Abstract
Huntington disease (HD) is an autosomal dominant neurodegenerative disorder caused by CAG-expansion in the huntingtin gene (HTT) that results in a toxic gain of function in the mutant huntingtin protein (mHTT). Reducing the expression of mHTT is therefore an attractive therapy for HD. However, wild-type HTT protein is essential for development and has critical roles in maintaining neuronal health. Therapies for HD that reduce wild-type HTT may therefore generate unintended negative consequences. We have identified single-nucleotide polymorphism (SNP) targets in the human HD population for the disease-specific targeting of the HTT gene. Using primary cells from patients with HD and the transgenic YAC18 and BACHD mouse lines, we developed antisense oligonucleotide (ASO) molecules that potently and selectively silence mHTT at both exonic and intronic SNP sites. Modification of these ASOs with S-constrained-ethyl (cET) motifs significantly improves potency while maintaining allele selectively in vitro. The developed ASO is potent and selective for mHTT in vivo after delivery to the mouse brain. We demonstrate that potent and selective allele-specific knockdown of the mHTT protein can be achieved at therapeutically relevant SNP sites using ASOs in vitro and in vivo.
Introduction
Unlike neurodegenerative diseases such as Alzheimer's or Parkinson's disease that are primarily idiopathic, every case of Huntington disease (HD) is associated with an expanded CAG tract in the HTT gene.1 In theory, the identification of this specific mutation allows for neuroprotective therapies in presymptomatic HD mutation carriers. However, therapies currently available to patients with HD only offer moderate symptom relief and have no effect on disease progression.2,3
The large number of proposed disease mechanisms in HD has hindered development of disease-modifying therapies. HTT is a 340-kDa protein that associates with many molecular partners including proteins, lipids, DNA, and mRNA.4 Wild-type HTT plays roles in critical processes including transcription, apoptosis, ER stress signaling, calcium homeostasis, axonal transport, endocytosis, and synaptic transmission.4 Mutant huntingtin protein (mHTT) leads to altered proteolysis, transcriptional dysregulation, altered intracellular trafficking, impaired metabolism, dysregulated calcium signaling, and altered synaptic activity.4 A wide variety of therapies aimed at these downstream events have been investigated, but results of these trials often show only modest benefit in mice and as yet no benefit in humans.
There is encouraging in vivo evidence that the reduction of HTT levels could be an effective therapy for human HD. The severity of signs and symptoms of HD in humans and mice is linked to mHTT expression5, and the postnatal reduction of HTT expression improves features of HD in transgenic mouse models.6,7,8,9,10,11 There are several distinct strategies being pursued to lower cellular levels of HTT including viral delivery of siRNA6,10,12 or miRNA,13 and small molecules.14,15 Preliminary short-term experiments suggest that decreasing both wild type and mHTT in the brain is tolerated and produces a clinical benefit in mouse models.
