Abstract
Myostatin, or GDF8, is an inhibitor of skeletal muscle growth. A non-functional myostatin mutation leads to a double muscling phenotype in some species, for example, mice, cattle and humans. Previous studies have indicated that there are loci in the genome that interact with myostatin to control backfat depth and other complex traits. We now report a quantitative trait loci (QTL) mapping study designed to identify loci that interact with myostatin to impact growth traits in mice. Body weight and average daily gain traits were collected on F2 progeny derived from a myostatin-null C57BL/6 strain by M16i cross. In all, 44 main effect QTL were detected above a 5% genome-wide significance threshold when an interval mapping method was used. An additional 37 QTL were identified to significantly interact with myostatin, sex or reciprocal cross. A total of 12 of these QTL interacted with myostatin genotype. These results provide a foundation for the further fine mapping of genome regions that harbor loci that interact with myostatin.
Keywords: QTL mapping, growth traits, epistasis, myostatin, mouse
Introduction
Growth traits are complex traits of economic importance to animal agriculture. A good understanding of mechanisms that control growth will benefit both meat production and human health. Therefore, growth traits have been widely studied in animal models (Corva and Medrano, 2001; Cheverud, 2005). Heritability studies of growth-related traits (McCarthy, 1982; Eisen, 1986) indicate that there are genetic mechanisms that partially control these traits. Previous publications have estimated the amount of phenotypic variation in growth traits that can be explained by different types of genetic effects in chicken (Carlborg et al., 2003) and mice (Brockmann et al., 2000). It has been demonstrated that, among these genetic effects, epistasis explains a large amount of phenotypic variation, as compared with additive and dominance effects. For example, Brockmann et al. (2000) estimated that about 33% of the phenotypic variation of body weight in mice could be attributed to epistasis. Another study indicated that there might be a time-dependent pattern for these genetic effects and epistasis might be more important for early growth traits (Carlborg et al., 2003).
Identification of key genes that control growth traits has shed light on the molecular mechanisms that regulate growth. For example, myostatin has been shown to inhibit muscle growth in cattle, dogs, mice, sheep and humans (Grobet et al., 1997; Kambadur et al., 1997; McPherron and Lee, 1997; McPherron et al., 1997; Schuelke et al., 2004; Clop et al., 2006; Mosher et al., 2007). As a member of the transforming growth factor-β family, myostatin was first identified in mice for its significant effect on skeletal muscle growth (McPherron et al., 1997). Interestingly, not all myostatin-null animals show an obvious difference in their muscle mass. For example, some South Devon cattle that are homozygous for myostatin-null mutations do not exhibit the double-muscling phenotype (Smith et al., 2000). In addition, Grobet et al. (1997) showed that Limousin and Blonde dAquitaine, which both possess wild-type myostatin alleles, exhibited a double muscling phenotype.
Other loci might epistatically interact with myostatin to control skeletal muscle growth. In cattle, only loci that interact with myostatin to control backfat depth and meat tenderness have been identified (Casas et al., 2000; Casas et al., 2001). Compared with cattle, there is more evidence of genetic loci that interact with myostatin to control muscle growth in mice (Varga et al., 2003; Bunger et al., 2004). As the main function of myostatin is to control skeletal muscle growth, it will be critical to map these loci. Furthermore, the knowledge gained will help elucidate the molecular details behind the genetic architecture of growth traits.
Here we developed an F2 population from myostatin-null C57BL/6 mice by M16i obese myostatin wild-type mice. M16i mice have been used to identify quantitative trait loci (QTL) that are associated with body weight, fat weight and body composition traits (Jerez-Timaure et al., 2005). As an obese mouse line, it is a good model to search for causal mutations that affect muscle and adipose development. As there was a large phenotypic difference of muscle and fat weight traits between myostatin-null C57BL/6 mice and M16i mice, we expected loci that controlled these traits would also have an effect on body weight and segregate in the F2 mice derived from these two lines. A whole-genome scan was performed to identify main effect QTL. Subsequently, a model comparison approach was applied to search for potential epistatic QTL. We further investigated the phenotypic and genetic variation that could be accounted for by these QTL.
Materials and methods
Mapping population
An F2 cross design was used for this study. We first derived the mapping population from reciprocal crosses between the two founder strains, myostatin-null C57BL/6 (McPherron et al., 1997) and M16i high body weight mice (Hanrahan et al., 1973; Eisen, 1986). Four myostatin-null male mice were mated with eight M16i females to produce 35 male and 37 female F1 offspring. In addition, two M16i males and seven myostatin-null females were crossed to generate 31 male and 55 female F1 offspring. Within each of these two crosses, F1 mice were intercrossed to produce the F2 mapping population.
Phenotyping and genotyping procedure
Each F2 litter was standardized to nine pups at 1 day after birth. At 1 week of age, mice were individually identified. Mice were weaned when they were 3 weeks old. Isolated DNA was used to determine the myostatin genotype by a standard PCR reaction. A total of 1000 F2 generation progeny that were either homozygous myostatin wild-type or homozygous myostatin-null were collected for genotype and phenotype information. Among these 1000 F2 mice, 552 mice were from the myostatin-null male by M16i female cross, whereas 448 mice were from the reciprocal cross. Each of the F2 individuals had its body weight recorded at 7, 14, 21, 28, 35 and 42 days of age. On the basis of the measurements of body weights, growth rate was calculated for different growth periods. For the pre-weaning period, average daily gain was calculated for the 2nd week and 3rd week, along with the 1st–3rd week interval. During the post-weaning period, average daily gain was calculated for the 4th week, 5th week, 6th week, and the 4th–6th week interval. Furthermore, average daily gain for the 1st–6th week interval was calculated and included as a measurement of overall growth rate. Genomic DNA from each of the F0, F1 and F2 generation mice was purified by a phenol chloroform method. All animal procedures were approved by the Iowa State University Animal Care and Use Committee before this study was conducted.
Linkage map
We genotyped 242 single-nucleotide polymorphisms on the Sequenom platform at GeneSeek (Lincoln, Nebraska, NE, USA). These SNPs were evenly distributed on 19 autosomes and the X chromosome. First, genotyping data were checked for genotyping errors on the basis of Mendelian inheritance and pedigree information. SNPs with low call rates (<80%) and genotyping errors were discarded. After this first selection, 152 SNPs were left and they were distributed on 17 autosomes (no informative SNPs were present on chromosome 15 and chromosome 16) and the X chromosome. Second, a χ2-test was used to evaluate the extent of segregation distortion in the F2 population. Most markers did not significantly deviate from the expected Mendelian segregation ratios, except for SNPs close to the myostatin locus, which was expected because only homozygotes at the myostatin locus were included. Therefore, the remaining 152 SNPs were used for our statistical analysis. Finally, a linkage map was constructed in Kosambi centimorgans with Cri-map (Green et al., 1990). Marker order and position in this map (Supplementary Table 4) was in reasonable agreement with those from the Wellcome-CTC Mouse Strain SNP Genotype Set (http://mus.well.ox.ac.uk/mouse/INBREDS/). Thus, we performed the following QTL mapping analysis based on our linkage map.
