Abstract
In the present paper, we report that transcription affects the location of a DNA target in Escherichia coli K-12. A strain whose chromosome had been engineered to encode a lac repressor–GFP (green fluorescent protein) fusion was used as a host for a low copy number plasmid that carries an array of five lac operator sites. Individual cells of this strain exhibited a diffuse fluorescence signal, suggesting that the plasmid is distributed throughout the cell cytoplasm. However, a derivative of this plasmid carrying a cloned constitutive promoter is targeted to a location at the edge of the nucleoid towards the pole of the host cell. We conclude that transcription from the cloned promoter is driving the location of the plasmid and that specific locations in bacterial cells may favour gene expression.
Keywords: bacterial nucleoid, Escherichia coli, promoter location, transcription
Abbreviations: GFP, green fluorescent protein; LB, Luria–Bertani
INTRODUCTION
Bacteria differ from higher organisms in their strategy for ‘managing’ their DNA. In cells of higher organisms with nuclear compartments, DNA is packaged into nucleosomes that are folded into chromosomes, and a complex network of proteins controls the activity of nucleosomes, which regulates transcription. In bacteria, where there is no nucleus and no nucleosomes, chromosomes are compacted and folded into a structure known as the nucleoid, which is located in the cytoplasm, where DNA replication, transcription and translation all occur in the same cell compartment. Although this is one of the major differences between ‘higher’ eukaryotic cells and ‘lower’ prokaryotic cells, both types of cell have the same fundamental problem of how to accommodate their DNA in a small space and yet ensure that it is accessible to the transcription machinery.
Most studies that address the issue of how location within a folded chromosome affects gene expression have been conducted by examination of eukaryotic cell nuclei. Visualization of active genes and sites of transcription has led to the discovery of transcription factories, where many active RNA polymerase molecules are concentrated and anchored to a nuclear substructure [1–3]. In bacteria, the situation is less clear, although it is known that the structure of the nucleoid changes with environmental conditions [4] and, under some conditions, RNA polymerase appears to be concentrated into transcription foci or factories [5,6]. In the present study, we set up a simple assay to determine whether the location of a plasmid-borne segment of DNA was affected by transcription. We show that transcription drives the plasmid to a location at the periphery of the bacterial nucleoid and argue that position within the folded chromosome may play a role in setting the expression of some genes.
MATERIALS AND METHODS
Bacterial strain and growth conditions
We used a derivative of Escherichia coli K-12 strain MG1655 that had been engineered, using the gene-doctoring method of Lee et al. [7], to fuse the gfp (green fluorescent protein) gene to the 3′-end of the lacI gene. During the construction of this strain, which expresses a LacI–GFP fusion protein under the control of the lacI gene promoter, the lac operon 01, 02 and 03 operators were eliminated. This strain was transformed with different pRW plasmids and transformants were grown aerobically in rich LB (Luria–Bertani) medium at 23°C to a D600 of 0.1–0.2. For induction of the cka promoter, 8.5 μg·ml−1 nalidixic acid was added to cultures.
Construction of the derivative pRW plasmids
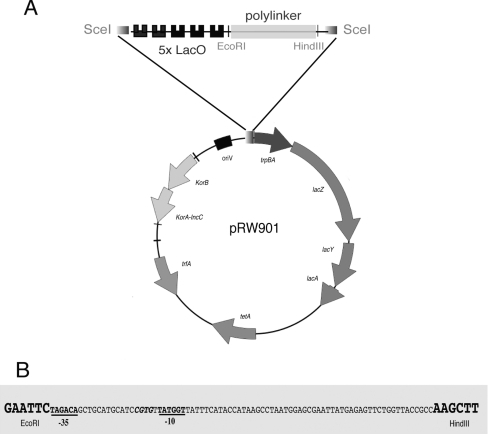
The pRW plasmids used are constructed from pRW50, a low copy number broad host range RK2 derivative [8]. This plasmid, derived from pRK2501, belongs to the IncP-1 incompatibility group and is present at five to eight copies per chromosome [9]. Briefly, the pRW plasmids contain the gene for the plasmid replication initiator protein (trfA), the origin for vegetative DNA replication (oriV), the gene encoding resistance to tetracycline (tetA), the promoter-less lacZYA operon, and an EcoRI/HindIII insert, cloned immediately downstream of five LacI operator sites, with flanking 18-bp target sites for the yeast meganuclease I-SceI (Figure 1A). The control region of the pRW plasmids carries genes encoding IncC and KorB proteins, but lacks the OB3-binding site for KorB that is essential for partitioning [10]. The pRW901, pRW902 and pRW904 plasmids used carry different inserts between the EcoRI and HindIII sites. Hence pRW901 carries a promoter-less fragment, whereas pRW904 carries the fragment illustrated in Figure 1(B) that contains the constitutively active KAB-CGTG promoter [11]. Plasmids pRW905 and pRW906 are derivatives of pRW901 and pRW904 respectively in which the trpBA and lacZYA genes were deleted (using HindIII and RsrII) in order to stop translation of RNA starting at the KAB promoter. Plasmid pRW902 carries an insert with the cka promoter [8].
