Abstract
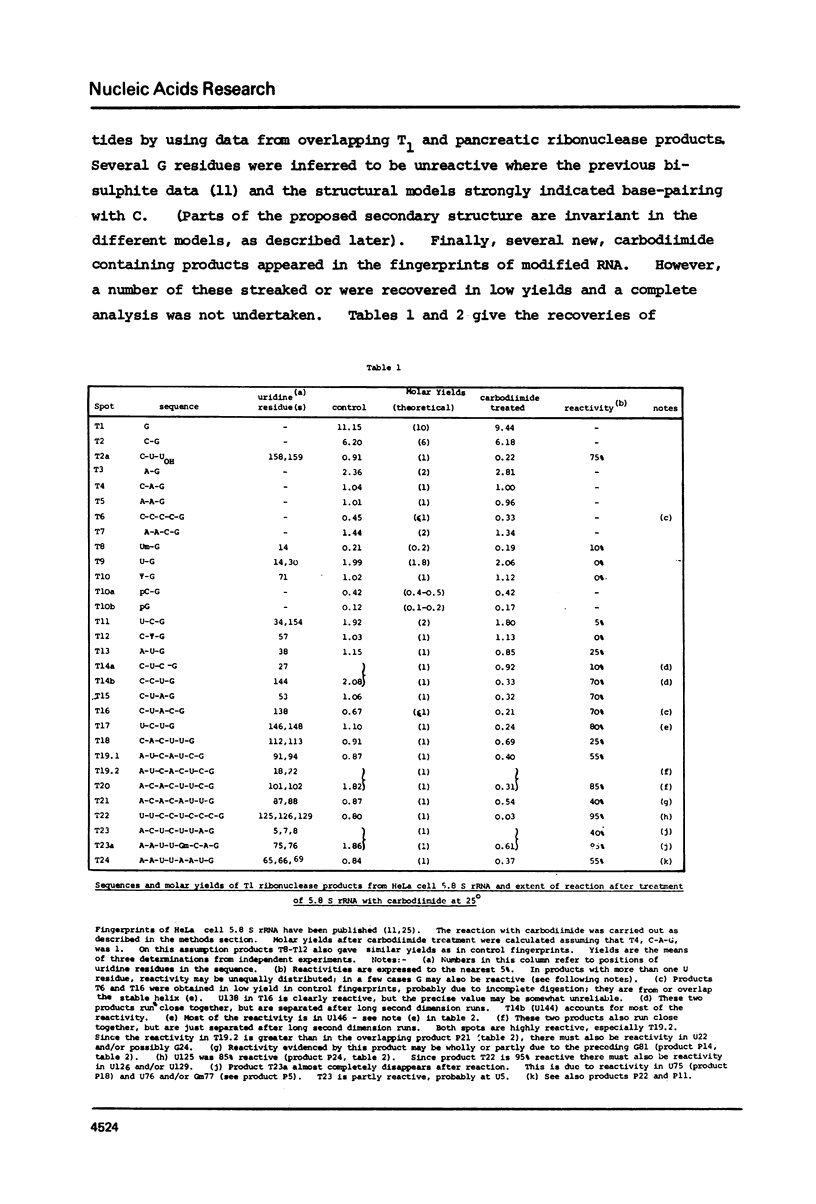
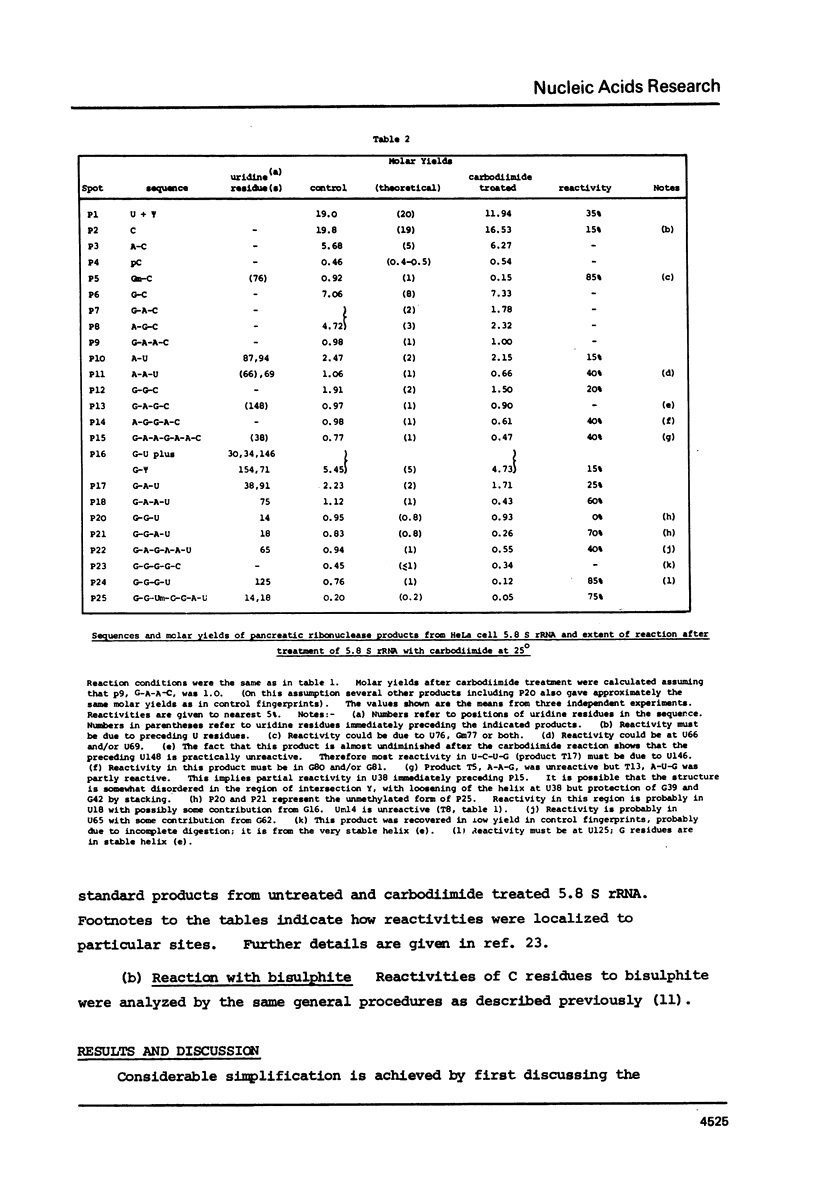
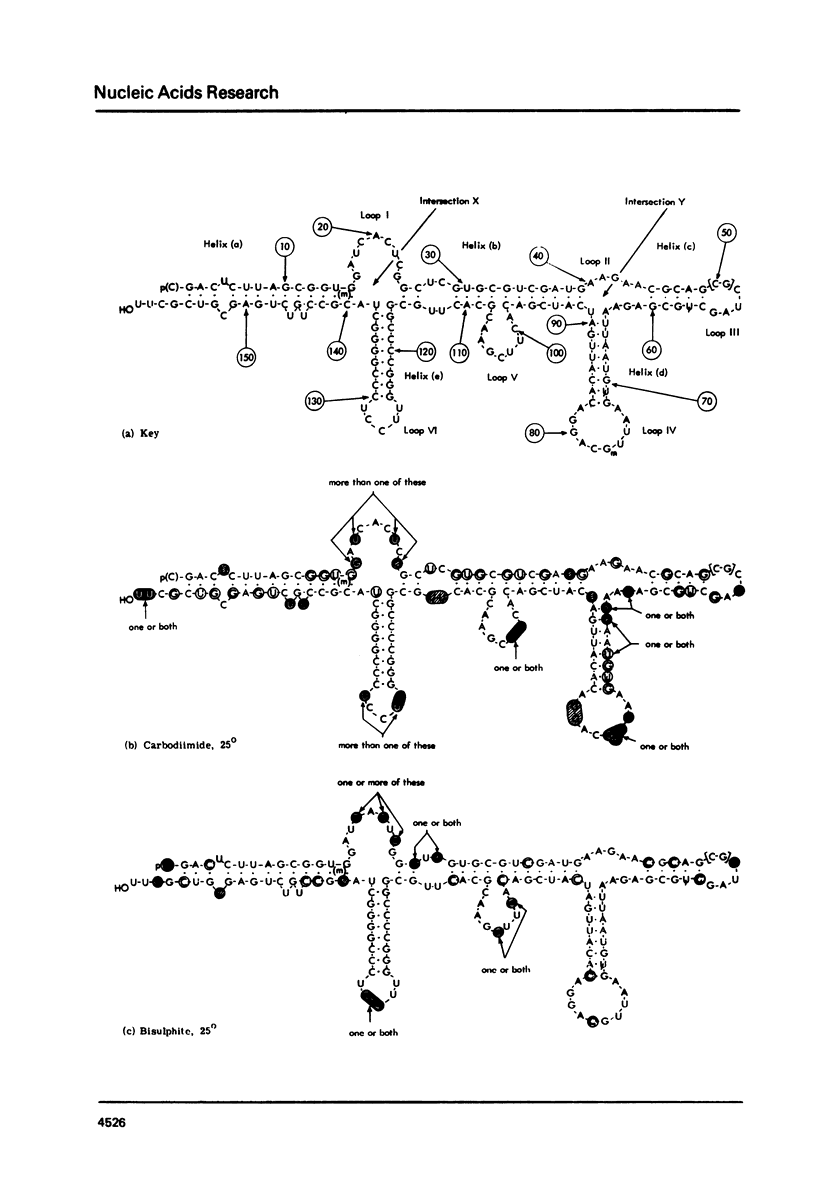
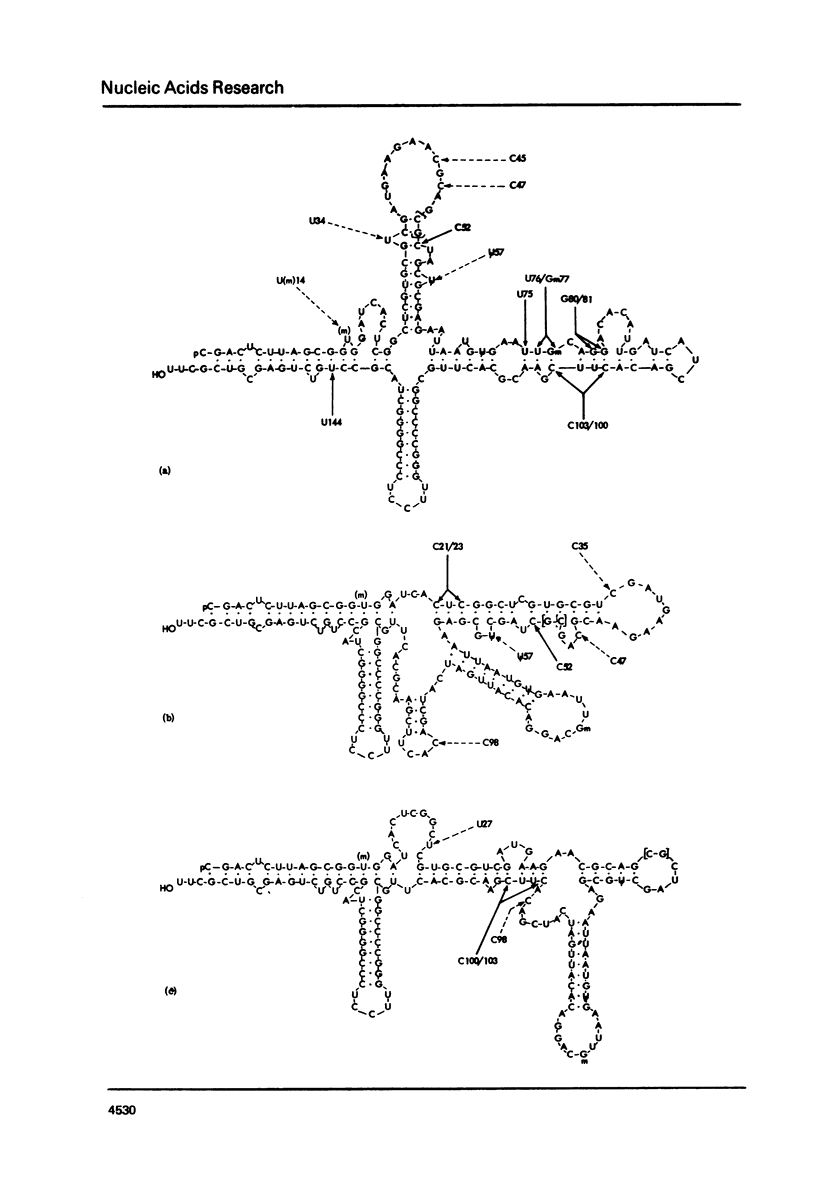
Various secondary structure models have been proposed for 5.8 S rRNA. In this paper HeLa cell 5.8 S rRNA is shown to possess several sites that are reactive to carbodiimide at 25 degrees, and other regions that are unreactive. Previous work has established the distribution of reactive and unreactive cytidine residues along the primary structure (11). The secondary structure model of Nazar et al. (7) is fully compatible with the chemical reactivity data whereas other models are partly incompatible. We conclude that the model of Nazar et al. provides the best approximation so far available to the conformation of isolated 5.8 S rRNA. Findings on the effect temperature on the chemical reactivity of different parts of the structure are summarized. The findings described in this paper should provide a basis for examining the specific interaction of 5.8 S rRNA with 28 s rRNA.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boseley P. G., Tuyns A., Birnstiel M. L. Mapping of the Xenopus laevis 5.8S rDNA by restriction and DNA sequencing. Nucleic Acids Res. 1978 Apr;5(4):1121–1137. doi: 10.1093/nar/5.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee G. G., Sanger F., Barrell B. G. The sequence of 5 s ribosomal ribonucleic acid. J Mol Biol. 1968 Jun 28;34(3):379–412. doi: 10.1016/0022-2836(68)90168-x. [DOI] [PubMed] [Google Scholar]