General statistical analysis
SAS 9.2. (SAS Institute Inc., Cary, NC, USA) was used to explore the data structure of the F2 phenotypic data. First, simple statistics were calculated on the 14 growth traits. Second, Q–Q plot and normality tests were used to verify that each trait was normally distributed. Third, the Proc General Linear Model procedure was applied to each trait to identify significant factors that need to be accounted for in the QTL mapping model. Specifically, effects of myostatin genotype, coat color, sex, reciprocal cross and interactions among these factors were evaluated. All these factors have been previously shown to impact growth traits. Finally, after adjusting fixed effects that were identified in the previous step, residual correlations were calculated and tested between each pair of traits by the Proc CORR procedure.
Main effect QTL analysis
GridQTL (Seaton et al., 2006), a web-based QTL analysis program, was used to identify QTL associated with the 14 growth traits by interval mapping. The QTL model included the main effects of myostatin genotype, coat color, sex and reciprocal cross, and the interaction effects of sex × myostatin genotype, sex × reciprocal cross, myostatin genotype × reciprocal cross, sex × coat color and sex × myostatin genotype × reciprocal cross along with the additive and dominance effect at the single QTL position. For each trait, both F-values and logarithm of the odds (LOD) values were provided by GridQTL. Genome-wide significance levels (1 and 5%) were determined by the genome-wide permutation procedure (Churchill and Doerge, 1994) implemented in GridQTL, using 1000 permutations. Additive and dominance effects, along with the corresponding standard errors, were estimated for each significant QTL peaks. The percentage of phenotypic variation accounted for by a QTL position was computed as the percentage of residual sum of squares explained by the additive and dominance effects at the QTL. For each trait, the total phenotypic variation of all QTL was calculated by summing the percentage of phenotypic variation together.
QTL interaction with myostatin genotype, sex and reciprocal cross effect
To test the potential interaction between a QTL and myostatin genotype, we first applied a forward QTL selection strategy. In this step, the dataset was split into two subsets by myostatin genotype. Interval mapping analysis was completed using the main effect QTL model without myostatin genotype effect within each subset. All positions that reached a 5% genome-wide significance level were identified. In the second step, the interaction between each of these positions and myostatin genotype was tested by comparing four models in the full data. Fixed effects in the following models were the same as before.
Model 1:
 |
Model 2:
 |
Model 3:
 |
Model 4:
 |
The F-value was computed with the following formula:
To test overall interaction:
 |
To test additive interaction:
 |
To test dominance interaction:
 |
The corresponding P-value was calculated from the F-distribution. A comparison-wise P-value less than 0.05 was considered significant.
To analyze the interaction effect between QTL and sex, the full dataset was split into two subsets according to gender, female F2 mice and male F2 mice data. A similar approach to the one described above was used by substituting the myostatin × additive effect with the sex × additive effect and the myostatin × dominance effect with the sex × dominance effect.
To analyze the interaction effect between QTL and reciprocal cross, the full mouse dataset was again split into two subsets according to reciprocal cross information: the M16i female × myostatin male cross and the M16i male × myostatin female cross data. Again, a similar approach as described above was used by substituting the myostatin × additive effect with the cross × additive effect and the myostatin × dominance effect with the cross × dominance effect.
For each of the detected QTL, the additive and dominance effects were estimated. The amount of phenotypic variation accounted for by each was calculated using the percentage of sum of squares explained as described above for main effect QTL. For each trait, the total variation explained by QTL was calculated as the sum of phenotypic variation from all main effect QTL and interaction QTL that were identified for the trait.
Results
General statistics
Our initial statistical analyses indicated that all 14 phenotype traits were normally distributed (Supplementary Figure 1). The histograms in Supplementary Figure 1 illustrate that the effect of myostatin-null genotype and male sex increased with age from 1st–6th week of age. The general statistics of these 14 traits are presented in Supplementary Table 1. Most of the body weight and average daily gain traits had significant correlations with one another (Table 1). Early body weights (1st–3rd week) were negatively correlated with body weights during the post-weaning growth period (4th–6th week). In addition, the 14 body weight traits were identified for significant main effects of myostatin genotype, sex and reciprocal cross. Interactions between main effects were significant for some traits (unpublished results).
Table 1. Phenotypic correlations among growth traits.
| Traits | BW7 | BW14 | BW21 | BW28 | BW35 | BW42 | ADG 7–14 | ADG 14–21 | ADG 21–28 | ADG 28–35 | ADG 35–42 | ADG 7–21 | ADG 28–42 | ADG 7–42 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BW7 | 1.00 | 0.78 | 0.81 | 0.68 | 0.48 | 0.31 | 0.15 | 0.62 | 0.33 | −0.14 | −0.20 | 0.56 | −0.20 | 0.16 |
| BW14 | — | 1.00 | 0.88 | 0.74 | 0.56 | 0.38 | 0.73 | 0.52 | 0.36 | −0.07 | −0.20 | 0.79 | −0.15 | 0.26 |
| BW21 | — | — | 1.00 | 0.86 | 0.63 | 0.43 | 0.52 | 0.86 | 0.43 | −0.11 | −0.23 | 0.94 | −0.19 | 0.31 |
| BW28 | — | — | — | 1.00 | 0.86 | 0.68 | 0.43 | 0.75 | 0.84 | 0.10 | −0.06 | 0.81 | 0.02 | 0.59 |
| BW35 | — | — | — | — | 1.00 | 0.92 | 0.37 | 0.54 | 0.82 | 0.60 | 0.22 | 0.62 | 0.47 | 0.87 |
| BW42 | — | — | — | — | — | 1.00 | 0.26 | 0.36 | 0.74 | 0.71 | 0.59 | 0.42 | 0.75 | 0.99 |
| ADG 7–14 | — | — | — | — | — | — | 1.00 | 0.15 | 0.20 | 0.04 | −0.10 | 0.64 | −0.03 | 0.25 |
| ADG 14–21 | — | — | — | — | — | — | — | 1.00 | 0.40 | −0.12 | −0.21 | 0.86 | −0.19 | 0.27 |
| ADG 21–28 | — | — | — | — | — | — | — | — | 1.00 | 0.28 | 0.14 | 0.42 | 0.24 | 0.71 |
| ADG 28–35 | — | — | — | — | — | — | — | — | — | 1.00 | 0.52 | −0.07 | 0.88 | 0.76 |
| ADG 35–42 | — | — | — | — | — | — | — | — | — | — | 1.00 | −0.22 | 0.87 | 0.65 |
| ADG 7–21 | — | — | — | — | — | — | — | — | — | — | — | 1.00 | −0.16 | 0.34 |
| ADG 28–42 | — | — | — | — | — | — | — | — | — | — | — | — | 1.00 | 0.81 |
| ADG 7–42 | — | — | — | — | — | — | — | — | — | — | — | — | — | 1.00 |
Phenotypic correlation coefficients have P-value less between 0.01 and 0.05 are in italic.
