Abstract
DNA topoisomerase I (Top1) and topoisomerase II (Top2) inhibitors are widely used to treat a variety of cancers. Their mechanism of action involves stabilization of otherwise transient (“cleavable”) complexes between Top1 or Top2 and DNA; collisions of DNA replication forks with such stabilized complexes lead to formation of DNA double-strand breaks (DSBs). In this study, using 5-ethynyl-2′deoxyuridine (EdU) as a DNA precursor, we directly assessed the relationship between DNA replication and induction of DSBs revealed as γH2AX foci in A549 cells treated with Top1 inhibitors topotecan (Tpt) or camptothecin (Cpt) and Top2 inhibitors mitoxantrone (Mxt) and etoposide (Etp). Analysis of cells by multiparameter laser scanning cytometry following treatment with Tpt or Cpt revealed that only DNA replicating cells showed induction of γH2AX and a strong correlation between DNA replication and formation of DSBs (r = 0.86). In cells treated with Mxt or Etp, the correlation was weaker (r = 0.52 and 0.64). In addition, both Mtx and Etp caused induction of γH2AX in cells not replicating DNA. Confocal imaging of nuclei of cells treated with Tpt revealed the presence of γH2AX foci predominantly in DNA replicating cells and close association and co-localization of γH2AX foci with DNA replication sites. In cells treated with Mxt or Etp, the γH2AX foci were induced in DNA replicating as well as non-replicating cells but the close association between a large proportion of γH2AX foci and DNA replication sites was also apparent. The data are consistent with the view that collision of DNA replication forks with cleavable Top1–DNA complexes stabilized by Tpt/Cpt is the sole cause of induction of DSBs. Additional mechanisms such as involvement of transcription and/or generation of oxidative stress may contribute to DSBs induction by Mxt and Etp. The confocal analysis of the association between DNA replication sites and the sites of DSBs (γH2AX foci) opens a new approach for mechanistic studies of the involvement of DNA replication in induction of DNA damage.
Keywords: S phase, cell cycle, EdU incorporation, DNA damage response, click chemistry, H2AX phosphorylation
DNA topoisomerases are essential enzymes that mediate changes in the topology of double helical DNA during replication, transcription, recombination, and chromatin remodeling. Their activity allows for transitions between supercoiling and relaxing, knotting and un-knotting, and decatenation versus catenation of superhelical DNA (reviews 1–5). Type I topoisomerases (Top1) cause relaxation of superhelical DNA by generating a transient single-strand nick followed by DNA religation. Type II topoisomerases (Top2) mediate ATP-dependent cleavage of both strands of the DNA double helix followed by crossing of the DNA double-strand (ds) through the transiently opened gap (1–5). Mammalian cells have two Top2 isoenzymes, Top2α and Top2β (2,6). While Top2α is associated with cell proliferation and its level is much higher in rapidly proliferating than in nondividing cells Top2β may play a role in transcription (7).
Top1 and Top2 proved to be attractive targets of anticancer agents and their inhibitors are among the most clinically effective drugs that are widely used to treat different kinds of cancer. Their mechanism of action involves stabilization of otherwise transient (“cleavable”) complexes formed between Top1 or Top2 and DNA (8,9). Collisions of the progressing DNA replication forks with such stabilized complexes lead to formation of double-strand breaks (DSBs) (8,9). Similarly, within the DNA region being transcribed, collisions of the progressing RNA polymerase molecule with the inhibitor-stabilized Top1 or Top2 “cleavable complexes” located on the transcribed template strand result in formation of DSBs (10). DSBs are recognized by the cell as lethal lesions and often induce apoptosis.
DNA damage, in particular when it involves induction of DSBs, triggers a complex series of molecular events which are broadly defined as the DNA damage response (DDR). The DDR events involve a post-translational modification of numerous proteins that activate many signaling pathways associated with cell cycle progression, DNA repair, apoptosis, and cell senescence (11). One of the early events of the DDR is activation of phosphatidyl inositol 3′ kinase-related kinases (PIKKs): Ataxia telangiectasia mutated (ATM), ATM and Rad3-related (ATR), and/or DNA-dependent protein kinase (DNA-PKcs) (12,13). The function of these protein kinases is to signal the presence of DNA damage by phosphorylating a multitude of proteins whose main function is to preserve genome integrity.
One of the key substrates phosphorylated by PIKKs is histone H2AX, a variant of the nucleosome core histone H2A (14,15). Its phosphorylation on Ser139 takes place on a large number of nucleosomes, along a megabase span of DNA flanking the DSB; the phosphorylated H2AX has been defined as γH2AX (15). Phosphorylation of several PIKKs and of H2AX upon induction of DNA damage can be immunocytochemically detected with the respective phospho-specific Abs and assessed by cytometry (16–20).
In our prior studies on the effects of the Top1 inhibitor topotecan (Tpt) and Top2 inhibitors mitoxantrone (Mxt) and etoposide (Etp), we observed a strong cell cycle-phase dependence in DNA damage signaling as revealed by ATM activation and induction of γH2AX (18,20–24). Specifically, exposure of either human leukemic TK6 or pulmonary adenocarcinoma A549 cells to Tpt, or its analogue camptothecin (Cpt), led to rapid phosphorylation of ATM and H2AX selectively in S-phase cells. In contrast, the exposure to Mxt or Etp induced ATM and H2AX phosphorylation in all phases of the cycle (20–24). Moreover, whereas DNA damage signaling induced by Mxt or Etp was significantly attenuated by the reactive oxygen species (ROS) scavenger N-acetyl-L-cysteine (NAC) (22), no similar attenuation was seen upon induction of DNA damage by Tpt or Cpt (22,25). The occurrence of apoptosis, which was subsequent to ATM activation and H2AX phosphorylation, was also cell cycle-phase specific, being selective to S-phase cells following treatment with Tpt and Mxt but not with Etp (20,21).
