 are different from the other two CC269 strains, M01-240013and M04-240196, and all strains in CC32, but identical with at least 1 outgroup genome. It is most likely that these genes resulted from recent horizontal transfer (via homologous recombination) from different CCs. Because this analysis considered identical genes and not intragenic recombination, the true number of transfers should be much higher than the estimates in figure 4. In table 3, the nearest neighbors of the 1,072 universal genes in M13399 were identified. Even though M13399 belongs to CC269, only 47.39% of its genes have their nearest neighbors in CC269. Of the genes, 11.29% have their nearest neighbors in CC32, whereas 28.45% of the genes have their nearest neighbors in a CC other than CC269 and CC32. It is noteworthy that the extensive recombination between CCs is not unique to the strain M13399. A significant number of horizontal transfer events were observed in several other strains (fig. 4). Recently, Budroni et al. (2011) reported that the Nm strains contain clade-associated restriction modification systems and suggested that these systems could serve as a barrier to DNA exchange between different populations. Our results, however, suggest that even with the proposed barrier generated by clade-associated restriction modification systems, a significant number of genes could still recombine between different CCs. Our results are consistent with the findings from several previous studies that indicate restriction modification systems may not only provide defense against foreign DNA but may also play a significant role in genetic exchange process. By providing recombinogenic ends for exonucleases or helicases to further act on (DuBose et al. 1988) and limiting the size of transferred fragments, such systems could facilitate small and multiple sequence replacements (McKane and Milkman 1995). The complete functional consequences of clade-associated restriction modification systems, therefore, demand further thorough investigation in future studies.
FIG. 4.—
A conservative estimate on genes horizontally transferred between CCs. Lineages A and B are required to belong to the same CC. Genes in A that are different from all members of B and C but identical to at least one outgroup genome were counted.
Table 3.
Distribution of the Nearest Neighbors of the 1,072 Universally Present Genes in M13399
| ST/CC | Results |
Bootstrapped Results |
||
| Genes | Percentage | Genes | Percentage | |
| CC269a | 508 | 47.39% | 50,050 | 46.69 |
| CC32 | 121 | 11.29% | 12,019 | 11.21 |
| CC41/44 | 57 | 5.32% | 4,935 | 4.60 |
| CC23 | 37 | 3.45% | 3,527 | 3.29 |
| CC8 | 35 | 3.26% | 3,124 | 2.91 |
| ST5 | 28 | 2.61% | 2,528 | 2.36 |
| ST751 | 25 | 2.33% | 2,317 | 2.16 |
| ST177 | 18 | 1.68% | 2,131 | 1.99 |
| CC11 | 17 | 1.59% | 1,682 | 1.57 |
| ST213 | 15 | 1.40% | 1,235 | 1.15 |
| CC22 | 14 | 1.31% | 1,339 | 1.25 |
| ST53 | 13 | 1.21% | 1,072 | 1.00 |
| ST7355 | 11 | 1.03% | 1,225 | 1.14 |
| CC167 | 10 | 0.93% | 872 | 0.81 |
| ST4 | 9 | 0.84% | 1,173 | 1.09 |
| ST1136 | 8 | 0.75% | 829 | 0.77 |
| ST4821 | 7 | 0.65% | 695 | 0.65 |
| Nlact | 1 | 0.09% | 160 | 0.15 |
| Ng | — | — | 15 | 0.01 |
| No signal | 138 | 12.87% | 16,272 | 15.18 |
| Total | 1,072 | — | 107,200 | — |
It is important to note that CC was assigned whenever the nearest neighbor was one of the genomes from a CC even through some genomes in a CC might be substantially different from the rest of the CC. For this reason, the percentage supporting M13399 to be CC269 here (47.39%) is much higher than the phylogenetic support (0.12) from individual gene trees in figure 1. Similar criteria also apply for other CCs.