Phenotypic correlation coefficients have P-value larger than 0.05 are in bold.
All other phenotypic correlation coefficients have P-value less than 0.01.
Main effect QTL
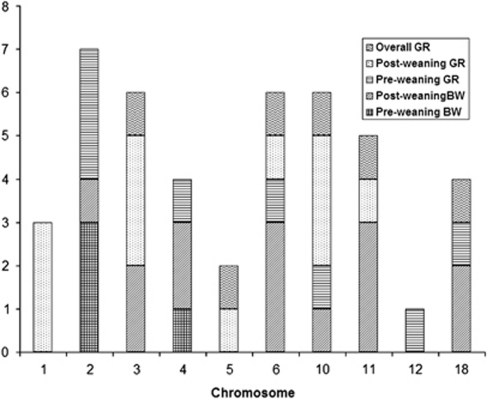
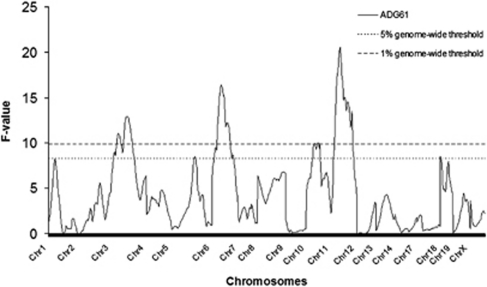
On the basis of genome-wide scan in the whole F2 population with the additive and dominance QTL model specified in the method section, we observed 26 and 44 QTL that were associated with these 14 growth traits at a 1% and 5% genome-wide significance level, respectively (Table 2). These 44 QTL were distributed on 10 autosomes (chromosome 1, 2, 3, 4, 5, 6, 10, 11, 12 and 18; Figure 1). Chromosome 2 harbored the greatest number of QTL, whereas chromosome 12 contained the least number of QTL. We identified four pre-weaning body weight QTL, 14 post-weaning body weight QTL, eight pre-weaning growth rate QTL, 12 post-weaning growth rate QTL and six overall growth rate QTL (Figure 1). The total phenotypic variation explained by these QTL was summed for each trait in Table 3, along with the number of QTL. The average phenotypic variation accounted for by each of the 44 QTL was about 2.5%. Average daily gain for the 1st–6th week period (Figure 2) had the most QTL identified and these QTL also explained the largest amount, about 15.6%, of the phenotypic variance (Table 3).
Table 2. Statistics of main effect quantitative trait loci detected in genome wide scan in the full data set.
| MMU | Traits | Peak (cM)a |
Flanking markersb |
Estimate |
%var | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Left | Right | F | LOD | a | s.ea | d | s.ed | ||||
| 1 | ADG35–42** | 22 | rs3696088 | rs13472794 | 15.83 | 6.766 | −0.107 | 0.020 | −0.027 | 0.026 | 3.11 |
| 1 | ADG28–42** | 23 | rs3696088 | rs13472794 | 25.91 | 10.968 | −0.108 | 0.015 | −0.027 | 0.021 | 4.99 |
| 1 | ADG28–35** | 24 | rs3696088 | rs13472794 | 10.63 | 4.568 | −0.110 | 0.024 | −0.027 | 0.032 | 2.11 |
| 2 | ADG14–21** | 71 | rs3144393 | rs13476878 | 26.27 | 11.115 | 0.061 | 0.009 | 0.028 | 0.012 | 5.05 |
| 2 | ADG7–21** | 79 | rs3144393 | rs13476878 | 15.18 | 6.491 | 0.024 | 0.006 | 0.035 | 0.009 | 2.98 |
| 2 | BW28** | 79 | rs3144393 | rs13476878 | 11.88 | 5.096 | 0.548 | 0.187 | 1.082 | 0.276 | 2.35 |
| 2 | BW21** | 81 | rs3144393 | rs13476878 | 14.21 | 6.083 | 0.427 | 0.118 | 0.708 | 0.179 | 2.80 |
| 2 | ADG7–14 | 96 | rs13476878 | rs3143843 | 8.13 | 3.500 | −0.018 | 0.006 | 0.024 | 0.009 | 1.62 |
| 2 | BW14** | 101 | rs13476878 | rs3143843 | 10.27 | 4.414 | −0.032 | 0.064 | 0.400 | 0.089 | 2.04 |
| 2 | BW7** | 103 | rs3143843 | rs6335805 | 10.46 | 4.497 | 0.077 | 0.044 | 0.269 | 0.063 | 2.08 |
| 3 | ADG28–35** | 25 | rs3663409 | rs13477132 | 13.32 | 5.708 | 0.040 | 0.013 | 0.075 | 0.019 | 2.63 |
| 3 | ADG28–42 | 30 | rs3663409 | rs13477132 | 10.55 | 4.534 | 0.027 | 0.009 | 0.045 | 0.013 | 2.09 |
| 3 | ADG7–42** | 58 | rs3663873 | rs13477430 | 12.94 | 5.546 | 0.027 | 0.006 | 0.026 | 0.009 | 2.55 |
| 3 | BW42** | 61 | rs3663873 | rs13477430 | 10.78 | 4.632 | 0.988 | 0.239 | 1.011 | 0.405 | 2.14 |
| 3 | BW35 | 64 | rs3663873 | rs13477430 | 8.95 | 3.851 | 0.999 | 0.246 | 0.713 | 0.438 | 1.78 |
| 3 | ADG21–28 | 120 | rs3724562 | CEL–3_159340478 | 8.85 | 3.811 | 0.057 | 0.014 | −0.011 | 0.027 | 1.76 |
| 4 | ADG7–21** | 63 | rs6324470 | rs3659226 | 10.15 | 4.363 | −0.023 | 0.006 | 0.015 | 0.008 | 2.02 |
| 4 | BW21 | 64 | rs6324470 | rs3659226 | 7.95 | 3.426 | −0.400 | 0.110 | 0.262 | 0.169 | 1.59 |
| 4 | BW28** | 64 | rs6324470 | rs3659226 | 10.64 | 4.572 | −0.738 | 0.175 | 0.486 | 0.269 | 2.11 |
| 4 | BW35 | 67 | rs6324470 | rs3659226 | 8.08 | 3.480 | −0.733 | 0.212 | 0.667 | 0.342 | 1.61 |