This study was designed to explore the cell cycle-phase selectivity in DNA signaling induced by Tpt/Cpt, Mxt, and Etp with more accuracy, particularly to study any correlation between the DDR signaling and S-phase progression as revealed by DNA replication. Toward this end we have labeled DNA replicating cells with a short pulse of the DNA precursor 5-ethynyl-2′deoxyuridine (EdU) and subsequently exposed them to the inhibitors. The detection of the EdU incorporation using the “click chemistry” approach (26) makes it possible to concurrently detect intracellular epitopes (27), including their modification by phosphorylation, with the use of phospho-specific Abs (28–32). Using multiparameter LSC, we have been able, therefore, to directly correlate DNA replication with the induction of DNA damage signaling triggered by topoisomerase inhibitors as revealed by H2AX phosphorylation. The use of confocal microscopy made it possible to relate the induction of γH2AX foci, likely reporting the presence of DSBs (15), with the discrete sites of DNA replication (“DNA replication factories”) made up of DNA associated with protein complexes involved in DNA replication within chromatin (33).
Materials and Methods
Cells, Cell Treatment
Human lung carcinoma A549 cells were obtained from American Type Culture Collection (ATCC CCL-185, Manassas, VA). The cells were cultured in Ham’s F12K medium with 2 mM l-glutamine adjusted to contain 1.5 g/L sodium bicarbonate (ATCC) supplemented with 10% fetal bovine serum (ATCC). Dual-chambered slides (Nunc Lab-Tek II) were seeded with 105 cells/mL suspended in 2 mL medium per chamber. During treatment with EdU and Tpt, Cpt, Mxt, or Etp, the cells were in exponential phase of growth. The cultures were treated with 20 µM EdU (Invitrogen/Molecular Probes, Eugene, OR) for 30 min and then, while still in the presence of EdU, either with 0.2 µM Tpt, or 0.2 µM Cpt, 0.2 µM Mxt, or 10 µM Etp (all from Sigma/Aldrich Chemical Co., St. Louis, MO) for another 120 min. The cells were then rinsed with phosphate-buffered salt solution (PBS) and fixed by transferring slides into Coplin jars containing 1% methanol-free formaldehyde (Poly-sciences, Warrington, PA) for 15 min. The slides were rinsed with PBS and kept at 4°C with a 1% (w/v) solution of bovine serum albumin (BSA; Sigma/Aldrich) in PBS until staining.
Detection of H2AX Phosphorylation and Incorporation of EdU
Slides in fixative were washed twice in PBS and the cells on the slides treated with 0.1% Triton X-100 (Sigma) in PBS for 15 min, and with a 1% (w/v) solution of BSA (Sigma) in PBS for 30 min (or overnight incubation for confocal imaging) to suppress nonspecific antibody (Ab) binding. The cells were then incubated in a 100 µL volume of 1% BSA containing a 1:300 dilution of phospho-specific (Ser139) γH2AX mAb (BioLegend, San Diego, CA) for 1.5 h at room temperature or overnight at 4°C. The secondary Ab was tagged with Alexa-Fluor 647 fluorochrome (Invitrogen/Molecular Probes, at a dilution of 1:100) and incubated for 45 min at room temperature. The Click-iT® EdU AlexaFluor® 488 imaging kit (Invitrogen/Molecular Probes) was used to detect EdU incorporation according to the Invitrogen protocol. Before measurement by LSC, the cells were counterstained with 2.8 µg/mL 4,6-diamidino-2-phenylindole (DAPI; Sigma/Aldrich) in PBS for 15 min. Each experiment was performed with an IgG control in which cells were labeled only with the secondary AlexaFluor 647 Ab, without primary antibody incubation to estimate the extent of nonspecific binding of the secondary antibody to the cells. Other details of cell incubations with the primary and secondary Ab have been previously described (30,32).
Measurement of Cell Fluorescence by LSC
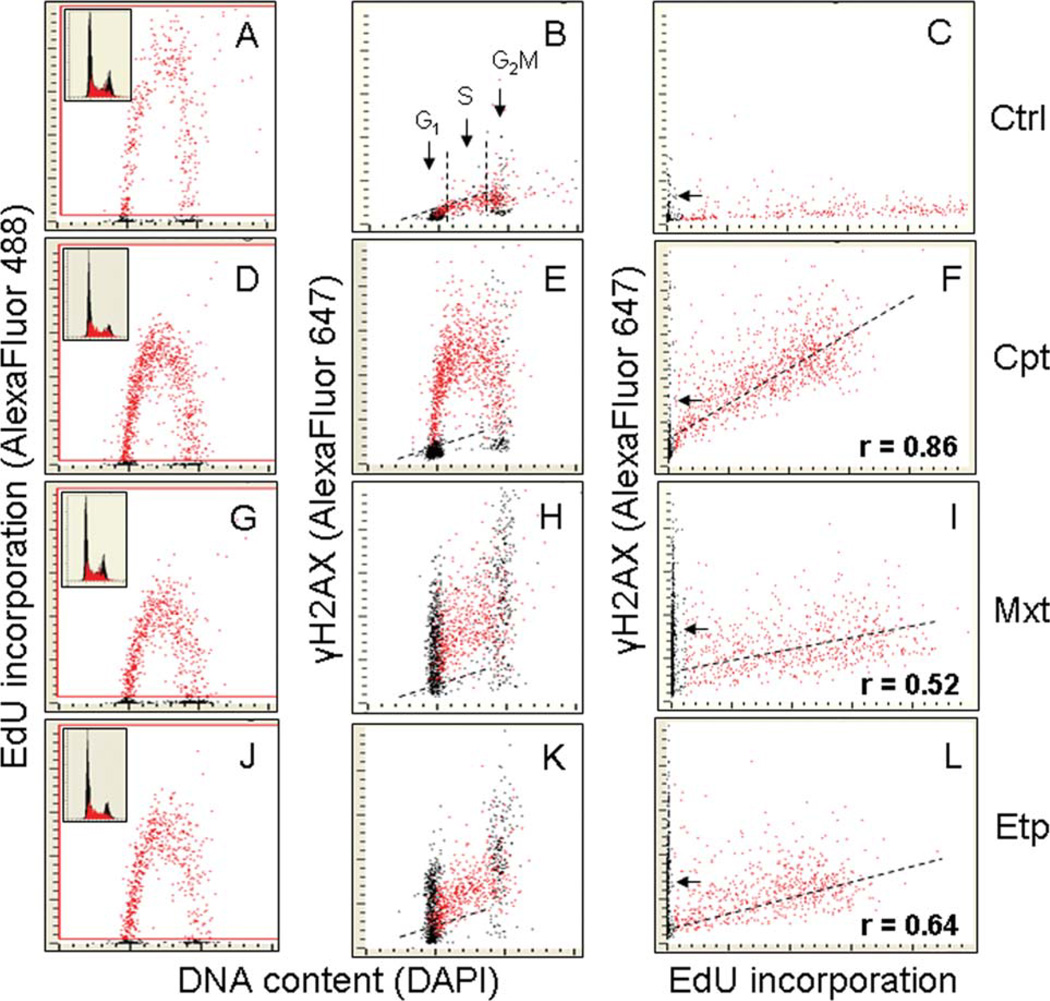