However, there are also lines of evidence to suggest that loss of wild-type HTT function has detrimental consequences. HTT is essential for embryogenesis; total excision of hdh causes loss of viability in early embryonic stages while reduction of hdh to ~33% results in perinatal lethality and altered neurodevelopment.16,17 Complete loss of HTT could lead to dysregulation of many cellular processes that are important to the ongoing health of neurons in adulthood, given its known roles in negatively regulating apoptosis and transcription of prosurvival genes.4,18,19,20,21,22 Many previous models of in vivo HTT silencing have reduced mHTT but maintained a partial or complete complement of wild-type Hdh, confounding our understanding of the relative importance of loss of HTT function in HD.6,7,8,9,10 However, it is clear that the complete inactivation of wild-type Hdh in the forebrain of mice postnatally causes neurodegeneration.23
Considering that neuroprotective therapies for HD will likely begin in early adulthood and continue throughout life, examining the potential for the allele-specific reduction of mHTT is prudent.24,25,26,27,28,29,30,31 Allele-specific silencing of HTT may be possible, either by targeting the expanded CAG-tract directly30,32 or by targeting genetic polymorphisms linked to CAG-expansion.28,33
Short synthetic antisense oligonucleotides (ASOs) silence gene expression post-transcriptionally by several mechanisms34 and can discriminate between single-nucleotide polymorphism (SNP) alleles at targeted sites.35 ASOs have been successfully used in the central nervous system (CNS) of rodents, nonhuman primates, and human patients with cytomegalovirus (CMV) retinitis and are being investigated for motor neuron diseases following intrathecal administration.36 They are therefore a viable option for allele-specific therapeutics in HD. Relative to viral delivery of siRNA, ASOs provide substantial benefits. Most importantly for allele-specific approaches, ASOs can target SNPs anywhere in the pre-mRNA, including introns, and therefore the number of potential SNP targets is extensive. Only 8% of the validated SNPs in the HTT pre-mRNA are coding, and therefore the number of targets accessible to siRNA is smaller (Supplementary Table S2). ASOs can also be effectively delivered to multiple cell types, including astrocytes and neurons, throughout the adult primate CNS via intracerebroventricular or intrathecal infusion.36 Because ASO delivery to the adult primate CNS can be achieved by simple infusion, viral delivery is not required and ASO dosing can be precisely controlled or stopped as needed. Based on these advantages, we have chosen to investigate the use of ASOs to target and silence SNP alleles of mutant HTT mRNA.
Results
SNP identification and genotyping
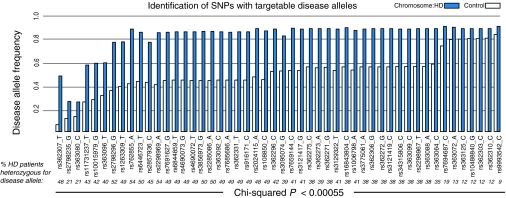
Using a custom SNP genotyping assay (“96SNP panel”), we analyzed 234 patients with HD at 91 SNP locations in the HTT gene (Supplementary Table S1). We have previously described 22 of these SNPs in a subset of these patients, genotyped using the SNPstream assay.33 After using family trio information to phase the SNP alleles to the CAG-tract size of each chromosome, we found that 50/91 SNPs are enriched on HD chromosomes (>36 CAG repeats) relative to control chromosomes (<27CAG repeats) (Bonferroni-corrected chi-squared P < 0.00055). The targets of allele-specific silencing are heterozygous SNPs, which allow sparing of the wild-type allele. Of the potential SNP targets, 41 alleles are individually heterozygous in greater than 30% of the sequenced HD population (Figure 1). Combinatorial analysis suggests that a panel of as few as three of these alleles, successfully targeted, could provide coverage for up to 85% of the HD population.28,33
Figure 1.
Identification of SNP alleles that are associated with HD chromosomes and can be used for allele-specific targeting in HD. A SNP allele was defined as a disease allele if it is significantly more common on HD than control chromosomes. In a population of 234 Caucasian patients with HD, we identify 50 SNPs with disease alleles (chi-squared, Bonferroni-corrected P value P < 0.00055) and present the heterozygosity rate for the disease allele in patients with HD (percentage heterozygosity in italics). The best targets for allele-specific therapy will be specific SNP alleles that maximize both the frequency on HD chromosomes and heterozygosity in patients with HD. HD, Huntington disease; SNP, single-nucleotide polymorphism.
ASO development pipeline and human fibroblast genotyping
In order to facilitate the identification, screening and optimization of therapeutics against disease-specific SNP targets, we describe the pipeline we have used for ASO development (Supplementary Figure S1). We propose that this pipeline will be useful for the continued development of therapeutic approaches in HD, and the methods useful for the allele-specific targeting of other dominant diseases using ASO or other silencing technology.