| 5 | ADG7–42 | 76 | rs3720626 | rs3706737 | 8.52 | 3.669 | 0.020 | 0.005 | −0.007 | 0.007 | 1.70 |
| 5 | ADG28–42 | 78 | rs3720626 | rs3706737 | 8.94 | 3.850 | 0.029 | 0.008 | −0.020 | 0.012 | 1.78 |
| 6 | ADG7–21 | 23 | rs13478727 | rs13478839 | 8.11 | 3.495 | 0.019 | 0.006 | 0.021 | 0.009 | 1.62 |
| 6 | BW28** | 27 | rs13478727 | rs13478839 | 14.07 | 6.026 | 0.885 | 0.177 | 0.544 | 0.268 | 2.77 |
| 6 | BW35** | 27 | rs13478727 | rs13478839 | 16.54 | 7.064 | 1.157 | 0.203 | 0.316 | 0.307 | 3.24 |
| 6 | BW42** | 28 | rs13478727 | rs13478839 | 15.88 | 6.789 | 1.124 | 0.203 | 0.428 | 0.304 | 3.12 |
| 6 | ADG7–42** | 29 | rs13478727 | rs13478839 | 16.40 | 7.009 | 0.030 | 0.005 | 0.010 | 0.008 | 3.22 |
| 6 | ADG21–28** | 30 | rs13478727 | rs13478839 | 16.65 | 7.112 | 0.076 | 0.013 | 0.026 | 0.019 | 3.26 |
| 10 | ADG7–14** | 32 | CEL-10_58149652 | rs13480647 | 10.82 | 4.647 | −0.026 | 0.006 | −0.020 | 0.010 | 2.14 |
| 10 | ADG28–42 | 32 | CEL-10_58149653 | rs13480648 | 10.65 | 4.578 | 0.038 | 0.008 | −0.007 | 0.013 | 2.11 |
| 10 | ADG28–35 | 36 | CEL-10_58149654 | rs13480649 | 8.77 | 3.777 | 0.056 | 0.013 | −0.010 | 0.021 | 1.75 |
| 10 | BW42 | 37 | CEL-10_58149655 | rs13480650 | 9.08 | 3.907 | 0.864 | 0.207 | −0.270 | 0.319 | 1.81 |
| 10 | ADG21–28 | 44 | rs3717445 | rs13480707 | 8.39 | 3.611 | 0.053 | 0.013 | −0.016 | 0.019 | 1.67 |
| 10 | ADG7–42** | 47 | rs13480707 | rs13480754 | 10.05 | 4.322 | 0.023 | 0.005 | −0.008 | 0.008 | 2.00 |
| 11 | ADG21–28** | 26 | rs6276300 | rs6199956 | 17.78 | 7.584 | 0.073 | 0.014 | 0.053 | 0.022 | 3.48 |
| 11 | ADG7–42** | 28 | rs6276300 | rs6199956 | 20.51 | 8.726 | 0.034 | 0.006 | 0.012 | 0.009 | 3.99 |
| 11 | BW42** | 28 | rs6276300 | rs6199956 | 18.86 | 8.038 | 1.258 | 0.215 | 0.439 | 0.335 | 3.68 |
| 11 | BW28** | 29 | rs6276300 | rs6199956 | 13.66 | 5.851 | 0.873 | 0.187 | 0.563 | 0.291 | 2.69 |
| 11 | BW35** | 29 | rs6276300 | rs6199956 | 17.53 | 7.482 | 1.176 | 0.214 | 0.573 | 0.333 | 3.43 |
| 12 | ADG7–14 | 61 | rs6288403 | rs6390948 | 8.91 | 3.836 | −0.022 | 0.006 | 0.016 | 0.008 | 1.77 |
| 18 | ADG7–42 | 8 | rs13483233 | rs3723947 | 8.53 | 3.673 | 0.021 | 0.005 | 0.002 | 0.007 | 1.70 |
| 18 | BW35 | 8 | rs13483233 | rs3723947 | 9.15 | 3.938 | 0.819 | 0.192 | 0.042 | 0.272 | 1.82 |
| 18 | BW42 | 8 | rs13483233 | rs3723947 | 8.61 | 3.707 | 0.802 | 0.193 | 0.043 | 0.275 | 1.71 |
| 18 | ADG14–21 | 36 | rs3670254 | rs3718618 | 8.59 | 3.698 | 0.038 | 0.009 | −0.001 | 0.014 | 1.71 |
Abbreviations: a, additive effect; d, dominance effect; MMU, mouse chromosome; s.ea, standard error of additive effect; s.ed, standard error of dominance effect; %var, percentage of phenotypic variance accounted for by quantitative trait loci.
Peak position of quantitative trait loci detected in Kosambi centimorgans.
Flanking markers (left and right) of the quantitative trait loci peak.
**Quantitative trait loci with an F-value that exceeded the 1% genome-wide permutation threshold.
Other quantitative trait loci exceeded the 5% threshold.
A positive estimate indicates that the effect of the recurrent quantitative trait loci genotype is larger than the effect of the heterozygous genotype.
Figure 1.
Chromosomal distribution of QTL for different growth periods. Pre-weaning BW: pre-weaning body weight trait, include BW7, BW14 and BW21; Late BW: late body weight traits, include BW28, BW35 and BW42; Pre-weaning GR: pre-weaning growth rate traits, include ADG7–14, ADG14–21 and ADG7–21; Late GR: late growth rate traits, include ADG21–28, ADG28–35, ADG35–42 and ADG28–42; Overall GR: overall growth rate trait, includes ADG7–42.