Figure 1. Schematic representation of pRW plasmid derivatives.
(A) Circular representation of pRW901 showing the most important features. The plasmid carries five DNA sites for LacI (LacO) upstream of a polylinker flanked by EcoRI and HindIII restriction sites. (B) DNA base sequence of the constitutively active KAB-CGTG promoter [11] inserted between the EcoRI and HindIII sites to give pRW904. The promoter −10 and −35 hexamer elements are in bold and underlined.
In vivo visualization of LacI–GFP-tagged plasmids
Strains containing the pRW plasmids derivatives were grown overnight at 23°C in LB medium, diluted 1:100 in fresh medium and grown to a D600 of 0.1–0.2. Samples of 1.5 ml from exponentially growing cultures were collected by centrifugation at 8000 g for 5 min. Cells were stained with FM4-64 and Hoechst 33258 (1 μg·ml−1 and 5 μg·ml−1 respectively) in 10 μl of mounting medium (40% glycerol in 0.02 M PBS, pH 7.5) and then fixed to a poly-lysine-treated coverslip. Images were captured with a Nikon Eclipse 90i microscope, equipped with a Hamamatsu Digital CCD (charge-coupled device) camera and a Plan Apochromat VC 100× oil-objective lens. Epi-FL filter blocks for DAPI (4′,6-diamidino-2-phenylindole) (340–380 nm excitation), FITC (465–495 nm excitation) and TRITC (tetramethylrhodamine β-isothiocyanate) (540–625 nm excitation) were used for DNA, LacI–GFP and cell membrane fluorescence imaging. Analysis was carried out with NIS-element AR software provided by Nikon. For time-lapse microscopy, cells were grown with FM4-64 (0.2 μg·ml−1) and concentrated 10-fold by centrifugation at 8000 g for 5 min. Cells were placed on an agarose slab (0.9% agarose/1% LB medium) and warmed to 23°C.
RESULTS
The starting point of the present study was a derivative of E. coli K-12 strain MG1655 in which the chromosomal lacI gene had been fused to the gfp gene [7]. This strain was transformed with pRW901, a previously constructed derivative of an RK2 broad host range low copy number plasmid that carries five lac operators adjacent to a polylinker (Figure 1). Our aim was to use fluorescence microscopy to visualize the distribution of this plasmid, tagged with LacI–GFP fusion protein, and to investigate the effect of introducing a constitutively expressed promoter into the plasmid.
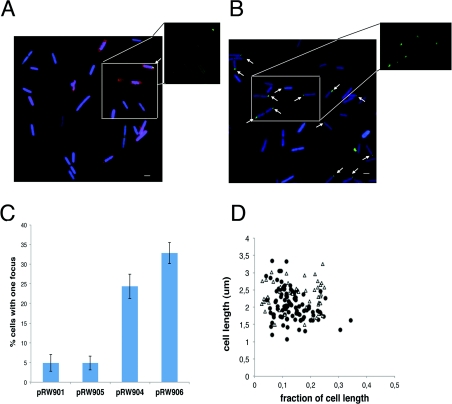
Many previous studies have used bacterial plasmids with cloned lac operators and LacI–GFP to visualize plasmid partition complexes [12,13]. However, pRW901 is defective in partitioning [10], and hence the GFP fluorescence of the MG1655 lacI::gfp strain carrying pRW901 appears diffuse in most cells (Figure 2A). Figure 2(B) shows a parallel experiment in which a short DNA fragment containing the constitutively active KAB-CGTG promoter was inserted into the polylinker of pRW901 (to give pRW904, Figure 1B). This promoter contains −10 and −35 elements that are close to the consensus. Results in Figure 2 show that ~25% of cells containing pRW904 display clear fluorescence foci. These were counted and their position was analysed (Figures 2C and 2D). Note that less than 0.5% of cells show more than one focus and these cells were not counted. Inspection of the images indicates that most foci are located near the cell pole and on the edge of the nucleoid, as revealed by staining with Hoechst blue stain (Figure 3). These foci must be due to clustering of individual plasmid molecules, and the prima facie conclusion is that transcription due to the promoter insert in pRW904 drives location of the plasmid. To check whether translation contributes to the formation of the foci, the experiments were repeated with pRW905 and pRW906 in which the trpBA and lacZYA genes downstream of the promoter were deleted. Cells carrying pRW906 (KAB-CGTG promoter and no translation) showed similar behaviour to that of cells carrying pRW904 (Figure 2C).