Detectable Functional Consequences of Recombination Events
Recombination and Mosaic Capsule Gene Cluster
The capsule gene cluster is located downstream of the dnaJ and ctrG genes. The galE-rfbBAC operon (for lipooligosaccharide biosynthesis) and the tex gene (putative regulator) are universally present in all Nm strains, including the α14 and Nm1140 that are incapable of capsule production (Claus et al. 2002). All serogroupable strains (A, B, C, Y, and W135) also contain the ctrABCD operon (capsule transport genes) and the methyltransferase operon hphIAB. The two A strains contain the sacABCD operon (for capsule biosynthesis), whereas B, C, Y, and W135 strains contain the siaABCD operon (for capsule biosynthesis). The siaD gene in serogroups Y and W135 shares no homology with other serogroups and is also called synF/G. The serogroups Y and W135 could be distinguished by a single amino acid substitution in synF/G (Claus et al. 2009). The B and C strains also contain the ctrG gene downstream of the siaABCD operon, and specifically in C strains, oatC that encodes polysialic acid-specific O-acetyltransferase is adjacent to ctrG (supplementary fig. S6, Supplementary Material online). Furthermore, the lipAB operon is often found downstream of the duplicated galE-rfbBAC operon (Beddek et al. 2009). The lipAB operon was not included in the supplementary figure S1, Supplementary Material online because the operon was absent from the seven genomes sequenced in this study.
It is notable that the distribution of different serogroups is not associated with the evolutionary relationship of the strains (supplementary fig. S6, Supplementary Material online) and different serogroups were even found in the same ST (e.g., B and C in ST11; supplementary table S1, Supplementary Material online). In other words, the entire capsule gene cluster could be involved in recombination, which is known as capsule switching (Swartley et al. 1997; Beddek et al. 2009; Wang et al. 2010). Gene movement is also evident within the capsule gene cluster (supplementary fig. S6, Supplementary Material online). Gene cluster inversion seemed to have taken place multiple times. For instance, the operons sacABCD, ctrABCD, and tex are in different orientations between the two A strains (Z2491 and WUE_2594), and different operon orientations were also found in serogroup B strains. A number of transposable elements (or IS elements) were observed at various locations in several capsule gene clusters. They are IS4 inserted between ctrG and siaD in M01-240149 and NZ-05/33; IS1301 between ctrG and siaD in G2136; IS1301 and IS1016 flanking synF/G in Nm3127 and Nm8187; IS1016 flanking synF/G in Nm6756; and IS1301 (between ctrA and siaA) and IS1655 (between rfbA and rfbC) in M04-240196 (supplementary fig. S6, Supplementary Material online). The recent occurrence of IS elements is consistent with the fast turnover of IS elements in the bacterial genomes (Wagner 2006).
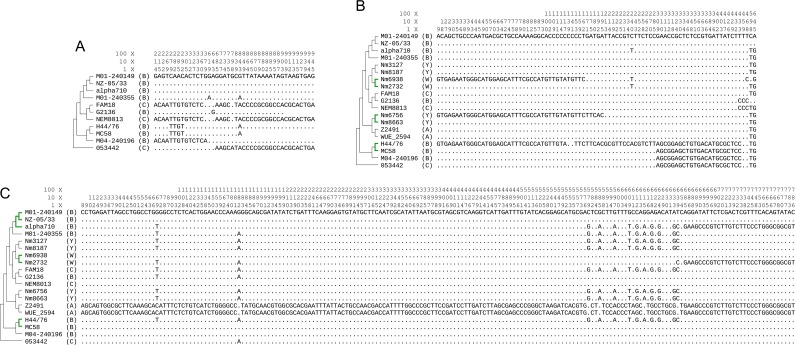
Furthermore, genes within the capsule gene cluster could undergo intragenic recombination beyond both CCs and serogroups. The 5′ end (1-360 nt) of ctrG in a serogroup B strain M04-240196 is identical with two C strains (FAM18 and NEM8013), whereas its 3′ end is virtually identical with other B strains. The third C strain 053442, however, has a 5′ end identical to most B strains and shares a very similar 3′ end (with only 1 nucleotide different) with FAM18 and NEM8013 (fig. 5A). The ctrD gene in a serogroup B strain H44/76 differs 57 nucleotides from another B strain MC58. It is likely that position 1-285 nt in H44/76 is a recent transfer from a fairly divergent genome. Such a divergent sequence is also present in a W135 strain Nm6938 and a Y strain Nm6756, but the breakpoints are different (fig. 5B). The two A strains, namely Z2491 and WUE_2594, contain a very divergent ctrC sequence compared with other strains. The 3′ end (685-786 nt) of this divergent-type sequence was also found in α710 and Nm2732. There is a third sequence type in position 579-639 nt. Position 579-639 in a CC41/44 strain α710 is different from the other two CC41/44 strains, and the same region in a CC32 strain H44/76 is also different from the other CC32 strain MC58 (fig. 5C). Similarly, mosaic structure was also found in the tex gene in multiple strains. The supplementary figure S7, Supplementary Material online shows origins of different regions based on breakpoints specific to the α710 strain. Position 1177-2271 in α710 is different by only 1 nucleotide, otherwise identical with other CC41/44 strains (M01-240149 and NZ-05/33), whereas position 1-1176 in α710 differs 67 nucleotides from both M01-240149 and NZ-05/33 and positions 1-148, 149-1176 in α710 yield two different origins (supplementary fig. S7, Supplementary Material online). Such frequent recombination in capsule gene cluster at both gene cluster level and intragenic level could potentially lead to the escape of vaccine-induced or natural protective immunity.