Cellular green (EdU) or red immunofluorescence (IF) representing the binding of the respective phospho-specific Abs as well as the blue emission of DAPI-stained DNA was measured using an LSC (iCys; CompuCyte, Westwood, MA) utilizing standard filter settings; fluorescence was excited with heliumneon (633 nm) argon ion (488 nm) and violet (405 nm) lasers. The intensities of maximal pixel and integrated fluorescence were measured and recorded for each cell. No electronic compensation for possible emission spectral overlap was applied. At least 3,000 cells were measured per sample. Gating analysis was carried out as described in legend of Figure 1. To measure the correlation between incorporation of EdU and expression of γH2AX, the subpopulations of DNA replicating cells were gated as shown in Figure 1 and the data transferred to Microsoft Excel which were used to analyze the correlation coefficient (Pearson’s r) within these subpopulations (Examples of the raw data of LSC analysis are presented in Supplemental Information).
Figure 1.
Correlation between induction of γH2AX by Cpt, Mxt, and Etp and DNA replication in A549 cells. Exponentially growing A549 cells were exposed to 20 µM EdU for 30 min and then (still in the presence of EdU) were treated either with 0.2 µM Cpt (D, E, F), 0.2 µM Mxt (G, H, I), or 20 µM Etp (J, K, L) for 2 h. Control cells (A—C) were exposed only to 20 µM EdU for 2.5 h. The cells were then fixed and their blue (DAPI), green (EdU), and red (γH2AX) fluorescence were measured by LSC. The cells that incorporated EdU were “paint-gated” and the data were plotted as bivariate distributions representing γH2AX versus DNA content (B, E, H, K) or γH2AX expression versus EdU incorporation (C, F, I, L). Note that Cpt induced γH2AX only in EdU incorporating cells. In contrast, in response to treatment with Mxt or Etp, H2AX was phosphorylated in both EdU incorporating and non-incorporating (G1 and G2M) cells. A correlation between EdU incorporation and induction of γH2AX by the Top inhibitors is shown in Panels F, I, and L; the correlation coefficient refers only to DNA replicating cells. Insets in A, D, G, and J panels present DNA content frequency histograms from the respective cultures, with EdU incorporating cells marked in red. The dashed skewed lines in B, E, H, and K show the upper threshold for 97% of G1 and S-phase cells expressing γH2AX in the untreated culture (Ctrl). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Mapping Replication and H2AX Phosphorylation by Confocal Microscopy
Cell cultures intended for imaging experiments were grown on round coverslips submerged in Petri dishes. Cells were incubated with EdU first (30 min; Click-iT® EdU Alexa-Fluor® 488 Imaging Kit cat. # C10337; Invitrogen/Molecular Probes) and the topoisomerase inhibitors were added at a final concentration of 0.15 µM of Tpt, 0.2 µM of Mxt or 10 µM of Etp for an additional 60 or 120 min in the presence of EdU. Cells were fixed with methanol-free formaldehyde (4%, 15 min) and treated with Triton X-100 (0.1%, 15 min). Blocking was done overnight in 3% (w/v) BSA; phospho-specific (Ser139) γH2AX mAb (Upstate Biotechnology, Lake Placid, NY) was used followed by the secondary antibody Alexa-Fluor® 568 goat anti-mouse IgG (H+L) (cat. # A11004; Invitrogen/Molecular Probes). Subsequently, detection of EdU was performed, according to the manufacturer’s instructions. Images were recorded using a Leica LSC SP5 confocal microscope (Leica Microsystems GmbH, Wetzlar, Germany). The following instrumental parameters were used: 63× HCX PL APO CS NA 1.4 oil immersion lens; excitation was at 488 (Ar) and 561 nm (HeNe); emission detection bands used were 500–550 nm for AlexaFluor488 (Click-iT EdU) and 560–700 nm for AlexaFluor568 (immunofluorescence, γH2A.X); registration was in sequential mode; and scanning was done at 8,000 Hz (resonant scanner), with 16–32 averaged frames. Images were deconvoluted using AutoDeblur software v.X2.2 (Media Cybernetics).