- Chang S. E., Ish-Horowicz D. Selective modification of cytidine, uridine, guanosine and pseudouridine residues in Escherichia coli leucine transfer ribonucleic acid. J Mol Biol. 1974 Apr 15;84(3):375–388. doi: 10.1016/0022-2836(74)90446-x. [DOI] [PubMed] [Google Scholar]
- Chang S. E. Selective modification of cytidine and uridine residues in Escherichia coli formylmethionine transfer ribonucleic acid. J Mol Biol. 1973 Apr 15;75(3):533–547. doi: 10.1016/0022-2836(73)90459-2. [DOI] [PubMed] [Google Scholar]
- Dawid I. B., Wellauer P. K. A reinvestigation of 5' leads to 3' polarity in 40S ribosomal RNA precursor of Xenopus laevis. Cell. 1976 Jul;8(3):443–448. doi: 10.1016/0092-8674(76)90157-4. [DOI] [PubMed] [Google Scholar]
- Ford P. J., Mathieson T. The nucleotide sequences of 5.8-S ribosomal RNA from Xenopus laevis and Xenopus borealis. Eur J Biochem. 1978 Jun 1;87(1):199–214. doi: 10.1111/j.1432-1033.1978.tb12367.x. [DOI] [PubMed] [Google Scholar]
- Goddard J. P. The structures and functions of transfer RNA. Prog Biophys Mol Biol. 1977;32(3):233–308. [PubMed] [Google Scholar]