We began our screening with the list of SNPs enriched in the target population identified above. We then obtained a number of publically available primary human fibroblast cell lines from the Coriell repository (http://www.coriell.org/). Cell lines were chosen for which sufficient family information was available to enable successful phasing of genotyping results. The cell lines were genotyped using our 96SNP panel assay (Supplementary Table S3). Cell lines homozygous for a target SNP allele are considered “on target” because both HTT alleles are completely complementary to ASOs targeting the SNP. Cell lines homozygous for the nontarget allele are “off target” because neither HTT allele is perfectly targeted by SNP-targeted ASOs. This “on/off target” approach allows us to perform our primary potency screen as a counter screen in primary cells, with endogenous levels of genetically accurate target transcript. This approach of using homozygous on/off target cell lines also avoids the need to quantify huntingtin alleles separately, which is possible but technically demanding for a large number of SNP targets.
Potency and selectivity: human fibroblasts
Following identification of SNP targets and cell lines with the appropriate genotypes, we synthesized ASOs targeting SNP alleles and screened them for potency and selectivity. For initial screening, the ASOs used were phosphorothioate substituted 19-mers containing five 2'-O-methoxy-ethyl ribose sugars in each wing and a string of nine DNA residues in the gap (“5-9-5 2'-O-Methoxyethyl (MOE) gapmers”). These ASOs bind to target RNA and support cleavage by RNase H of target RNA opposite the DNA gap. Data shown are from ASOs with the target SNP positioned opposite the DNA gap.37 A potent non-allele-specific positive control 5-10-5 MOE gapmer was tested in each experiment (“non-allele-specific positive control”).
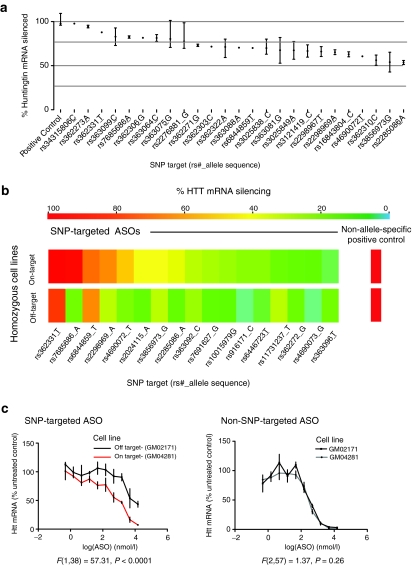
ASOs were screened for activity at a single dose in “on target” cell lines, to enable direct comparisons to non-allele-specific positive control. This primary screen identified a number of ASOs that effectively reduce HTT mRNA (Figure 2a). ASOs whose silencing was not superior to off target negative control ASOs were discarded at this step. We next counter-screened ASOs at a single dose in “off target” cell lines homozygous for the nontargeted allele. Potency of HTT mRNA knockdown in on versus off target cell lines is summarized in Figure 2b for those ASOs that silence HTT more effectively in on target cell lines than off target cell lines. Given these results, 19 ASOs targeting 14 alleles were chosen for detailed dose–response studies of HTT mRNA knockdown in fibroblasts to confirm selectivity. Sample data for an allele-specific ASO targeting rs7685686_A in Figure 2c demonstrate increased potency in on versus off target cell lines (left), while non-allele-specific positive control ASO is equally potent in both cell lines (right). These potency, selectivity, and dose–response studies enabled the prioritization of ASOs. In prioritizing ASOs for protein silencing experiments, we also considered relative coverage of the HD patient population for the targeted SNP. We proceeded with validation of protein knockdown of ASOs targeting our top four ranked SNPs (rs7685686, rs4690072, rs2024115, and rs363088). This panel of four SNPs does not provide combinatorial advantage as they are all in high linkage disequilibrium with each other, but rather represents a panel of leads from which to select our top ASO.
Figure 2.