FIG. 5.—
Chimeric genes (A, ctrG; B, ctrD; and C, ctrC) in the capsule gene cluster. Only nucleotide changes are shown in the alignment. The suggested topoplogy based on the concatenated 1,072 genes (fig. 1) is shown along with each alignment. Branches involved in the same CC are colored in green, when the members of the CC don't share the same type of sequence. Serogroups are shown as one letter in parentheses (W for W135). Spaces in the ctrG alignment signify gene truncation in G2136 and MC58.
Nonvertical Evolution and Antibiotic Resistance
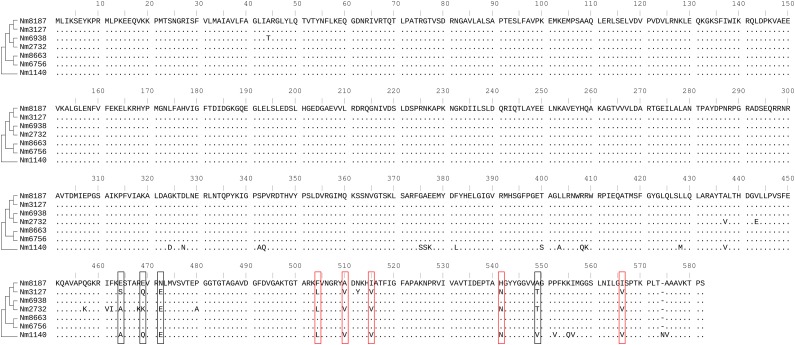
The seven sequenced strains showed different levels of antibiotic susceptibility (table 1). Among the three closely related genome pairs, two pairs have different penicillin MIC values. That is, Nm3127 (MIC = 0.50) has lower susceptibility to penicillin than Nm8187 (MIC = 0.12), and Nm2723 (MIC = 0.12) has lower susceptibility to penicillin than Nm6938 (MIC ≤ 0.06). It is important to note that antibiotic susceptibility played no role in choosing isolates to be sequenced. In fact, antibiotic susceptibility results were reviewed after the whole genome sequencing data were analyzed. The observed decrease of susceptibility in recent isolates could result from bias in small sample size and/or genuinely decreased susceptibility to penicillin in Ontario, Canada (Brown et al. 2001). Different penA sequences were found to be associated with the difference in susceptibility within these two pairs. There are 10 amino acids different between Nm3127 and Nm8187, and 16 amino acids different between Nm2723 and Nm6938 (fig. 6). Nine of these amino acid sites have been previously linked to the decreased susceptibility to beta-lactam antibiotics, such as penicillin and cephalosporins (Thulin et al. 2006; Taha et al. 2007; du Plessis et al. 2008). Furthermore, mutations in some other genes (e.g., mtrR, porB, and ponA) also contribute to changes in antibiotic susceptibility in Neisseria species, and the degree of antibiotic susceptibility is determined by the combination of all these genes (Orus and Vinas 2001; Lindberg et al. 2007; Lee et al. 2010). Such an association was also found in this study. Nm8187 exhibits intermediate resistance (MIC = 0.12), whereas Nm8663 and Nm6756 are penicillin susceptible (MIC ≤ 0.06), yet all have identical penA sequences. The decreased susceptibility to penicillin in Nm8187 could be explained by differences in the mtrR and porB genes. Similarly, the highly susceptible Nm1140 shares 8 nonsynonymous penA changes with the less susceptible Nm3127 and Nm2732 strains. Although changes in penA do not correlate with the low MIC observed in Nm1140, analysis reveals mtrR and porB alleles that are highly similar to sequences from the penicillin-susceptible strains Nm8663 and Nm6756. The three closely related genome pairs all have nearly identical mtrR, porB, and ponA sequences (supplementary fig. S8, Supplementary Material online). Only 1 nonsynonymous difference was observed in porB between Nm8187 and Nm3127, and only two differences in ponA were observed between Nm6938 and Nm2732. This supports the conclusion that differences between the penA genes are most likely responsible for the different penicillin susceptibility profiles of Nm3127 and Nm8187 and between Nm2723 and Nm6938.