Figure 2. Location of pRW plasmids in E. coli MG1655 carrying a LacI–GFP fusion.
(A) Fluorescence micrographs of cells containing pRW901 (no promoter). The inset shows GFP fluorescence only of selected cells. (B) Fluorescence micrographs of cells containing pRW904 (KAB promoter). The inset shows GFP fluorescence only of selected cells. (C) Percentage of cells containing pRW901, pRW904, pRW905 and pRW906 that show fluorescence foci. The number of cells counted range from 330 to 984 from several repeat experiments. Results are means±S.D. (D) The position of fluorescence foci with respect to the cell length (fraction of cell length from the nearest pole, x-axis) was plotted as a function of the cell length (μm, y-axis) for 105 cells containing pRW904 (●) and 46 cells containing pRW901 (Δ). Cell membranes were stained with FM4-64 (red) and DNA was stained with Hoechst 33258 (blue). Scale bars, 1 μm.
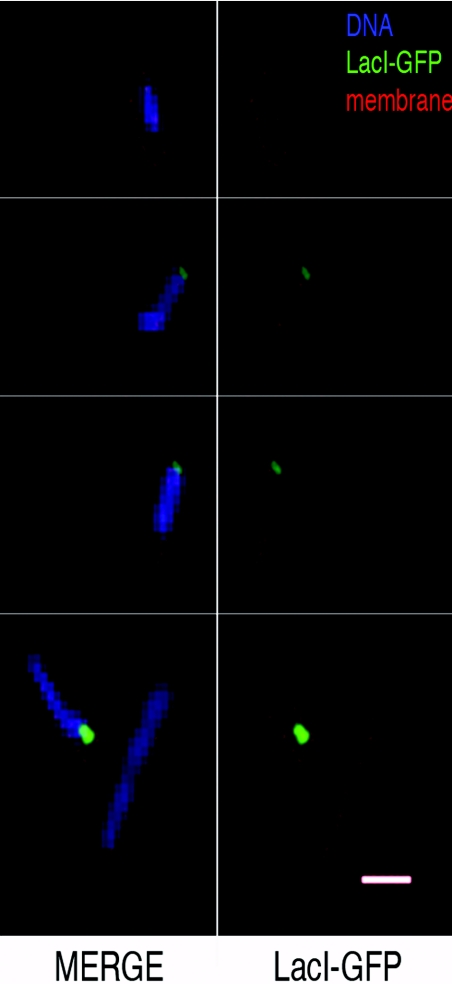
Figure 3. Intracellular location of pRW904 plasmid foci.
Magnified fluorescence micrographs of selected cells carrying pRW904, showing the juxtaposition of GFP foci and cell membranes stained with FM4-64 (red) and DNA stained with Hoechst 33258 (blue). Scale bar, 1 μm.
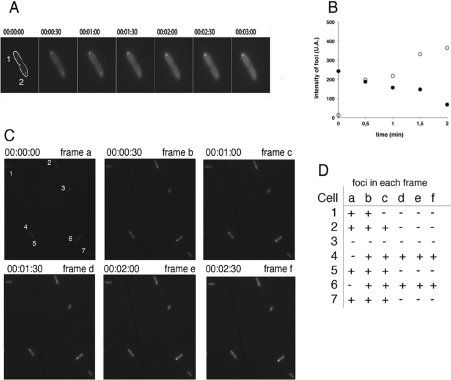
We next used time-lapse microscopy to observe the dynamics of the foci by capturing images of live cells every 30 s. Results in Figure 4(A) and 4(B) show that foci due to pRW904 appear in some cells (cell 1) and disappear from others (cell 2). Analysis of a larger field of view containing many cells over a longer time (Figures 4C and 4D) confirmed that, whereas the majority of cells do not exhibit fluorescence foci, most cells show a focus at some point during the time sequence. This shows that the foci are not permanent structures and that most cells that are focus-free in a single frame do have the ability to form foci.
Figure 4. Time-lapse microscopy of GFP foci.