Selectivity and potency of lead ASOs in primary human HD patient fibroblasts. (a) Forty-eight ASOs targeting SNPs reduce total HTT mRNA in a panel of on target human HD fibroblasts (black bars = range). (b) Summary of single dose cell line counter screen demonstrating selectivity of ASOs targeting SNPs in homozygous on (top panel) and off (bottom panel) target human HD fibroblasts. (c) Sample dose–response data in primary on and off target fibroblasts. Left—An ASO targeting rs7685686_A is more potent in on target cell lines versus off target. Right—A non-SNP-targeted positive control ASO shows equivalent silencing in both cell lines. F-values in c are the results of an extra sum of squares test after fitting a log(inhibitor) versus concentration curve. ASO, antisense oligonucleotide; HD, Huntington disease; SNP, single-nucleotide polymorphism.
Potency and selectivity: mouse neurons
Neurons are one of the primary cell types of interest for HD therapeutics, and ASOs effectively target neurons in the adult CNS.36 Because neurons efficiently take up ASO in vivo, we considered the possibility that they might do so in vitro. We examined the response of murine primary cortical neurons in culture to ASO exposure in culture media. In neurons derived from YAC128 HD mice,38 a single application of nonallele-targeted positive control ASO reduces transgenic HTT protein expression in neurons, with a clear dose response (Figure 3a, left, IC50 = 27.0 nmol/l, 95% CI 22.7-32.2 nmol/l). We examined other full-length transgenic mouse models (the YAC1839 mice and BACHD40 mice) and found that ASO-induced silencing of transgenic htt was similar in both lines at 500 and 1,500 nmol/l Figure 3a, right). HTT silencing in primary neurons exposed to ASO is rapid (maximal by 5 days) and lasts for at least 20 days after a single treatment.
Figure 3.
Development and use of a primary cortical neuron counter screen utilizing YAC and BACHD transgenic mice. (a) Left—Development of primary neuron assay in cortical neurons from the YAC128 mice. Bath application at DIV 2 with a non-allele-specific positive control ASO leads to clear dose-dependent silencing of HTT (IC50 = 27.03 nmol/l, 95% CI 22.66–32.24 nmol/l) after 6 days of treatment. Right—Treatment of primary cortical neurons from YAC18 and BACHD mice with 500 or 1,500 nmol/l of non-allele-specific positive control ASO leads to potent reduction of transgenic human HTT, detected with a human HTT-specific antibody (N = 6 YAC18 cultures, 10 BACHD cultures; 2-way ANOVA genotype F(1,42) = 23.08, P < 0.0001; treatment F(1,42) = 1170, P < 0.0001; interaction F(1,42) = 6.2, P = 0.0042). (b) Paired primary cortical neurons from YAC18 or BACHD mice were treated after 2 days in vitro with the indicated doses of ASO for 6 days and HTT detected with a human-specific HTT antibody. ASOs targeting SNP alleles selectively knockdown BACHD, but not YAC18, HTT transgene. rs7685686: N = 6 YAC18 cultures, 6 BACHD cultures; 2-way ANOVA genotype F(1,30) = 67.24, P < 0.0001; treatment F(2,30) = 24.9, P < 0.0001; interaction F(2,30) = 17.5, P <0.0001. rs4690072: N = 6 YAC18 cultures, 5 BACHD cultures; 2-way ANOVA genotype F(1,24) = 29.6, P < 0.0001; treatment F(2,24) = 18.3, P < 0.0001; interaction F(2,24) = 10.0, P <0.0001. rs2024115: N = 6 YAC18 cultures, 4 BACHD cultures; 2-way ANOVA genotype F(1,33) = 66.2, P < 0.0001; treatment F(2,33) = 7.5, P = 0.0021; interaction F(2,33) = 17.0, P <0.0001. rs363088: N = 6 YAC18 cultures, 4 BACHD cultures; 2-way ANOVA genotype F(1,24) = 62.3, P < 0.0001; treatment F(2,24) = 9.3, P = 0.0010; interaction F(2,24) = 16.2, P <0.0001.(*, **** Represent Bonferroni multiple comparison P values of P < 0.05 and P < 0.0001, respectively).