Table 3. Percentage of phenotypic variance accounted for by detected QTL.
| Traits |
Main effect QTLa |
Interaction QTLb |
||
|---|---|---|---|---|
| Number of QTLc | Total variation (%)d | Number of QTLc | Total variation (%)d | |
| BW7 | 1 | 2.08 | 1 | 1.18 |
| BW14 | 1 | 2.04 | — | — |
| BW21 | 2 | 4.39 | 3 | 3.34 |
| BW28 | 4 | 9.92 | 3 | 3.12 |
| BW35 | 5 | 11.88 | 2 | 1.47 |
| BW42 | 5 | 12.46 | 4 | 3.73 |
| ADG7–14 | 3 | 5.53 | — | — |
| ADG14–21 | 2 | 6.76 | 5 | 6.36 |
| ADG21–28 | 4 | 10.17 | 4 | 3.08 |
| ADG28–35 | 3 | 6.49 | 2 | 2.68 |
| ADG35–42 | 1 | 3.11 | 1 | 0.87 |
| ADG7–21 | 3 | 6.62 | 3 | 3.16 |
| ADG28–42 | 4 | 10.97 | 4 | 6.11 |
| ADG7–42 | 6 | 15.16 | 5 | 4.4 |
Abbreviations: QTL, quantitative trait loci; —, no QTL was identified for this trait.
Main effect QTL.
QTL that interact with myostatin, sex or reciprocal cross.
Total number of QTL that were associated with each trait.
Total percentage of phenotypic variance accounted for by QTL for each trait.
Figure 2.
QTL scan for average daily gain of 1st week and 6th week by interval mapping.
Epistasis and interaction effect
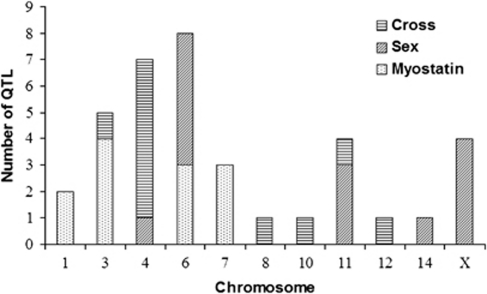
We identified 12 QTL that significantly interacted (P-value <0.05) with myostatin genotype (Table 4). These QTL represent a statistical epistatic effect, which means that QTL genotype significantly impacted the effect of myostatin. Similarly, 11 and 14 QTL were detected with a significant interaction with reciprocal cross and sex, respectively (Supplementary Tables 2 and 3). These 37 QTL were distributed on chromosomes 1, 3, 4, 6, 7, 8, 10, 11, 12, 14 and X chromosome (Figure 3). In all, 21 of these additive or dominant QTL were also detected when the genome scan was performed on the full dataset. However, the other 16 QTL did not have a significant QTL effect when the whole F2 population was analyzed. The phenotypic variation accounted for by these additive and dominance interaction effects are summarized in Table 4, Supplementary Tables 2 and 3, along with the corresponding F- and P-values. The amount of total phenotypic variation of each trait that was explained by these interaction effects ranged from 1.18–6.36% (Table 3). Epistatic interaction effects accounted for the greatest amount of phenotypic variation for 3rd week average daily gain. Sex × QTL interactions were detected during both the pre-weaning and post-weaning periods. However, no myostatin × QTL or cross × QTL interaction effects were detected for pre-weaning body growth traits (Table 5).
Table 4. Statistics of quantitative trait loci with significant interactions with myostatin genotype.
| MMU | Traitsa | Position |
Flanking markersc |
AI+DI |
AI |
DI |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| (cM)b | Left | Right | %var | P-value | %var | P-value | %var | P-value | ||
| 1 | ADG35–42 | 23 | rs3696088 | rs13472794 | 0.87 | 1.18E-02 | 0.21 | 1.81E-01 | 0.66 | 4.17E-03 |
| 1 | ADG28–42 | 24 | rs3696088 | rs13472794 | 0.86 | 1.11E-02 | 0.37 | 6.16E-02 | 0.50 | 1.99E-02 |
| 3 | ADG28–35 | 31 | rs3663409 | rs13477132 | 0.80 | 1.71E-02 | 0.63 | 9.10E-04 | 0.17 | 2.55E-01 |
| 3 | BW42** | 31 | rs3663409 | rs13477132 | 1.12 | 3.43E-03 | 0.75 | 1.22E-02 | 0.38 | 3.92E-03 |
| 3 | ADG7–42** | 32 | rs3663409 | rs13477132 | 1.15 | 2.93E-03 | 0.86 | 8.42E-03 | 0.29 | 1.04E-02 |
| 3 | ADG28–42** | 41 | rs13477174 | rs3670634 | 2.07 | 2.74E-05 | 1.41 | 7.93E-04 | 0.66 | 2.41E-03 |
| 6 | ADG7–42 | 53 | rs4226048 | mCV24115224 | 0.90 | 1.07E-02 | 0.30 | 9.89E-04 | 0.59 | 2.35E-02 |
| 6 | BW42 | 54 | rs4226048 | mCV24115224 | 0.83 | 1.52E-02 | 0.28 | 1.46E-03 | 0.55 | 4.07E-02 |
| 6 | BW35 | 55 | rs4226048 | mCV24115224 | 0.73 | 2.51E-02 | 0.15 | 7.76E-04 | 0.58 | 4.10E-02 |
| 7 | ADG14–21** | 12 | mCV22975338 | rs3719256 | 1.25 | 1.92E-03 | 1.25 | 1.08E-03 | 0.01 | 1.45E-02 |
| 7 | ADG28–35** | 43 | rs3676254 | rs3656205 | 1.87 | 8.81E-05 | 1.59 | 7.17E-05 | 0.28 | 5.19E-02 |
| 7 | ADG28–42** | 43 | rs3676254 | rs3656205 | 2.29 | 1.07E-05 | 1.74 | 3.91E-05 | 0.56 | 1.39E-02 |
Abbreviations: AI, additive interaction; DI, dominance interaction; MMU, mouse chromosome; %var, percentage of phenotypic variance accounted for by quantitative trait loci.
Trait name of quantitative trait loci with significant interaction.
Peak position of quantitative trait loci with significant interaction in Kosambi centimorgans.
Flanking markers (left and right) of the quantitative trait loci peak.
**Interactions with P-value <0.01.
Others have P-value <0.05.
Figure 3.
Chromosomal distribution of QTL that interact with different main effect.
Table 5. Total percentage of phenotypic variance accounted for by QTL interactions.
| Interactiona | Growth periods | QTLb | Total %var |
|---|---|---|---|
| Cross | Pre-weaning BW | — | — |
| Post-weaning BW | 3 | 2.44 | |
| Pre-weaning GR | 3 | 2.16 | |
| Post-weaning GR | 3 | 2.66 | |
| Overall GR | 2 | 1.67 | |
| Myostatin | Pre-weaning BW | — | — |
| Post-weaning BW | 3 | 2.68 | |
| Pre-weaning GR | 1 | 1.25 | |
| Post-weaning GR | 6 | 8.78 | |
| Overall GR | 2 | 2.05 | |
| Sex | Pre-weaning BW | 4 | 4.61 |
| Post-weaning BW | 3 | 3.19 | |
| Pre-weaning GR | 4 | 6.1 | |
| Post-weaning GR | 2 | 1.3 | |
| Overall GR | 1 | 0.69 |
Abbreviations: BW, body weight; GR, growth rate (average daily gain); QTL, quantitative trait loci; %var, total percentage of phenotypic variance accounted for by QTL interaction.