- Grunberg-Manago M., Dessen P., Pantaloni D., Godefroy-Colburn T., Wolfe A. D., Dondon J. Light-scattering studies showing the effect of initiation factors on the reversible dissociation of Escherichia coli ribosomes. J Mol Biol. 1975 May 25;94(3):461–478. doi: 10.1016/0022-2836(75)90215-6. [DOI] [PubMed] [Google Scholar]
- Harada F., Dahlberg J. E. Specific cleavage of tRNA by nuclease S1. Nucleic Acids Res. 1975 Jun;2(6):865–871. doi: 10.1093/nar/2.6.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho N. W., Gilham P. T. Reaction of pseudouridine and inosine with N-cyclohexyl-N'-beta-(4-methylmorpholinium)ethylcarbodiimide. Biochemistry. 1971 Sep 28;10(20):3651–3657. [PubMed] [Google Scholar]
- Ho N. W., Gilham P. T. The reversible chemical modification of uracil, thymine, and guanine nucleotides and the modification of the action of ribonuclease on ribonucleic acid. Biochemistry. 1967 Dec;6(12):3632–3639. doi: 10.1021/bi00864a002. [DOI] [PubMed] [Google Scholar]
- Kelly J. M., Goddard J. P., Maden E. H. Evidence on the conformation of HeLa-cell 5.8S ribosomal ribonucleic acid from the reaction of specific cytidine residues with sodium bisulphite. Biochem J. 1978 Aug 1;173(2):521–532. doi: 10.1042/bj1730521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King H. W., Gould H. Low molecular weight ribonucleic acid in rabbit reticulocyte ribosomes. J Mol Biol. 1970 Aug;51(3):687–702. doi: 10.1016/0022-2836(70)90017-3. [DOI] [PubMed] [Google Scholar]