Genotyping transgenic HTT in YAC18 (wild type) and BACHD (CAG-expanded) mice using our 91SNP assay revealed genotype differences at a number of target SNP allele sites (Supplementary Table S2). Sequencing of target sites of interest verified that targeted SNP alleles were the only genetic variation in the region targeted by ASOs. In the case of the top four ranked ASOs, the BACHD transgene shares target alleles with the majority of HD chromosomes, while the YAC18 transgene does not. This enabled us to establish a primary neuron counter screen in which human HTT knockdown was compared in paired cortical neuron cultures from YAC18 and BACHD neurons (Figure 3b). At 1.5 µmol/l ASO BACHD human HTT knockdown after treatment with allele-specific ASO varies between 39% (rs4690072, Figure 3b red line, t = 4.291, P < 0.01) and 68% (rs363088 Figure 3b red line, t = 7.37, P < 0.001). None of the allele-specific ASOs tested significantly reduced HTT levels in YAC18 neurons (Figure 3b, black lines), while treatment with non-allele-specific positive control ASO robustly reduced HTT levels (Figure 3a, right).
Structure activity relationship (SAR)
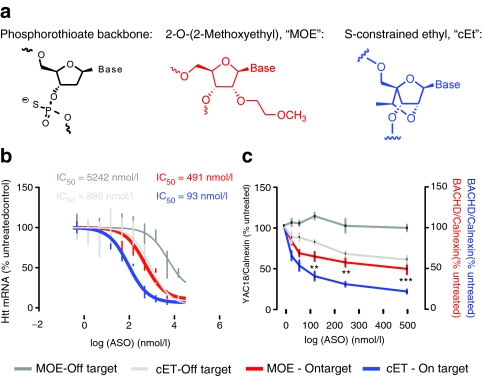
ASO modifications have been used to increase affinity and potency toward target mRNA, reduce in vivo toxicity, and improve pharmacokinetic and pharmacodynamic properties.34 Modifications can be made to the backbone chemistry by altering either the backbone itself, or the base sugar moiety, generally at the 2'-position.34 In particular, a 2'–4' constraint in the furanose ring improves hybridization to target mRNA by enforcing a N-type sugar conformation.41 To improve potency of our lead ASOs, a high affinity bicyclic nucleoside41 S-constrained ethyl (cEt) was substituted in the 5' and 3' “wings” of ASOs targeting alleles of SNP rs7685686 (Figure 4a). cEt-modified ASOs were electroporated into human HD patient fibroblasts on target or off target for the disease allele (A) at SNP rs7685686. Addition of cEt motifs to wing sequences increased potency of ASOs toward HTT mRNA, while sparing selectivity for the targeted allele (Figure 4b, compare red and blue lines, MOE IC50 = 491 nmol/l, cEt IC50 = 93 nmol/l). Selectivity was spared as the ratio of IC50 in on versus off cell lines remained similar between MOE and cEt ASOs (Figure 4b, IC50 ratio MOE = 10.7, cET = 9.5). We then examined the potency of a cEt-modified ASO in primary neurons from the YAC18 and BACHD mice. cEt-modified ASO was more potent than the paired MOE at this SNP allele (Figure 4c, 500 nmol/l ASO htt protein remaining = 45.3 ± 14.8% MOE, 16.6 ± 5.6% cEt), while selectivity is maintained.
Figure 4.
An ASO with S-constrained ethyl motif (cET) backbone has increased potency and equivalent allele-selectivity to a traditional MOE backbone. (a) Motifs used in ASO backbones. (b) Dose–response curves for HTT silencing after electroporation of human HD fibroblasts with MOE or cEt-modified ASOs targeting rs7685686_A. ASOs were, in each case, more potent toward HTT in cell lines homozygous for the targeted SNP allele versus homozygous for the untargeted allele. ASO potency is increased with no loss of specificity by addition of cEt motifs to wing structures, relative to 5-9-5 MOE design (IC50's indicated). Error bars indicate range, and lines indicate fit curves. (c) Verification of increased potency and preserved specificity of cEt-modified ASOs targeting rs7685686_A (blue) toward HTT protein in BACHD neurons, relative to the equivalent MOE ASO (red). (Two-way ANOVA effect of ASO modification in BACHD neurons P < 0.0001. **, **** Represent Bonferroni multiple comparisons between MOE ASO and cET ASO P values of P < 0.01 and P < 0.0001, respectively). ASO, antisense oligonucleotide; MOE, 2'-O-Methoxyethyl.