Results
Figure 1 illustrates the correlation between the induction of γH2AX by treatment of A549 cells with Cpt, Mxt, or Etp vis-à-vis DNA replication. To directly assess a relationship between the expression of γH2AX induced by the inhibitors and the extent of EdU incorporation, these topoisomerase inhibitors were included into the cultures at the time when the cells were incorporating the DNA precursor EdU. In the course of multiparameter analysis by LSC, the cells incorporating EdU were identified using “paint-a-gate” analysis which made it possible to evaluate the expression of γH2AX by them and compare it with that of the cells that did not replicate DNA during exposure to EdU. The evidence is quite straight-forward that upon exposure to Cpt γH2AX was induced exclusively in DNA replicating cells. In cells treated with Mxt or Etp on the other hand, both DNA replicating as well as non-replicating cells demonstrated elevated level of γH2AX (e.g., above that of the maximal threshold of γH2AX of untreated cells; Fig. 1, Panel B). A relatively strong correlation between EdU incorporation and induction of γH2AX was observed in cells treated with Cpt (r = 0.86). The correlation was of a lesser degree in cells treated with Mxt (r = 0.52) or Etp (r = 0.64). The data shown in Figure 1 also indicate that treatment of cells with Cpt, Mxt, or Etp decreased the intensity of their labeling with EdU as is evident by the lower level of EdU incorporation in the D, G, and J panels compared with A.
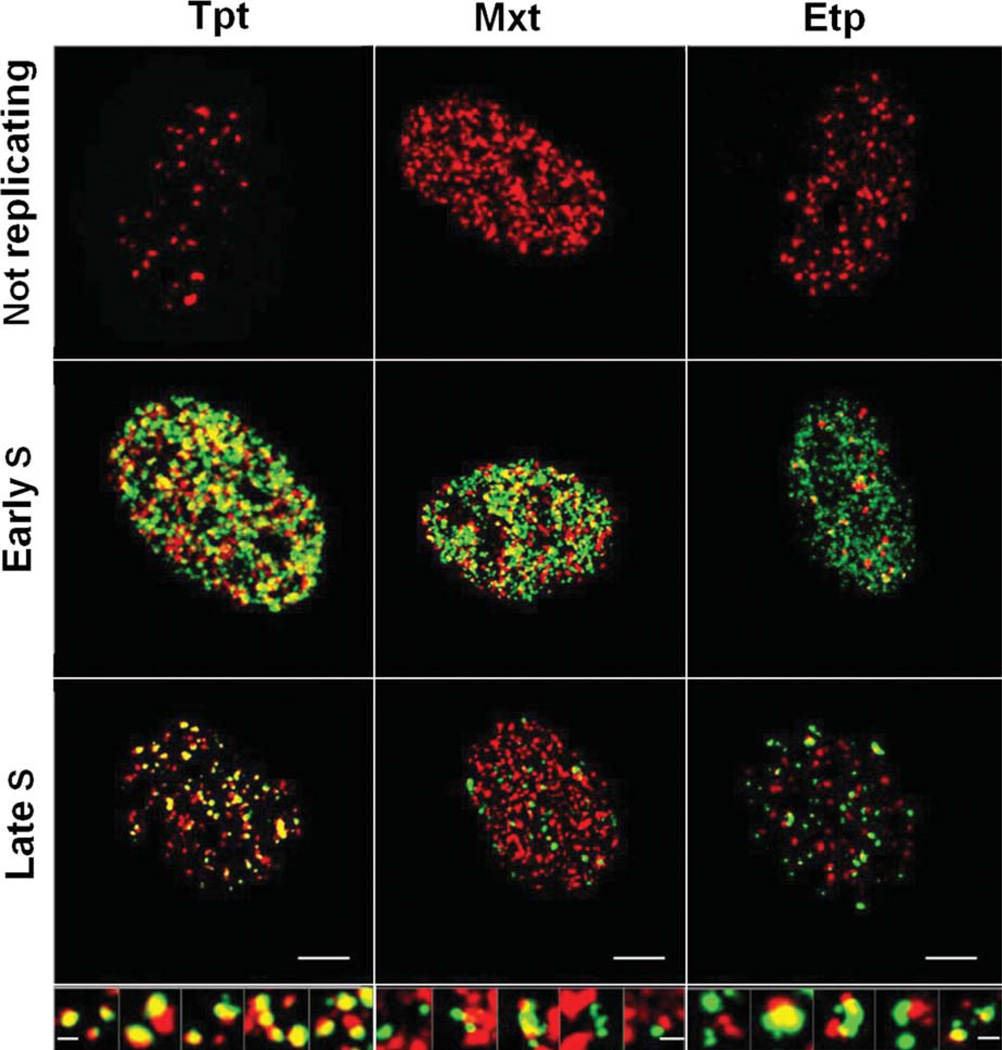
The spatial relationship in chromatin between the sites of DNA replication and the sites of H2AX phosphorylation (γH2AX foci) induced by treatment with Tpt is shown in the confocal image of A549 cells nuclei (Fig. 2; left column). The cells were exposed to EdU for 30 min and subsequently treated with Tpt for an additional 60 or 120 min. The incorporated EdU was detected using the green-fluorochrome (AlexaFluor 488)-tagged azide, whereas γH2AX was detected immunocytochemically using a secondary Ab conjugated to a red fluorescing dye (AlexaFluor 568). The most conspicuous observation was that all cells showing EdU incorporation (DNA replication) sites also contained a multitude of γH2AX foci. On the other hand, most cells that were EdU negative had low numbers (1–5) of γH2AX foci. However, a few EdU unlabeled cells had a slightly higher number of foci (Fig. 2; left column, top panel). As it is quite apparent from Figure 2, while there were numerous DNA replication sites that were not associated with γH2AX foci and there were some γH2AX foci alone, with no distinct association with sites of EdU incorporation, a significant proportion of the γH2AX foci were associated with the replication sites. In fact, in numerous sites, a distinct co-localization of EdU and γH2AX was apparent, revealed by yellow fluorescence, a result of the red plus green fluorescence overlap.