- Luoma G. A., Marshall A. G. Laser Raman evidence for new cloverleaf secondary structures for eukaryotic 5.8S RNA and prokaryotic 5S RNA. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4901–4905. doi: 10.1073/pnas.75.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E., Robertson J. S. Demonstration of the "5-8 S" ribosomal sequence in HeLa cell ribosomal precursor RNA. J Mol Biol. 1974 Aug 5;87(2):227–235. doi: 10.1016/0022-2836(74)90145-4. [DOI] [PubMed] [Google Scholar]
- Metz D. H., Brown G. L. The investigation of nucleic acid secondary structure by means of chemical modification with a carbodiimide reagent. I. The reaction between N-cyclohexyl-N'-beta-(4-methylmorpholinium)ethylcarbodiimide and model nucleotides. Biochemistry. 1969 Jun;8(6):2312–2328. doi: 10.1021/bi00834a012. [DOI] [PubMed] [Google Scholar]
- Nazar R. N., Sitz T. O., Busch H. Structural analyses of mammalian ribosomal ribonucleic acid and its precursors. Nucleotide sequence of ribosomal 5.8 S ribonucleic acid. J Biol Chem. 1975 Nov 25;250(22):8591–8597. [PubMed] [Google Scholar]
- Pace N. R., Walker T. A., Schroeder E. Structure of the 5.8S RNA component of the 5.8S-28S ribosomal RNA junction complex. Biochemistry. 1977 Nov 29;16(24):5321–5328. doi: 10.1021/bi00643a025. [DOI] [PubMed] [Google Scholar]