Potency and selectivity: in vivo
Because intracerebroventricular infusion of ASO into the murine CNS has been shown lead to transgene silencing,36 we considered the possibility that intraparenchymal bolus delivery of ASO directly to the striatum of mice would lead to effective silencing. Using BACHD and YAC18 mice, which express the A and T alleles of rs7685686, respectively, allows us to examine allele specificity in a manner analogous to our primary neuron counter screen (Figures 3 and 4).
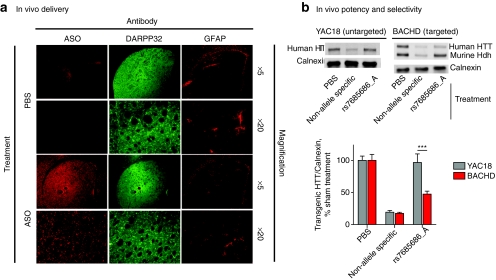
Adult mice were injected with either vehicle or 50 µg of non-allele-specific ASO that inhibits mouse and human HTT expression or allele-specific MOE ASO targeting rs7685686_A. To examine potential toxic effects resulting from the use of ASOs in the CNS, we examined gliosis in injected striata using an antibody reactive to glial fibrillary acidic protein (GFAP) and a novel antibody reactive to the chemical backbone of the ASO molecule. The ASO-reactive antibody shows clear immunoreactivity in injected striata, while sham-treated mice do not (Figure 5a). DARPP-32, a striatally enriched gene, shows strong immunoreactivity in striata, confirming that the injected ASO is being delivered to the striatum (Figure 5a). Staining with GFAP-reactive antibodies reveals mild gliosis immediately proximal to the injection site in both sham- and ASO-injected animals, which is not qualitatively worse in the ASO-injected animals (Figure 5a).
Figure 5.
ASOs are potent and selective after acute delivery to the murine central nervous system (CNS). (a) YAC18 and BACHD mice were injected intrastriatally with PBS vehicle (sham) or 50 µg ASO (non-allele-specific positive control or allele-specific targeting rs7685686). Injected striata were examined with antibodies reactive to the ASO backbone, DARPP-32, and GFAP. ASO immunoreactivity is observed throughout the DARPP-32 positive striatum in injected animals. GFAP immunoreactivity is limited proximally to the needle tract in both sham- and ASO-injected animals. (b) Transgenic HTT levels were examined by western blot 14 days post-injection. Both YAC18 and BACHD transgenes are reduced after injection with non-allele-specific ASO, while the ASO targeting rs7685686_A is only potent in the BACHD striatum (YAC18 versus BACHD Bonferroni post-test t = 8.4, P < 0.0001). ASO, antisense oligonucleotide; GFAP, glial fibrillary acidic protein; PBS, phosphate-buffered saline.
Western blots from injected striata demonstrate that non-allele-specific positive control ASO silence transgenic human HTT in both YAC18 and BACHD mice (Figure 5b, YAC18: 19.23 ± 2.77 % HTT remaining; BACHD: 17.44 ± 1.75 % HTT remaining). Human allele-specific ASO targeting rs7685686_A is potent against human transgenic HTT in BACHD, but not YAC18 mice (Figure 5a, YAC18: 96.69 ± 13.33 % HTT remaining; BACHD: 47.44 ± 4.61 % HTT remaining; Bonferroni post-test t = 8.4; P < 0.0001).
Discussion
We have identified 50 viable SNP targets in the human HTT gene (Figure 1) and established a unique set of tools and strategies to develop SNP-targeted ASOs that potently and selectively silence mHTT. This allele-specific targeting pipeline (Supplementary Figure S1) allows for the rapid prioritization of allele-specific compounds based on potency, selectivity, and population coverage for patients with HD. The pipeline uses genotyped and phased HD patient fibroblasts and two lines of transgenic HD mice with different human HD haplotypes to efficiently identify allele-specific lead compounds.