Figure 2.
Relationship between the sites of EdU incorporation (replication factories) and the induction of γH2AX foci in A549 cells treated with Tpt (left column), Mxt (middle column), or Etp (right column). Confocal images of A549 nuclei that were exposed to 10 µM EdU for 30 min and then for additional 60 or 120 min treated with 0.15 µM Tpt, 0.2 µM Mxt, or 10 µM Etp. The incorporation of EdU was detected using the “click chemistry” methodology [refs. 26—30] utilizing AlexaFluor 488-tagged azide (green fluorescence) while γH2AX was detected immunocytochemically using a secondary Ab labeled with AlexaFluor 568 (red fluorescence). The size marker = 10 µm. The displayed images are 3D maximum intensity projections of all the confocal planes of a given nucleus for non-replicating, and late-S phase cells, or a stack of sections from the equatorial region of the nucleus for early S-phase cells, where the frequency of foci was much greater. The top row shows nuclei of EdU non-incorporating cells, the middle—nuclei of cells in early S phase, the bottom—nuclei of late S phase. The distinction between early- and late-S phase is based on the characteristic differences in the pattern of DNA replication sites [ref. 33]. Enlarged images of selected foci/sites of DNA replication (late S phase) are shown at the bottom of the respective columns; the size marker = 0.5 µm). Nuclei of cells treated with Tpt demonstrate the sites of EdU incorporation (“replication factories”) often in close proximity or co-localized (green and red overlap = yellow fluorescence) with γH2AX foci. Fewer phosphorylation foci co-localize with replication sites in cells treated with Mxt or Etp. The co-localization coefficients are shown in Table 1. In Tpt-treated and EdU-negative cell, the frequency of γH2AX foci was similar to that of untreated controls. In Mxt and Etp treated cells, the number of γH2AX foci in EdU-negative cells was greater than in control cells.
The center column in Figure 2 shows confocal images of A549 cell nuclei treated with Mxt and EdU. Unlike in Tpt-treated cells, well over 50% of the MTX-treated nuclei showed the presence of large numbers of γH2AX foci in the absence of DNA replication sites. However, all nuclei containing DNA replication sites also had numerous γH2AX foci. As was the case with Tpt treated cells, such nuclei had individual DNA replication sites not in association with γH2AX foci, relatively few γH2AX foci alone and large number γH2AX foci co-localized with DNA replication foci.
The pattern of EdU and γH2AX localization in nuclei of cells treated with Etp resembled that of cells treated with Mxt (Fig. 2 right column). Thus, there was a conspicuous presence of cells having only γH2AX foci in the absence of DNA replication sites as well as large number of nuclei with both, γH2AX foci and DNA replication sites. Here also, the replication sites were seen either alone or co-localized with the γH2AX foci.
Discussion
These data confirm and provide additional information in support of our prior observations on the cell cycle phase specificity of Ctp/Tpt, Mxt, and Etp in terms of induction of DNA damage signaling (20–25,34). In these earlier studies, we observed that the Top1 inhibitors Cpt and Tpt selectively affected S-phase cells triggering their ATM and Chk2 activation as well as H2AX and p53 phosphorylation and had no effect on G1 or G2M phase cells. In contrast, Mxt and Etp triggered DNA damage signaling in G1 and G2M phase in addition to S-phase cells. In these earlier studies, we have identified these cell cycle phases by cellular DNA content. Such identification has a bias since the cells just initiating- or completing-DNA replication may have DNA content so close to that of G1 or G2M cells, respectively, that they can be erroneously classified as in G1 or G2M phase. In this study, we were able to correlate the induction of DDR by these drugs as revealed by H2AX phosphorylation directly with DNA replication, detected within the same cells. Thus, the identification of the S-phase cells was more accurate and revealed actual DNA replication. Furthermore, we were able to correlate the extent of DNA replication during the exposure to EdU based on the quantity of the incorporated precursor with the extent of phosphorylation of H2AX measured as the intensity of γH2AX immunofluorescence (Fig. 1, C, F, I, L). In addition, confocal microscopy enabled us to observe the spatial relationship between γH2AX foci and DNA replication sites in chromatin (Fig. 2). The presence of discrete γH2AX foci is considered to represent formation of DSBs (14,15). It was possible therefore to directly correlate the induction of DSBs with DNA replication.
The Top1 inhibitors Cpt/Tpt bind DNA by intercalation between the −1 and +1 bases of DNA where the −1 base is the nucleotide covalently bound to Top1, thereby stabilizing DNA–Top1 complexes (1–4). The Top1 inhibitors Cpt and its analogue Tpt were previously shown to selectively induce apoptosis of S-phase cells having no effect on cells in either G1 or G2M (35). These data provide additional information that the induction of γH2AX by these Top1 inhibitors was strongly associated with DNA replication. Thus, the low frequency of γH2AX foci (1–5) seen in most cells not replicating DNA was similar to that of untreated cells and has previously been shown to represent constitutive DNA damage signaling, induced primarily by endogenous, metabolically generated oxidants (36,37). Occasionally, some cells not replicating DNA in cultures treated with Tpt had a larger number of foci (Fig. 2, left top panel). We had observed in prior studies (36,37) that cells with elevated levels of constitutive DNA damage signaling were predominantly in late G2 or in prophase. On the other hand, among DNA replicating cells, a rather strong correlation was seen between the extent of DNA replication revealed by the amount of incorporated EdU and degree of H2AX phosphorylation (r = 0.86). Furthermore, the confocal images of Tpt-treated cells showed a high frequency of DNA replication sites in association and co-localized with γH2AX foci (Fig. 2). All this evidence, combined with the prior observation that the induction of DDR by Top1 inhibitors was not attenuated by the ROS scavenger NAC (22), points out that the induction of DSBs by Tpt/Cpt was solely a consequence of the collisions of DNA replications forks with “cleavable complexes” stabilized by the inhibitors, as originally proposed by Hsiang et al (7). In fact, among a large variety of the anticancer drugs studied, the Top1 inhibitors were shown to be the most effective agents in triggering apoptotic cell death selectively of DNA replicating cells (38).