Main effect that QTL interact with. Cross: reciprocal cross; myostatin: myostatin genotype; sex: sex.
Total number of QTL that significantly interact with cross or myostatin or sex.
Pre-weaning, 1st–3rd week; post-weaning, 4th–6th week.
Partitioning the total genetic variation
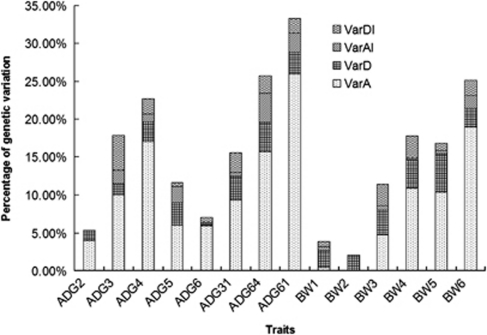
The total genetic variation accounted for by the detected QTL was partitioned into four genetic components, for example, additive, dominance, additive interaction and dominance interaction. The phenotypic variation explained by these four types of effects was summarized in Figure 4. Additive and dominance effects accounted for more genetic variation than interaction effects in body weights as age increased. This difference ranged from 1 to 18% of the total genetic variation. No interaction effects were detected for 2nd week body weight and average daily gain traits. For most of the growth traits, the variation explained by dominance interactions is about four times larger than the variation explained by additive interactions.
Figure 4.
Partitioning of the genetic variation explained by QTL mapped at a 5% genome-wide significance level.
Discussion
Comparison with other growth trait QTL studies
In this study, we analyzed 14 highly correlated growth traits in an F2 population derived from a myostatin-null C57BL/6 by M16i high body weight mouse line cross. Compared with previous QTL mapping studies in the mouse, some of the QTL identified in our study were coincident to QTL positions that had been previously discovered (Supplementary Table 5). In particular, the QTL with the largest F-value detected in our study, which was associated with ADG14–21 on chromosome 2, was very close to the largest QTL identified by Rocha et al. (2004). Although it was shown to be associated with week 10 body weight, it was very possible that these two QTL are actually the same and the causal mutation in this genomic region has a pleiotropic effect. In addition, these regions on chromosomes 1, 14 and X that contain QTL in our study were not shown to be associated with body weight traits in mice before, although other regions on the same chromosomes are known to harbor body weight QTL, for example, the X chromosome (Rance et al., 1997). This again confirms that there are some major QTL on the X chromosome that can affect body weight. In addition to chromosomes 1, 2, 14 and X, we also detected QTL in several regions on chromosomes 3, 6, 10 and 18. Interestingly, these regions were not detected in the Rocha et al. (2004) growth study. Similar to our study, their study also used M16i as one of the founders for its mapping population. This indicated that the unique QTL we identified was most likely from the C57BL/6 genetic background, and these loci might not be polymorphic in other mouse strains, which makes the F2 progeny derived from a C57BL/6 by M16i cross a valuable mapping population. However, we need to point out that although we used a genome-wide significance level to control for multiple testing, the usage of multiple models to test a large number of traits increases the possible false-positive rate. Moreover, some of the QTL that had been identified in previous studies were associated with different traits in our studies. As trait names and their definition are not very consistent between studies, it is difficult to conclude whether or not these QTL really control the same traits. Even for our own study, many of the QTL that we detected for different traits were localized to the same chromosomal region. One reason might be that some traits were highly correlated. Moreover, this might indicate the presence of pleiotropic QTL. Without testing for pleiotropy, we cannot tell whether this is caused by pleiotropic QTL or multiple QTL that are tightly linked. Further investigation, for example, fine mapping, can help to increase the mapping resolution and provide more information.
Epistatic QTL interactions with myostatin gene
During skeletal muscle growth, it is known that many molecular signals might be involved through multilevel interactions (Kambadur et al., 2004). This information was consistent with the fact that significant myostatin × QTL interactions were detected in our study. A study of mapping myostatin modifiers that impact muscle growth in the Compact mouse line was previously reported (Varga et al., 2003). Although the phenotypic measurements (qualitative) of Varga et al. (2003) may be less precise than the quantitative measurements used here, they did measure muscularity, which is directly affected by myostatin. In contrast, we evaluated body weight measurements. Their work indicated the presence of some QTL that segregate in myostatin-deficient mice that affect musclarity. Unfortunately, the QTL identified by Varga et al. were only observed in myostatin-deficient mice. Thus, it is unknown if these QTL have different effects in the presence and absence of functional myostatin. Furthermore, Bunger et al. (2004) reported that myostatin deficiency may result in varying phenotypes in different genetic backgrounds. Taken together, these results indicate the possible presence of QTL that epistatically interact with myostatin to control variation in growth phenotypes.
Most of the QTLs that were detected in this study were associated with post-weaning (3–6 week after birth) traits, which correspond with the onset of puberty. This indicated that these QTL might interact with sexual hormones. Moreover, it has been suggested that myostatin effects might be age dependent (Lee, 2004). Therefore, these QTL regions could provide more information about how this kind of time-dependent mechanism is regulated. On the other hand, body weight is composed of multiple organ weights. Therefore, it will be of further interest to dissect whole body weight gain into single organ and tissue weights, for example, adipose and skeletal muscle. It has been shown that myostatin is also expressed in adipose tissue of the adult mouse (McPherron et al., 1997). Other studies have indicated that QTL by myostatin interactions can influence some adipose traits in cattle (Casas et al., 2000) and muscle mass in mice (Varga et al., 2003).
Gene interaction with sex effect and cross effect
Sexual dimorphism in QTL mapping has been studied previously (Kenney-Hunt et al., 2006; Fawcett et al., 2008). These QTL might truly function differently between male and female. However, they can be a false-positive result in QTL analysis. In our study, we identified 14 QTL that had a significant sex × QTL interaction. Compared with QTL that exhibited a myostatin × QTL interaction, we did not observe a clear age-dependent trend in sex × QTL interactions (Table 5). We found that among these 14 QTL, 4 were located on chromosome X. These QTL associations can be a result of X chromosome dosage compensation or sex-specific genes. Previously, main effect body weight QTL have been detected on the X chromosome in several studies (Lembertas et al., 1996; Rance et al., 1997; Liu et al., 2001). Our results not only confirmed that the X chromosome contains QTL associated with body weight, it also indicates that these QTL may be sex-specific or have significant interaction with sex. In mammals, X chromosome dosage compensation results in equal gene expression between different sexes. Further, fine-mapping studies could be conducted to narrow these regions, and causal mutations could be identified by measuring the expression profiles of candidate genes. One successful example of this approach was the identification of Glypican-3 as the causal gene for a major growth QTL in mice (Oliver et al., 2005). The most significant sex × QTL interaction controlled growth traits during the pre-weaning period. Functional studies of genes that underlie these QTL could be important to elucidate the role that sex has in growth.