- Pene J. J., Knight E., Jr, Darnell J. E., Jr Characterization of a new low molecular weight RNA in HeLa cell ribosomes. J Mol Biol. 1968 May 14;33(3):609–623. doi: 10.1016/0022-2836(68)90309-4. [DOI] [PubMed] [Google Scholar]
- Robertson J. S., Maden B. E. Nucleotide sequences from the non-conserved region of HeLa cell 32-S ribosomal precursor RNA. Biochim Biophys Acta. 1973 Nov 26;331(1):61–70. doi: 10.1016/0005-2787(73)90419-x. [DOI] [PubMed] [Google Scholar]
- Rubin G. M. The nucleotide sequence of Saccharomyces cerevisiae 5.8 S ribosomal ribonucleic acid. J Biol Chem. 1973 Jun 10;248(11):3860–3875. [PubMed] [Google Scholar]
- Van N. T., Nazar R. N., Sitz T. O. Comparative studies on the secondary structure of eukaryotic 5.8S ribosomal RNA. Biochemistry. 1977 Aug 23;16(17):3754–3759. doi: 10.1021/bi00636a004. [DOI] [PubMed] [Google Scholar]
- Walker T. A., Pace N. R. Transcriptional organization of the 5.8S ribosomal RNA cistron in Xenopus laevis ribosomal DNA. Nucleic Acids Res. 1977 Mar;4(3):595–601. doi: 10.1093/nar/4.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshaw M. M., Tinoco I., Jr Absorption and optical rotatory dispersion of six dinucleoside phosphates. J Mol Biol. 1965 Aug;13(1):54–64. doi: 10.1016/s0022-2836(65)80079-1. [DOI] [PubMed] [Google Scholar]