FIG. 6.—
Alignment of the PenA protein sequences from the 7 sequenced strains. The suggested topology based on the concatenated 1,072 genes (fig. 1) is shown along with the alignment. Alterations in the nine amino acid sites (in boxes) have been shown to be associated with intermediate resistance to penicillins (Thulin et al. 2006; Taha et al. 2007). Among these nine amino acid sites, five alterations (F504L, A510V, I515V, H541N, and I566V; shown in red boxes) were found in all examined intermediate resistance isolates in (Taha et al. 2007; Vázquez et al. 2007; du Plessis et al. 2008).
Intragenic Recombination of Vaccine Target Gene NHBA (GNA2132)
Currently, a new universal vaccine (4CMenB) against the serogroup B meningococcal disease is being tested (Toneatto et al. 2011). The new vaccine is based on surface protein antigens rather than capsular polysaccharide antigens and is composed of five targets, GNA1870 (fHbp), GNA1994 (NadA), GNA2132 (NHBA), GNA1030, and GNA2091. Among the five genes, we found significant phylogenetic incongruence between the NHBA gene tree and the concatenated gene tree. Recombination has been observed in CC11, CC41/44, and CC269 (fig. 7). Strikingly, the NHBA in OX99-30304 (serogroup B) yields 99 amino acids different from the other three CC41/44 strains (M01-240149, NZ-05133, and M0579) but is identical with the NHBA in a serogroup C and CC11 strain FAM18 and serogroup A strain Z2491 (fig. 7 and supplementary fig. S9, Supplementary Material online). It is noteworthy that the NHBA sequences in the 23 strains are of significantly shorter length than those in the remaining 15 strains. Furthermore, after applying the recombination detection programs, we found strong evidence of intragenic recombination in the NHBA gene supported by various tests (PNSS = 0, PMax χ2 = 0, Pphi = 0, PRDP = 2.69 × 10−16, and POnePop = 0). The occurrence of intragenic recombination in the NHBA gene has introduced multiple amino acid substitutions. These could potentially have functional consequences, such as vaccine escape, because existing vaccines are less likely to be effective against distant variants of the antigenic proteins (Brunelli et al. 2011) and extensive recombination and gene conversion have been suggested to be associated with evasion of the host immune response (Andrews and Gojobori 2004; Helm and Seifert 2010). On the other hand, given the high frequency of recombination between the different serogroups, and even between species, the antigens recognized by the new 4CMenB vaccine are unlikely to be serogroup B specific. Thus, strains from other serogroups than B, or may be even from Ng, could potentially be preventable by the 4CMenB vaccine, if they contain 4CMenB-recognized antigen genes.
FIG. 7.—
Maximum likelihood tree constructed using the NHBA (GNA2132) DNA sequences. Bootstrap values, when ≥75, are shown. When a CC is not monophyletic, all members are colored. The three Ng strains are also colored due to their close relationship with Nm strains. Sequences in the shaded strains are of significantly shorter length than the rest of the sequences (for details, see supplementary fig. S9, Supplementary Material online).
Conclusion
We have found extensive recombination during the genome evolution of Nm. More than 92% of all possible gene pairs display significant phylogenetic incongruence and more than 48% of genes exhibit significant signals for intragenic recombination. These findings confirm that, within a CC determined by the 7 MLST loci, many genes do not evolve in a clonal manner, and in the strains of the same ST, up to 28% of genes may be nonvertical. A significant amount of recombination between CCs has been observed, even though the Nm strains contain restriction modification systems that have been suggested to generate barriers to recombination between clades (Budroni et al. 2011). Extensive recombination, especially changes in virulence genes and capsule genes, could lead to important functional consequences in strains that appear identical by standard classification methods, such as MLST. In this study, we found that recombination events altered penicillin susceptibility in 2 of our 3 closely matched genome pairs. Furthermore, we discovered intragenic recombination in the vaccine target NHBA, which raises the concern that recombination events could impair vaccine efficacy and lead to vaccine escape of Nm
Supplementary Material
Supplementary figures S1–S9 and tables S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We would like to acknowledge P. Rawte, S. Lo, and H. Siebert for their assistance with the Nm strains and susceptibility testing. This work was supported by funds from Sanofi-Pasteur to D.E.L. and a Natural Sciences and Engineering Research Council of Canada (NSERC) fellowship to W.H.