Top2 inhibitors Mxt and Etp both generate formation of stabilized Top2-DNA covalent complexes, but by different mechanisms (1–5). While the anthracycline analogue Mxt binds DNA by intercalation (39) Etp is not an intercalator and binds stoichiometrically to Top2 (40). We have observed that Mxt, similar to Cpt, induces apoptosis preferentially in S-phase cells (20,41), whereas Etp is not so selective and the cells exposed to it undergo apoptosis at all phases of the cycle (25). Furthermore, while the induction of DNA damage by Mxt and Etp to a large extent appears to be mediated by ROS, no such involvement of ROS was seen in the cells treated with Cpt/Tpt (22,25). In this study, the induction of γH2AX by Mxt and Etp was seen both in DNA replicating as well as in non-replicating cells. The correlation in the extent of induction of γH2AX and EdU incorporation was of a lesser degree (r = 0.52, Mxt; r = 0.64, Etp) compared with Cpt. However, the presence of γH2AX foci in association with and co-localizing with DNA replication sites was clearly evident (Fig. 2). Thus, our present data are compatible with the view that while some DSBs induced by Mxt or Etp were caused by collisions of DNA replication forks with the stabilized complexes of Top2-DNA (7), other DSBs could have originated from collisions of the DNA transcription machinery with these complexes (9,10) and/or were caused by ROS induced by treatment with these inhibitors (22,25). As we have shown recently, the oxidative stress-induced γH2AX foci are predominant in S-phase cells and do co-localize with DNA replication sites (32).
Our confocal studies on the association between DNA replication sites and the sites of DSBs (γH2AX foci) opened a new approach for mechanistic studies on the involvement of DNA replication with the genotoxicity of different agents. As shown in Figure 2, there were (i) DNA replication sites alone with no apparent association with γH2AX foci. Their presence is expected when the replication takes place at DNA sections not being affected by the drug (with no DSBs); (ii) DNA replication sites in association with or co-localized with γH2AX foci. The incidence of such sites points out that DNA replication may have a contributory role in DSBs formation; and (iii) γH2AX foci alone, which may indicate that DSBs were induced with no contribution of DNA replication. However, it is also possible that these γH2AX foci are at the sites of DNA replication at which the amount of incorporated EdU at the moment of the DSBs formation (e.g., collision of the replication forks) was as yet minimal and thus undetectable.
In an attempt to quantitatively express a correlation between H2AX phosphorylation foci (red) and DNA replication sites (green) (Fig. 2) we calculated the overlap between these sites using a JaCoP plugin (42) for ImageJ (43) approach. The co-localization coefficients, calculated according to the method of Manders et al. (44), are presented in Table 1. The data show the greatest degree of co-localization for Tpt (0.655, 0.475) and the lowest for Etp (0.155; 0.19). In the case of Tpt and Mxt, the co-localization was more pronounced in early- than in late-S phase cells.
Table 1.
Co-localization coefficient for the fraction of γH2AX foci (red fluorescence) overlapping DNA replication sites (green fluorescence). R→G = fraction of red overlapping green
| MANDERS’ COEFFICIENTS | |||
|---|---|---|---|
| TOPOTECAN | MITOXANTRONE | ETOPOSIDE | |
| Early S | R→G = 0.655 | R→G = 0.419 | R→G = 0.155 |
| Late S | R→G = 0.475 | R→G = 0.02 | R→G = 0.19 |
In the case of Tpt (Fig. 1, Panel E), the DSBs (γH2AX foci) are expected to be formed almost solely at the DNA replication sites (collapse of “cleavable complexes”; Ref. 7). Thus, the co-localization coefficient is expected to be close to 1.0. However, the γH2AX foci are relatively large (megabase domains of DNA at both flanks of DSB; Refs. 14,15) and may extend beyond the areas marked by the incorporated EdU. Moreover, translocation of particular areas of chromatin may occur due to local chromatin relaxation (decondensation) that occurs shortly after induction of DNA damage (45). Thus, at the time the cells were fixed some γH2AX foci, even if initially were at the replication sites, may not fully overlap with the latter at the time of fixation. This may explain why the co-localization coefficient is in the range 0.475–0.655 (Table 1) rather than be close to 1.0.
It is reasonable to expect that lower values of the coefficient represent the situation where DNA damage was inflicted not only at the sites of replication forks but also outside of them as well. The damage induced by Mxt or Etp falls into this category (Table 1). This is consistent with the LSC data (Fig. 1). The difference in the co-localization coefficients between the early- and late- S cells may be due to the fact that the rate of DNA replication in these sections of S phase is markedly different (33,46).
This initial analysis of the representative images as shown in Fig. 2 requires refinement, which will include measurements of the distances between the regions of replication and phosphorylation and a selection of a larger number of cells at earlymid- and late- sections of S phase. Such analysis, embracing a large body of new data is currently in progress. The data may provide further information on the role of DNA replication in formation of DSBs induced by these agents.
Conclusions
This study demonstrates the utility of the combination of: (i) “click chemistry” to detect DNA replication, (ii) multi-parameter analysis by laser scanning cytometry, and (iii) confocal microscopy, in mechanistic studies of DNA damage induced by widely used anticancer drugs. While this methodology can be used to study other cell types and other drugs, the aim of this study was to show the feasibility and advantages of such an approach.
Supplementary Material
Acknowledgments
Grant sponsor: NCI CA RO1 28 704 and Jagiellonian University (DS, BW).