(A) Seven time-lapse images of cells carrying pRW904 were collected 30 s apart, showing the appearance and disappearance of foci in two cells (denoted cell 1 and cell 2) shortly after division. (B) Time course of intensity profiles of GFP foci in cell 1 (○) and cell 2 (●). (C) Fluorescence time-lapse micrographs of E. coli carrying pRW904 collected 30 s apart. (D) Formation and de-formation of foci in individual cells from (C). The presence or absence of foci in individual cells is denoted by + and by − respectively.
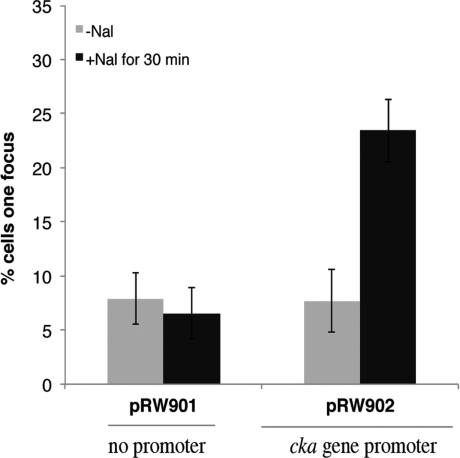
Our experiments demonstrate that a reporter plasmid relocates in response to an active promoter. To investigate this further, we next studied the inducible colicin K gene (cka) promoter that is repressed by the LexA global regulator [8,14], but can be induced by DNA damage. Thus a short DNA fragment carrying the cka regulatory region was inserted into pRW901 to give pRW902. With MG1655 lacI::gfp cells carrying pRW902, the number of cells showing foci of GFP fluorescence increased after induction with nalidixic acid, whereas there was no increase with pRW901 (Figure 5).
Figure 5. Plasmid relocation following induction of the cloned cka promoter.
The histogram illustrates the measured percentage of cells carrying pRW901 or pRW902 that show foci before or after induction with 8.5 μg·ml−1 nalidixic acid (Nal) for 30 min. Results are means±S.D. for over 200 cells from several repeat experiments.
DISCUSSION
In the present study, we used a derivative of E. coli K-12 strain MG1655 that expresses a LacI–GFP fusion, and we exploited the fact that, using a standard epifluorescence microscope, neither individual LacI–GFP molecules nor LacI–GFP bound to single plasmid molecules with five lac operators can be visualized. We observed that cloning of fragments carrying either a constitutively active or an inducible promoter into the plasmid results in the formation of fluorescence foci close to the surface of the nucleoid near the cell poles. These foci must be due to the congregation of several plasmids at the same locus, which appears to be driven by high levels of transcriptional activity rather than translation. Our results argue that the transcription-driven location of plasmids is transient with foci forming and de-forming (Figure 4). We suppose that the fluorescence foci form when several plasmids congregate in an area of high transcriptional activity, due, for example, to a cluster of RNA polymerase molecules, and this suggests the existence of favoured locations for transcription. Although the endogenous promoters of RK2 plasmids are repressed during normal growth [15], and the foci formation is due to the introduction of a transcriptionally active promoter, we did observe a small number of fluorescence foci with the control plasmid, pRW901, which lacks a constitutively active promoter (Figures 2A and 2C). This is consistent with the work of others who have reported polar congregation of partition-defective plasmids [16–19]. We reason that this effect is due to the very low levels of transcription of essential plasmid functions [15].
Little is understood about how bacterial nucleoids are organized and how their organization affects access to the transcription machinery. The present study supports the idea that bacteria can spatially organize transcription, moving promoters to specific locations. Hence transcription is located to the space surrounding the nucleoid and this facilitates the coupling between transcription and translation. This must have implications for cell organization and for our understanding of gene expression in bacteria. This is especially important because most systems-level analyses of bacterial transcription ignore gene location, and focus on base sequence and trans-acting factors as the primary determinants of promoter activity.
AUTHOR CONTRIBUTION
María-Antonia Sánchez-Romero performed experiments, designed experiments, and drafted the paper. David Lee and Eugenio Sánchez-Morán conceived and designed experiments, and helped to draft the paper. Stephen Busby conceived the study, its design and co-ordination, and drafted the paper. All authors read and approved the final paper.
ACKNOWLEDGEMENTS
We are grateful to Matej Butala, Jennie Mitchell and Chris Thomas for encouragement and helpful discussions.
FUNDING
This work was funded by a Wellcome Trust programme grant to S.J.W.B., a postdoctoral fellowship (to M.-A.S.-R.) from Junta de Extremadura (Spain), and a David Phillips Fellowship (to E.S.-M.).