Among the 11 QTL that had a significant reciprocal cross × QTL interaction, 6 were located on chromosome 4. This type of interaction indicated a strain-dependent gene expression pattern. There are several possible ways to explain this kind of reciprocal cross interaction. First, there might be an interaction between the mitochondrial and nuclear genome, as F2 individuals only inherited mitochondrial DNA from one of the F0 founder lines. Different alleles of these QTL might interact directly with mitochondrial DNA or its protein product to affect body growth. Given the importance of mitochondria in energy and metabolism processes, it was not surprising that such interactions were observed. Similarly, differential reciprocal cross effects have been documented in other QTL mapping studies (Smith Richards et al., 2002; Lagerholm et al., 2009). Another explanation of this reciprocal cross effect could be a result of some effects from the Y chromosome. As F2 mice within each cross only inherited their Y chromosome from either M16i or C57BL/6 founders. These QTL loci might interact with genes on the Y chromosome to affect growth. Statistically speaking, any effect resulting from the Y chromosome was confounded with the effect of mitochondrial DNA. Therefore, further investigation is needed to identify the real reason behind thess cross × QTL interactions.
Variation explained by epistatic interactions
In spite of the fact that we identified 37 QTL with significant interactions with myostatin, sex or reciprocal cross effect, the total phenotypic variation accounted for by these QTL was not very large (Table 3). The QTL with the largest effect accounted for only about 6.36% of the phenotypic variance in ADG14–21. This is much smaller than that estimated by Brockmann et al. (2000) and Carlborg et al. (2003). They reported that epistasis could account for approximately 33–36% of the phenotypic variance observed in body weight and fat accumulation in mice. However, both these studies and our study show that there might be a time-dependent trend for epistatic or interaction effects. Gene interactions seem to be more important for early growth (Table 5). In addition, dominance QTL interactions with myostatin genotype explained more phenotypic variation than did additive QTL interactions. Dominance interaction effects might be a preferred mechanism for the myostatin-associated growth pathway. There are several possible explanations for the difference between Carlborg et al. (2003) and our estimates of genetic variation. One obvious reason could be that we only considered the interactions between the myostatin locus and QTL. This is a small proportion of overall pair-wise gene interactions that might be involved in the control of body weight. A further analysis of the interaction between these QTL and interaction between nonsignificant genome locations could possibly explain more phenotypic variance. In addition, Carlborg et al. used a 20% genome wide significant threshold to identify epistatic QTL, whereas we used a 5% level. The difference of significance threshold might lead to a difference in the total number of QTL detected. Furthermore, the allele segregation status in M16i and C57BL/6 genome can also be a reason for this. Some QTL that were associated with body weight traits might not be segregating in the F2 population derived from these two strains, which limits the number of QTL that can be detected. One possible way to solve this problem is to use a mapping population that has more genetic variation. For example, the collaborative cross is a good candidate model (Threadgill et al., 2002) to accomplish this. The collaborative cross is generating more than 1000 recombinant inbred mouse lines derived from an eight-way intercross, and some of these lines are still in the breeding stage. The eight founder lines were carefully selected to represent most of the genetic variation in the mouse genome. Because a large number of recombination events occurred in the genome during the development of the collaborative cross, they are expected to be an ideal QTL fine mapping population. After the completion of collaborative cross project, QTL mapping studies in these mice will provide a higher statistical power and better resolution.
Conclusions
We identified a substantial number of QTL that control body weight and growth rate traits using an F2 mouse population derived from M16i obese myostatin wild-type and C57BL/6 myostatin-null mouse lines. In addition, we also detected QTL that significantly interacted with myostatin genotype, sex and reciprocal cross. Further investigation of individual tissues will help to elucidate more details on how myostatin regulates growth. In future studies, these QTL regions could be used to search for candidate genes that affect the myostatin signaling pathway.
Acknowledgments
We thank Dr Daniel Pomp for providing the M16i mice used in our study and technical insights. This research was supported by grants from the USDA CSREES 2006-35205-16696.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Brockmann GA, Kratzsch J, Haley CS, Renne U, Schwerin M, Karle S. Single QTL effects, epistasis, and pleiotropy account for two-thirds of the phenotypic F(2) variance of growth and obesity in DU6i x DBA/2 mice. Genome Res. 2000;10:1941–1957. doi: 10.1101/gr.gr1499r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger L, Ott G, Varga L, Schlote W, Rehfeldt C, Renne U, et al. Marker-assisted introgression of the Compact mutant myostatin allele MstnCmpt-dl1Abc into a mouse line with extreme growth effects on body composition and muscularity. Genet Res. 2004;84:161–173. doi: 10.1017/s0016672304007165. [DOI] [PubMed] [Google Scholar]
- Carlborg O, Kerje S, Schütz K, Jacobsson L, Jensen P, Andersson L. A global search reveals epistatic interaction between QTL for early growth in the chicken. Genome Res. 2003;13:413–421. doi: 10.1101/gr.528003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas E, Shackelford SD, Keele JW, Stone RT, Kappes SM, Koohmaraie M. Quantitative trait loci affecting growth and carcass composition of cattle segregating alternate forms of myostatin. J Anim Sci. 2000;78:560–569. doi: 10.2527/2000.783560x. [DOI] [PubMed] [Google Scholar]