References
- Andrews TD, Gojobori T. Strong positive selection and recombination drive the antigenic variation of the PilE protein of the human pathogen Neisseria meningitidis. Genetics. 2004;166:25–32. doi: 10.1534/genetics.166.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambini S, et al. Distribution and genetic variability of three vaccine components in a panel of strains representative of the diversity of serogroup B meningococcus. Vaccine. 2009;27:2794–2803. doi: 10.1016/j.vaccine.2009.02.098. [DOI] [PubMed] [Google Scholar]
- Beddek AJ, Li MS, Kroll JS, Jordan TW, Martin DR. Evidence for capsule switching between carried and disease-causing Neisseria meningitidis strains. Infect Immun. 2009;77:2989–2994. doi: 10.1128/IAI.00181-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett JS, et al. Independent evolution of the core and accessory gene sets in the genus Neisseria: insights gained from the genome of Neisseria lactamica isolate 020-06. BMC Genomics. 2010;11:652. doi: 10.1186/1471-2164-11-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Riley G, Jamieson F. Neisseria meningitidis with decreased susceptibility to penicillin in Ontario, Canada 1997-2000. Can Commun Dis Rep. 2001;27:73–75. [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelli B, et al. Influence of sequence variability on bactericidal activity sera induced by Factor H binding protein variant 1.1. Vaccine. 2011;29:1072–1081. doi: 10.1016/j.vaccine.2010.11.064. [DOI] [PubMed] [Google Scholar]
- Buckee CO, et al. Role of selection in the emergence of lineages and the evolution of virulence in Neisseria meningitidis. Proc Natl Acad Sci U S A. 2008;105:15082–15087. doi: 10.1073/pnas.0712019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budroni S, et al. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc Natl Acad Sci U S A. 2011;108:4494–4499. doi: 10.1073/pnas.1019751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus H, Maiden MC, Maag R, Frosch M, Vogel U. Many carried meningococci lack the genes required for capsule synthesis and transport. Microbiology. 2002;148:1813–1819. doi: 10.1099/00221287-148-6-1813. [DOI] [PubMed] [Google Scholar]
- Claus H, Stummeyer K, Batzilla J, Mühlenhoff M, Vogel U. Amino acid 310 determines the donor substrate specificity of serogroup W-135 and Y capsule polymerases of Neisseria meningitidis. Mol Microbiol. 2009;71:960–971. doi: 10.1111/j.1365-2958.2008.06580.x. [DOI] [PubMed] [Google Scholar]
- Claus H, et al. Genetic analysis of meningococci carried by children and young adults. J Infect Dis. 2005;191:1263–1271. doi: 10.1086/428590. [DOI] [PubMed] [Google Scholar]
- CLSI. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Seventh edition: approved standard M7-A7. Wayne (PA): Clinical and Laboratory Standards Institute; 2006. [Google Scholar]
- CLSI. Performance standards for antimicrobial susceptibility testing; twenty-first informational supplement CLSI document M100-S21. Wayne (PA): Clinical and Laboratory Standards Institute; 2011. [Google Scholar]
- Corless CE, et al. Simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J Clin Microbiol. 2001;39:1553–1558. doi: 10.1128/JCM.39.4.1553-1558.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Urwin R, Maiden MC, Falush D. Genealogical typing of Neisseria meningitidis. Microbiology. 2009;155:3176–3186. doi: 10.1099/mic.0.031534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin G, Prat F, Ell M, Clarke GD. Detecting and characterizing gene conversions between multigene family members. Mol Biol Evol. 1999;16:1369–1390. doi: 10.1093/oxfordjournals.molbev.a026047. [DOI] [PubMed] [Google Scholar]
- du Plessis M, et al. Neisseria meningitidis intermediately resistant to penicillin and causing invasive disease in South Africa in 2001 to 2005. J Clin Microbiol. 2008;46:3208–3214. doi: 10.1128/JCM.00221-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBose RF, Dykhuizen DE, Hartl DL. Genetic exchange among natural isolates of bacteria: recombination within the phoA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1988;85:7036–7040. doi: 10.1073/pnas.85.18.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning Hotopp JC, et al. Comparative genomics of Neisseria meningitidis: core genome, islands of horizontal transfer and pathogen-specific genes. Microbiology. 2006;152:3733–3749. doi: 10.1099/mic.0.29261-0. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, et al. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc Natl Acad Sci U S A. 2001;98:182–187. doi: 10.1073/pnas.98.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (phylogeny inference package). Version 3.2. Cladistics. 1989;5:164–166. [Google Scholar]