References
- 1.Cook P. R. A model for all genomes: the role of transcription factories. J. Mol. Biol. 2010;395:1–10. doi: 10.1016/j.jmb.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 2.Sutherland H., Bickmore W. A. Transcription factories: gene expression in unions? Nat. Rev. Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- 3.Xu M., Cook P. R. Similar active genes cluster in specialized transcription factories. J. Cell Biol. 2008;181:615–623. doi: 10.1083/jcb.200710053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin D. J., Cabrera J. E. Coupling the distribution of RNA polymerase to global gene regulation and the dynamic structure of the bacterial nucleoid in Escherichia coli. J. Struct. Biol. 2006;156:284–291. doi: 10.1016/j.jsb.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Cabrera J. E., Jin D. J. The distribution of RNA polymerase in Escherichia coli is dynamic and sensitive to environmental cues. Mol. Microbiol. 2003;50:1493–1505. doi: 10.1046/j.1365-2958.2003.03805.x. [DOI] [PubMed] [Google Scholar]
- 6.Davies K. M., Dedman A. J., van Horck S., Lewis P. J. The NusA:RNA polymerase ratio is increased at sites of rRNA synthesis in Bacillus subtilis. Mol. Microbiol. 2005;57:366–379. doi: 10.1111/j.1365-2958.2005.04469.x. [DOI] [PubMed] [Google Scholar]
- 7.Lee D. J., Bingle L. E., Heurlier K., Pallen M. J., Penn C. W., Busby S. J., Hobman J. L. Gene doctoring: a method for recombineering in laboratory and pathogenic Escherichia coli strains. BMC Microbiol. 2009;9:252. doi: 10.1186/1471-2180-9-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butala M., Busby S. J., Lee D. J. DNA sampling: a method for probing protein binding at specific loci on bacterial chromosomes. Nucleic Acids Res. 2009;37:e37. doi: 10.1093/nar/gkp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas C. M., Meyer R., Helinski D. R. Regions of broad-host-range plasmid RK2 which are essential for replication and maintenance. J. Bacteriol. 1980;141:213–222. doi: 10.1128/jb.141.1.213-222.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams D. R., Macartney D. P., Thomas C. M. The partitioning activity of the RK2 central control region requires only incC, korB and KorB-binding site OB3 but other KorB-binding sites form destabilizing complexes in the absence of OB3. Microbiology. 1998;144:3369–3378. doi: 10.1099/00221287-144-12-3369. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell J. E., Zheng D., Busby S. J., Minchin S. D. Identification and analysis of ‘extended −10’ promoters in Escherichia coli. Nucleic Acids Res. 2003;31:4689–4695. doi: 10.1093/nar/gkg694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pogliano J. Dynamic cellular location of bacterial plasmids. Curr. Opin. Microbiol. 2002;5:586–590. doi: 10.1016/s1369-5274(02)00370-3. [DOI] [PubMed] [Google Scholar]
- 13.Pogliano J., Ho T. Q., Zhong Z., Helinski D. R. Multicopy plasmids are clustered and localized in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4486–4491. doi: 10.1073/pnas.081075798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butala M., Zgur-Bertok D., Busby S. J. The bacterial LexA transcriptional repressor. Cell. Mol. Life Sci. 2009;66:82–93. doi: 10.1007/s00018-008-8378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herman D., Thomas C, Stekel D. Global transcription regulation of RK2 plasmids: a case study in the combined use of dynamical mathematical models and statistical inference for integration of experimental data and hypothesis exploration. BMC Syst. Biol. 2011;5:119. doi: 10.1186/1752-0509-5-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolatka K., Witosinska M., Pierechod M., Konieczny I. Bacterial partitioning proteins affect the subcellular location of broad-host-range plasmid RK2. Microbiology. 2008;154:2847–2856. doi: 10.1099/mic.0.2008/018762-0. [DOI] [PubMed] [Google Scholar]
- 17.Yao S., Helinski D. R., Toukdarian A. Localization of the naturally occurring plasmid ColE1 at the cell pole. J. Bacteriol. 2007;189:1946–1953. doi: 10.1128/JB.01451-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao S., Toukdarian A., Helinski D. R. Inhibition of protein and RNA synthesis in Escherichia coli results in declustering of plasmid RK2. Plasmid. 2006;56:124–132. doi: 10.1016/j.plasmid.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Verheust C., Helinski D. R. The incC korB region of RK2 repositions a mini-RK2 replicon in Escherichia coli. Plasmid. 2007;58:195–204. doi: 10.1016/j.plasmid.2007.03.004. [DOI] [PubMed] [Google Scholar]