- Casas E, Stone RT, Keele JW, Shackelford SD, Kappes SM, Koohmaraie M. A comprehensive search for quantitative trait loci affecting growth and carcass composition of cattle segregating alternative forms of the myostatin gene. J Anim Sci. 2001;79:854–860. doi: 10.2527/2001.794854x. [DOI] [PubMed] [Google Scholar]
- Cheverud JM. Genetics of Growth in the Mouse. Imperial College Press: London; 2005. [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibé B, Bouix J, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- Corva PM, Medrano JF. Quantitative trait loci (QTLs) mapping for growth traits in the mouse: a review. Genet Sel Evol. 2001;33:105–132. doi: 10.1186/1297-9686-33-2-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen EJ. Maturing patterns of organ weights in mice selected for rapid postweaning gain. Theor Appl Genet. 1986;73:148–157. doi: 10.1007/BF00273732. [DOI] [PubMed] [Google Scholar]
- Fawcett GL, Roseman CC, Jarvis JP, Wang B, Wolf JB, Cheverud JM. Genetic architecture of adiposity and organ weight using combined generation QTL analysis. Obesity (Silver Spring) 2008;16:1861–1868. doi: 10.1038/oby.2008.300. [DOI] [PubMed] [Google Scholar]
- Green P, Falls K, Crooks S. Documentation for CRIMAP, Version 2.4. Washington University School of Medicine: St Louis, MO, USA; 1990. [Google Scholar]
- Grobet L, Martin LJ, Poncelet D, Pirottin D, Brouwers B, Riquet J, et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet. 1997;17:71–74. doi: 10.1038/ng0997-71. [DOI] [PubMed] [Google Scholar]
- Hanrahan JP, Eisen EJ, Lagates JE. Effects of population size and selection intensity of short-term response to selection for postweaning gain in mice. Genetics. 1973;73:513–530. doi: 10.1093/genetics/73.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerez-Timaure NC, Eisen EJ, Pomp D. Fine mapping of a QTL region with large effects on growth and fatness on mouse chromosome 2. Physiol Genomics. 2005;21:411–422. doi: 10.1152/physiolgenomics.00256.2004. [DOI] [PubMed] [Google Scholar]
- Kambadur R, Bishop A, Salerno MS, Mccubbin JA, Sharma M.2004Role of myostatin in muscle growthIn: Te Pas MFW, Everts ME, Haagsman HP (eds).Muscle Development of Livestock Animals: Physiology, Genetics, and Meat Quality CABI Publishing: Cambridge [Google Scholar]
- Kambadur R, Sharma M, Smith TP, Bass JJ. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 1997;7:910–916. doi: 10.1101/gr.7.9.910. [DOI] [PubMed] [Google Scholar]
- Kenney-Hunt JP, Vaughn TT, Pletscher LS, Peripato A, Routman E, Cothran K, et al. Quantitative trait loci for body size components in mice. Mamm Genome. 2006;17:526–537. doi: 10.1007/s00335-005-0160-6. [DOI] [PubMed] [Google Scholar]
- Lagerholm S, Li LS, Jiao H, Park HB, Ohlsson C, Akesson K, et al. Genetic regulation of bone traits is influenced by sex and reciprocal cross in F(2) progeny from GK and F344 rats. J Bone Miner Res. 2009;24:1066–1074. doi: 10.1359/jbmr.081252. [DOI] [PubMed] [Google Scholar]
- Lee SJ. Regulation of muscle mass by myostatin. Annu Rev Cell Dev Biol. 2004;20:61–86. doi: 10.1146/annurev.cellbio.20.012103.135836. [DOI] [PubMed] [Google Scholar]
- Lembertas AV, Fisher JS, Warden CH, Wen PZ, Xia YR, Lusis AJ. A locus on the X chromosome is linked to body length in mice. Mamm Genome. 1996;7:171–173. doi: 10.1007/s003359900048. [DOI] [PubMed] [Google Scholar]
- Liu X, Bunger L, Keightley PD. Characterization of a major X-linked quantitative trait locus influencing body weight of mice. J Hered. 2001;92:355–357. doi: 10.1093/jhered/92.4.355. [DOI] [PubMed] [Google Scholar]
- McCarthy JC.1982The laboratory mouse as a model for animal breeding: a review of selection for increased body weight and litter size Proceedings of the 2nd World Congress on Genetics Applied to Livestock Productionvol. 5. Madrid, Spain; 66–83. [Google Scholar]
- McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA. 1997;94:12457–12461. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher DS, Quignon P, Bustamante CD, Sutter NB, Mellersh CS, Parker HG, et al. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet. 2007;3:e79. doi: 10.1371/journal.pgen.0030079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver F, Christians JK, Liu X, Rhind S, Verma V, Davison C, et al. Regulatory variation at glypican-3 underlies a major growth QTL in mice. PLoS Biol. 2005;3:e135. doi: 10.1371/journal.pbio.0030135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rance KA, Hill WG, Keightley PD. Mapping quantitative trait loci for body weight on the X chromosome in mice. I. Analysis of a reciprocal F2 population. Genet Res. 1997;70:117–124. doi: 10.1017/s0016672397002917. [DOI] [PubMed] [Google Scholar]
- Rocha JL, Eisen EJ, Van Vleck LD, Pomp D. A large-sample QTL study in mice: I. Growth. Mamm Genome. 2004;15:83–99. doi: 10.1007/s00335-003-2312-x. [DOI] [PubMed] [Google Scholar]
- Schuelke M, Wagner KR, Stolz LE, Huber C, Riebel T, Komen W, et al. Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med. 2004;350:2682–2688. doi: 10.1056/NEJMoa040933. [DOI] [PubMed] [Google Scholar]
- Seaton G, Hernandez J, Grunchec JA, White I, Allen J, De Koning DJ, et al. 2006GridQTL: a grid portal for QTL mapping of compute intensive datasets Proceedings of the 8th World Congress on Genetics Applied to Livestock Production13–18 August 2006. Belo Horizonte, Brazil [Google Scholar]
- Smith JA, Lewis AM, Wiener P, Williams JL. Genetic variation in the bovine myostatin gene in UK beef cattle: allele frequencies and haplotype analysis in the South Devon. Anim Genet. 2000;31:306–309. doi: 10.1046/j.1365-2052.2000.00521.x. [DOI] [PubMed] [Google Scholar]
- Smith Richards BK, Belton BN, Poole AC, Mancuso JJ, Churchill GA, Li R, et al. QTL analysis of self-selected macronutrient diet intake: fat, carbohydrate, and total kilocalories. Physiol Genomics. 2002;11:205–217. doi: 10.1152/physiolgenomics.00037.2002. [DOI] [PubMed] [Google Scholar]
- Threadgill DW, Hunter KW, Williams RW. Genetic dissection of complex and quantitative traits: from fantasy to reality via a community effort. Mamm Genome. 2002;13:175–178. doi: 10.1007/s00335-001-4001-Y. [DOI] [PubMed] [Google Scholar]
- Varga L, Müller G, Szabó G, Pinke O, Korom E, Kovács B, et al. Mapping modifiers affecting muscularity of the myostatin mutant (Mstn(Cmpt-dl1Abc)) compact mouse. Genetics. 2003;165:257–267. doi: 10.1093/genetics/165.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.