Footnotes
Additional Supporting Information may be found in the online version of this article.
Literature Cited
- 1.Salerno S, Da Settimo F, Taliani S, Simorini F, La Motta C, Fornaciari G, Marini A. Recent advances in the development of topoisomerase I and II inhibitors as anticancer drugs. Curr Med Chem. 2010;17:4270–4290. doi: 10.2174/092986710793361252. [DOI] [PubMed] [Google Scholar]
- 2.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer drugs. Chem Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heisig P. Type II topoisomerases—Inhibitors, repair mechanmisms and mutations. Mutagenesis. 2009;24:465–469. doi: 10.1093/mutage/gep035. [DOI] [PubMed] [Google Scholar]
- 4.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Z, Deibler RW, Chan HS, Zechiedrich L. The why and how of DNA unlinking. Nucleic acids Res. 2009;37:661–671. doi: 10.1093/nar/gkp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase IIα rapidly relaxes positively supercoiled DNA: Implications for enzyme action ahead of replication forks. J Biol Chem. 2005;280:39337–39345. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 7.Hsiang YH, Lihou MG, Liu LF. Arrest of replication forks by drug stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 1989;49:5077–5082. [PubMed] [Google Scholar]
- 8.Ju BG, Lunyak VV, Perissi V, Garcia-Bessets L, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIβ-mediated dsDNA break required for regulation of transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 9.D’Arpa P, Beardmore C, Liu LF. Involvement of nucleic acid synthesis in cell killing mechanisms of topoisomerase poisons. Cancer Res. 1990;50:6916–6924. [PubMed] [Google Scholar]
- 10.Wu J, Liu LF. Processing of topoisomerase I cleavable complexes into DNA by transcription. Nucleic Acids Res. 1997;25:4181–4186. doi: 10.1093/nar/25.21.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kastan MB. DNA damage responses: Mechanisms and roles in human disease. 2007 G.H.A. Cloves Memorial Award Lecture. Mol Cancer Res. 2008;6:517–524. doi: 10.1158/1541-7786.MCR-08-0020. [DOI] [PubMed] [Google Scholar]
- 12.Lovejoy CA, Cortez D. Common mechanisms of PIKK regulation. DNA Repair. 2009;8:1004–1008. doi: 10.1016/j.dnarep.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuadrado M, Martinez-Pastor B, Fernandez-Capetillo O. ATR activation in response to ionizing radiation: still ATM territory. Cell Division. 2006;1:7. doi: 10.1186/1747-1028-1-7. Available at: http://www.celldiv.com/content/1/1/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 15.Sedelnikova OA, Rogakou EP, Panuytin IG, Bonner W. Quantitative detection of 125IUdr-induced DNA double-strand breaks with γ-H2AX antibody. Radiat Res. 2002;158:486–492. doi: 10.1667/0033-7587(2002)158[0486:qdoiid]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 16.Huang X, Traganos F, Darzynkiewicz Z. DNA damage induced by DNA topoisomerase I- and topoisomerase II- inhibitors detected by histone H2AX phosphorylation in relation to the cell cycle phase and apoptosis. Cell Cycle. 2003;2:614–619. [PubMed] [Google Scholar]
- 17.Banath JP, Olive PL. Expression of phosphorylated histone H2AX as a surrogate of cell killing by drugs that create DNA double-strand breaks. Cancer Res. 2003;63:4347–4350. [PubMed] [Google Scholar]
- 18.Huang X, Okafuji M, Traganos F, Luther E, Holden E, Darzynkiewicz Z. Assessment of histone H2AX phosphorylation induced by DNA topoisomerase I and II inhibitors topotecan and mitoxantrone and by DNA crosslinking agent cisplatin. Cytometry A. 2004;58A:99–110. doi: 10.1002/cyto.a.20018. [DOI] [PubMed] [Google Scholar]
- 19.Olive PL, Banath JP, Sinnott LT. Phosphorylated histone H2AX in spheroids, tumors, and tissues of mice exposed to etoposide and 3-amino-1,2,4-benzotriazine-1,3-dioxide. Cancer Res. 2004;64:5363–5369. doi: 10.1158/0008-5472.CAN-04-0729. [DOI] [PubMed] [Google Scholar]
- 20.Kurose A, Tanaka T, Huang X, Halicka HD, Traganos F, Dai W, Darzynkiewicz Z. Assessment of ATM phosphorylation on Ser-1981 induced by DNA topoisomerase I and II inhibitors in relation to Ser-139-histone H2AX phosphorylation, cell cycle phase and apoptosis. Cytometry A. 2005;68A:1–9. doi: 10.1002/cyto.a.20186. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka T, Kurose A, Huang X, Dai W, Darzynkiewicz Z. ATM kinase activation and histone H2AX phosphorylation as indicators of DNA damage by DNA topoisomerase I inhibitor topotecan and during apoptosis. Cell Prolif. 2006;39:49–60. doi: 10.1111/j.1365-2184.2006.00364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang X, Kurose A, Tanaka T, Traganos F, Dai W, Darzynkiewicz Z. Activation of ATM and histone H2AX phosphorylation induced by mitoxantrone but not by topotecan is prevented by the antioxidant N-acetyl-L-cysteine. Cancer Biol Ther. 2006;5:959–964. doi: 10.4161/cbt.5.8.2878. [DOI] [PubMed] [Google Scholar]
- 23.Zhao H, Traganos F, Darzynkiewicz Z. Kinetics of histone H2AX phosphorylation and Chk2 activation in A549 cells treated with topotecan and mitoxantrone in relation to the cell cycle phase. Cytometry A. 2008;73A:480–489. doi: 10.1002/cyto.a.20574. [DOI] [PubMed] [Google Scholar]