Using this pipeline, we developed ASOs that specifically and potently reduce mHTT levels in primary human cells (Figure 2), cultured primary neurons (Figures 3 and 4), and the adult mouse CNS (Figure 5). The ASOs used in this study are clinically relevant for the human HD population—ASOs with similar chemical composition have a proven track record in human clinical trials.42 Furthermore, ASOs can be improved with backbone modifications, and we now demonstrate that these modifications result in improved potency without loss of allele specificity (Figure 4). Critically, the ASOs we have developed maintain potency and specificity after in vivo delivery, confirming their therapeutic relevance (Figure 5).
Genetics and population coverage
Our primary goal in this study was to develop the most effective possible single ASO with high population coverage. Our ASOs targeting rs7685686_A, for example, could be used in 49% of the HD patient population (Figure 1). However, our genetic approach and large number of genotyped SNPs allows the discovery of targets that have combinatorial advantage to maximize relative coverage of the HD patient population.28,33 The efforts described here are the first step in an iterative process of building a panel of ASOs that provide allele-specific silencing to the majority of patients with HD. The specific ASO best suited to any one individual can be determined using SNP-CAG phasing methods recently described.43,44
ASO SAR
A major benefit of ASOs is the mature set of chemical tools for modulating ASO activities via structure activity relationship (SAR) studies. ASO modifications have been used to increase affinity and potency toward target mRNA, reduce in vivo toxicity and improve pharmacokinetic/pharmacodynamic properties.34 Modifications can be made to the backbone chemistry by altering either the backbone itself, or the base sugar moiety, generally at the 2'-position.34
We have primarily used chimeric MOE gapmer designs for the current study (Figures 2 and 3). In an attempt to improve potency while maintaining selectivity, we substituted a bicyclic nucleoside cEt in the wings of the ASO.41 This motif has been shown to increase affinity of ASOs to their target mRNAs, but it was unclear whether allele selectivity could be maintained. We find that cEt-modified ASOs targeting rs7685686_A are fivefold more potent than the parental MOE ASO, while allele specificity is preserved (Figure 4). This suggests that significant improvements can be made to ASOs by manipulating backbone chemistry.
ASO potency in neurons in vitro and in vivo
The ultimate target of therapeutics for HD is the adult CNS, which is a notoriously difficult tissue compartment to access and treat.45 We first tested our ASO compounds with cultured neurons and found that neurons efficiently absorb ASO from culture media without transfection reagents. This phenomenon will be valuable for future experiments using gene silencing in primary neurons.
Further, using adult YAC18 and BACHD mice, we demonstrate that ASOs delivered to the striatum are potent, and selective for targeted alleles (Figure 5). The striatum is the most vulnerable region of the brain in HD and is the likely first target of HTT silencing approaches in humans.46 The fact that we are able to selectively silence full-length transgenic HTT in the adult striatum suggests that the ASOs we have developed are of direct therapeutic relevance.
Path to the clinic
We envision that leads developed here will be complemented by the development of additional ASOs providing more complete population coverage. A majority of the human HD population can be represented in a panel of as few as three SNP-targeting reagents.28,33 With such a panel of ASOs in hand, we foresee that patients would be genotyped at subject SNPs using a method that links the SNP genotypes to CAG-size.43,44 With these phased genotypes, an ASO from the panel would be chosen to preferentially target mHTT in each patient.
Assuming the outlined allele-specific silencing approach is possible, the question remains whether it is preferable to non-allele-specific HTT silencing. Available evidence23,47 indicates that HTT has vital roles in the mature CNS and that silencing both endogenous and mutant HTT is associated with significant transcriptional alterations.9,10 We speculate that mild phenotypes observed in mice living less than a year with partially reduced huntingtin levels may predict more severe complications in the human CNS after decades of treatment. This consideration argues for the most judicious and targeted approach possible to HTT silencing in humans. Following successful delivery and toxicity studies, allele-specific HTT ASOs represent such an option for the majority of the HD population.