- Gordon D, Desmarais C, Green P. Automated finishing with autofinish. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Ferguson N, Anderson RM. Chaos, persistence and the evolution of strain structure in populations of antigenically variable infectious agents. Science. 1998;240:912–915. doi: 10.1126/science.280.5365.912. [DOI] [PubMed] [Google Scholar]
- Gupta S, et al. The maintenance of strain structure in populations of recombining infectious agents. Nat Med. 1996;2:437–442. doi: 10.1038/nm0496-437. [DOI] [PubMed] [Google Scholar]
- Hao W. OrgConv: detection of gene conversion using consensus sequences and its application in plant mitochondrial and chloroplast homologs. BMC Bioinformatics. 2010;11:114. doi: 10.1186/1471-2105-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W, Golding GB. High rates of lateral gene transfer are not due to false diagnosis of gene absence. Gene. 2008;421:27–31. doi: 10.1016/j.gene.2008.06.015. [DOI] [PubMed] [Google Scholar]
- Helm RA, Seifert HS. Frequency and rate of pilin antigenic variation of Neisseria meningitidis. J Bacteriol. 2010;192:3822–3823. doi: 10.1128/JB.00280-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC, Urwin R, Maiden MC. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol Biol Evol. 1999;16:741–749. doi: 10.1093/oxfordjournals.molbev.a026159. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Jolley KA, Maiden MC. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian protein metabolism. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- Koski LB, Golding GB. The closest BLAST hit is often not the nearest neighbor. J Mol Evol. 2001;52:540–542. doi: 10.1007/s002390010184. [DOI] [PubMed] [Google Scholar]
- Lee SG, et al. Various penA mutations together with mtrR, porB and ponA mutations in Neisseria gonorrhoeae isolates with reduced susceptibility to cefixime or ceftriaxone. J Antimicrob Chemother. 2010;65:669–675. doi: 10.1093/jac/dkp505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg R, Fredlund H, Nicholas R, Unemo M. Neisseria gonorrhoeae isolates with reduced susceptibility to cefixime and ceftriaxone: association with genetic polymorphisms in penA, mtrR, porB1b, and ponA. Antimicrob Agents Chemother. 2007;51:2117–2122. doi: 10.1128/AAC.01604-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden MC. Multilocus sequence typing of bacteria. Annu Rev Microbiol. 2006;60:561–588. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- Maiden MC, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marri PR, et al. Genome sequencing reveals widespread virulence gene exchange among human Neisseria species. PLoS One. 2010;5:e11835. doi: 10.1371/journal.pone.0011835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- McKane M, Milkman R. Transduction, restriction and recombination patterns in Escherichia coli. Genetics. 1995;139:35–43. doi: 10.1093/genetics/139.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothershed EA, et al. Use of real-time PCR to resolve slide agglutination discrepancies in serogroup identification of Neisseria meningitidis. J Clin Microbiol. 2004;42:320–328. doi: 10.1128/JCM.42.1.320-328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Itoh T, Matsuda H, Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet. 2004;36:760–766. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- Orus P, Vinas M. Mechanisms other than penicillin-binding protein-2 alterations may contribute to moderate penicillin resistance in Neisseria meningitidis. Int J Antimicrob Agents. 2001;18:113–119. doi: 10.1016/s0924-8579(01)00362-4. [DOI] [PubMed] [Google Scholar]