- 24.Smart DJ, Halicka HD, Schmuck G, Traganos F, Darzynkiewicz Z, Williams GM. Assessment of DNA double-strand breaks and γH2AX induced by the topoisomerase II poisons etoposide and mitoxantrone. Mutat Res. 2008;641:43–47. doi: 10.1016/j.mrfmmm.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka T, Halicka HD, Traganos F, Seiter K, Darzynkiewicz Z. Induction of ATM activation, histone H2AX phosphorylation and apoptosis by etoposide: Relation to the cell cycle phase. Cell Cycle. 2007;6:371–376. doi: 10.4161/cc.6.3.3835. [DOI] [PubMed] [Google Scholar]
- 26.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diermeier-Daucher S, Clarke ST, Hill D, Vollmann-Zwerenz A, Bradford JA, Brockhoff G. Cell type specific applicability of 5-ethynyl-2-deoxyuridine (EdU) for dynamic proliferation assessment in flow cytometry. Cytometry A. 2009;75A:535–546. doi: 10.1002/cyto.a.20712. [DOI] [PubMed] [Google Scholar]
- 28.Hamelik RA, Krishan A. Click-iT assay with improved DNA frequency histograms. Cytometry A. 2009;75:862–865. doi: 10.1002/cyto.a.20780. [DOI] [PubMed] [Google Scholar]
- 29.Darzynkiewicz Z, Traganos F, Zhao H, Halicka HD, Li J. Cytometry of DNA replication and RNA synthesis: Historical perspective and recent advances based on “click chemistry”. Cytometry A. 2011;79A:328–337. doi: 10.1002/cyto.a.21048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao H, Li J, Traganos F, Halicka HD, Zarebski M, Dobrucki J, Darzynkiewicz Z. Cell fixation in zinc salt solution (ZBF) is compatible with DNA damage response detection by phospho-specific antibodies. Cytometry A. 2011;79A:470–476. doi: 10.1002/cyto.a.21060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao H, Traganos F, Darzynkiewicz Z. Kinetics of the UV-induced DNA damage response in relation to cell cycle phase. Correlation with DNA replication. Cytometry A. 2010;77A:285–293. doi: 10.1002/cyto.a.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao H, Dobrucki J, Rybak P, Traganos F, Halicka HD, Darzynkiewicz Z. Induction of DNA damage signaling by oxidative stress in relation to DNA replication as detected using “click chemistry”. Cytometry A. 2011;79A:897–902. doi: 10.1002/cyto.a.21137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leonhardt H, Rahn HP, Weinzierl P, Sporbert A, Cremen T, Zink D, Cardoso MC. Dynamics of DNA replication factories in living cells. J Cell Biol. 2000;149:271–279. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao H, Traganos F, Darzynkiewicz Z. Phosphorylation of p53 on Ser15 during cell cycle caused by Topo I and Topo II inhibitors in relation to ATM and Chk2 activation. Cell Cycle. 2008;7:3048–3055. doi: 10.4161/cc.7.19.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Del Bino G, Lassota P, Darzynkiewicz Z. The S-phase cytotoxicity of camptothecin. Exp Cell Res. 1991;193:27–35. doi: 10.1016/0014-4827(91)90534-2. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka T, Halicka HD, Huang X, Traganos F, Darzynkiewicz Z. Constitutive histone H2AX phosphorylation and ATM activation, the reporters of DNA damage by endogenous oxidants. Cell Cycle. 2006;5:1940–1945. doi: 10.4161/cc.5.17.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka T, Kajstura M, Halicka HD, Traganos F, Darzynkiewicz Z. Constitutive histone H2AX phosphorylation and ATM activation are strongly amplified during mitogenic stimulation of lymphocytes. Cell Prolif. 2007;40:1–13. doi: 10.1111/j.1365-2184.2007.00417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorczyca W, Gong J, Ardelt B, Traganos F, Darzynkiewicz Z. The cell cycle related differences in susceptibility of HL-60 cells to apoptosis induced by various antitumor agents. Cancer Res. 1993;53:3186–3192. [PubMed] [Google Scholar]
- 39.Kapuscinski J, Darzynkiewicz Z. Interactions of antitumor agents ametantrone and mitoxantrone (novantrone) with double-stranded DNA. Biochem Pharmacol. 1985;34:4203–4213. doi: 10.1016/0006-2952(85)90275-8. [DOI] [PubMed] [Google Scholar]
- 40.Bender RP, Jablonksy MJ, Shadid M, Romaine I, Dunlap N, Anklin C, Graves DE, Osheroff N. Substituents on etoposide that interact with human topoisomerase IIalpha in the binary enzyme-drug complex: Contributions to etoposide binding and activity. Biochemistry. 2008;47:4501–4509. doi: 10.1021/bi702019z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka T, Huang X, Halicka HD, Zhao H, Traganos F, Albino AP, Dai W, Darzynkiewicz Z. Cytometry of ATM activation and histone H2AX phosphorylation to estimate extent of DNA damage induced by exogenous agents. Cytometry A. 2007;71A:648–661. doi: 10.1002/cyto.a.20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolte S, Coldelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 43.Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with ImageJ. Biophoton Int. 2004;7:36–42. [Google Scholar]
- 44.Manders EEM, Verbeek FJ, Aten JA. Measurement of co-localization on objects in dual confocal images. J Microsc. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 45.Halicka HD, Zhao H, Podhorecka M, Traganos F, Darzynkiewicz Z. Cytometric detection of chromatin relaxation, an early reporter of DNA damage response. Cell Cycle. 2009;8:2233–2237. doi: 10.4161/cc.8.14.8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Housman D, Huberman JA. Changes in the rate of DNA replication fork movement along the S phase in mammalian cells. J Mol Biol. 1975;94:173–176. doi: 10.1016/0022-2836(75)90076-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.