- Parkhill J, et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- Perez-Losada M, et al. Population genetics of microbial pathogens estimated from multilocus sequence typing (MLST) data. Infect Genet Evol. 2006;6:97–112. doi: 10.1016/j.meegid.2005.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. Evaluation of methods for detecting recombination from DNA sequences: empirical data. Mol Biol Evol. 2002;19:708–717. doi: 10.1093/oxfordjournals.molbev.a004129. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci U S A. 2001;98:13757–13762. doi: 10.1073/pnas.241370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. The effect of recombination on the accuracy of phylogeny estimation. J Mol Evol. 2002;54:396–402. doi: 10.1007/s00239-001-0034-9. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2009. [Google Scholar]
- Rusniok CD, et al. NeMeSys: a biological resource for narrowing the gap between sequence and function in the human pathogen Neisseria meningitidis. Genome Biol. 2009;10:R110. doi: 10.1186/gb-2009-10-10-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen C, et al. Whole-genome comparison of disease and carriage strains provides insights into virulence evolution in Neisseria meningitidis. Proc Natl Acad Sci U S A. 2008;105:3473–3478. doi: 10.1073/pnas.0800151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Smith JM, Smith NH, O'Rourke M, Spratt BG. How clonal are bacteria? Proc Natl Acad Sci U S A. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder LA, Saunders NJ. The majority of genes in the pathogenic Neisseria species are present in non-pathogenic Neisseria lactamica, including those designated as ‘virulence genes’. BMC Genomics. 2006;7:128. doi: 10.1186/1471-2164-7-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich JE, Hahn MW. Disentangling the effects of demography and selection in human history. Mol Biol Evol. 2005;22:63–73. doi: 10.1093/molbev/msh252. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stephens DS, Greenwood B, Brandtzaeg P. Epidemic meningitis, meningococcaemia and Neisseria meningitidis. Lancet. 2007;369:2196–2210. doi: 10.1016/S0140-6736(07)61016-2. [DOI] [PubMed] [Google Scholar]
- Strimmer K, von Haeseler A. Quartet puzzling: a quartet maximum-likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- Swartley JS, et al. Capsule switching of Neisseria meningitidis. Proc Natl Acad Sci U S A. 1997;94:271–276. doi: 10.1073/pnas.94.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha MK. Simultaneous approach for nonculture PCR-based identification and serogroup prediction of Neisseria meningitidis. J Clin Microbiol. 2000;38:855–857. doi: 10.1128/jcm.38.2.855-857.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha MK, et al. Target gene sequencing to characterize the penicillin G susceptibility of Neisseria meningitidis. Antimicrob Agents Chemother. 2007;51:2784–2792. doi: 10.1128/AAC.00412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulin S, Olcén P, Fredlund H, Unemo M. Total variation in the penA gene of Neisseria meningitidis: correlation between susceptibility to β-lactam antibiotics and penA gene heterogeneity. Antimicrob Agents Chemother. 2006;50:3317–3324. doi: 10.1128/AAC.00353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toneatto D, et al. The first use of an investigational multicomponent meningococcal serogroup B vaccine (4CMenB) in humans. Hum Vaccin. 2011;7:646–653. doi: 10.4161/hv.7.6.15482. [DOI] [PubMed] [Google Scholar]
- Vázquez JA, et al. Antibiotic resistant meningococci in Europe: any need to act? FEMS Microbiol Rev. 2007;31:64–70. doi: 10.1111/j.1574-6976.2006.00049.x. [DOI] [PubMed] [Google Scholar]
- Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 2009;3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- Wagner A. Periodic extinctions of transposable elements in bacterial lineages: evidence from intragenomic variation in multiple genomes. Mol Biol Evol. 2006;23:723–733. doi: 10.1093/molbev/msj085. [DOI] [PubMed] [Google Scholar]
- Wang Q, et al. Genetic study of capsular switching between Neisseria meningitidis sequence type 7 serogroup A and C strains. Infect Immun. 2010;78:3883–3888. doi: 10.1128/IAI.00363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiuf C, Christensen T, Hein J. A simulation study of the reliability of recombination detection methods. Mol Biol Evol. 2001;18:1929–1939. doi: 10.1093/oxfordjournals.molbev.a003733. [DOI] [PubMed] [Google Scholar]
- Yazdankhah SP, et al. Distribution of serogroups and genotypes among disease-associated and carried isolates of Neisseria meningitidis from the Czech Republic, Greece, and Norway. J Clin Microbiol. 2004;42:5146–5153. doi: 10.1128/JCM.42.11.5146-5153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]