Materials and Methods
Genotyping. We designed a genotyping panel of 96 SNPs using a Goldengate assay on the Illumina BeadArray platform. The LD patterns of 900 SNPs known to exist in the htt region were compiled by gathering data from Hapmap, DBSNP, and direct sequencing of the htt region performed at the CMMT; 96 top priority SNPs were selected. The 96SNP assay provided genotyping information on 91 SNPs in the HTT gene region (5 SNPs failed quality control). The 96 SNP genotyping assay was performed on a total of 1,151 different DNA samples from individuals in 390 different HD pedigrees. The genotyping results for each patient with HD were phased by incorporating genotype information of family trios into PHASE 2.0 software.
Ethics statement. DNA samples were taken from the Huntington Disease BioBank at the University of British Columbia. Consent and access procedures were in accordance with institutional ethics approval for human research (UBC certificate H05-70532).
ASO treatment in vitro. ASOs were transfected into fibroblasts by electroporation using the Electro Square PoratorTM ECM 830 (BTX Harvard Apparatus). Cells were seeded in 96-well electroporation plates (2 mm) at 2.5 × 105 cells/ml and electroporated (130 V, 6.9 ms). For neuronal treatments, ASOs were resuspended in sterilized phosphate-buffered saline (PBS) to a concentration of 1–5 mmol/l and stock added to supplementary media on DIV2 to the indicated final concentration.
SNP sequencing in transgenic mice. SNP sequencing was performed in BACHD and YAC mice using the GoldenGate 96 SNP assay outlined above (see genotyping section). Several lines of both BAC HD and YAC HD mice were tested to confirm the results.
Primary neuron screen. YAC18 cultures were prepared as described48 from E16.5 pregnant female YAC18 mice. BACHD E16.5 embryos were isolated from pregnant FVB WT mice bred with BACHD (+/-) male mice. Each 6-well culture plate consisted of two untreated wells, and two duplicate ASO treatments. The quantification of transgenic HTT protein was performed using immunoblotting with an antibody (HD650) specific to human htt (amino acids 650–663 VLRDEATEPGDQEN),38 normalized to calnexin levels (Supplementary Materials and Methods).
ASO treatment in vivo. Mice were anesthetized with ketamine or xylazine and placed in a stereotaxic frame. After shaving and disinfection of the scalp, an incision was made along the midline. A burr hole was made at 0.75 mm anterior and 2 mm lateral to Bregma, and 50 µg ASO in 4 µl total volume or sterile PBS vehicle was injected at a rate of 0.5 µl/minute and a depth of 3.5 mm using a 5-µl Hamilton syringe. The needle was left in place for 5 minutes and slowly withdrawn. Mice were killed 14 days post-injection and processed for immunohistochemistry or protein. For immunohistochemistry, brain tissue was fixed by perfusion of 4% paraformaldehyde in PBS. Brains were cut into 25-µm free-floating coronal sections. Sections were blocked for 30 minutes in 3% bovine serum albumin, 10% normal goat serum, 0.1% Triton X-100 in PBS. Primary antibodies: rabbit anti-ASO (1:7500), rabbit anti-GFAP (1:1000 Dako, Carpinteria, CA), or rat anti-dopamine- and cyclic AMP-regulated phosphoprotein (DARPP-32) (1:1000 R&D Systems, Minneapolis, MN) were diluted in blocking solution and incubated on sections overnight at 4 °C. Secondary Alexa-fluro 488-conjugated goat anti-mouse or Alexa-fluor 568 conjugated goat anti-rabbit antibodies (1:250 Invitrogen, Carlsbad, CA) were incubated on sections for 1.5 hours at room temperature. Sections were then mounted on slides with ProLong Gold antifade reagent with DAPI (Invitrogen). For protein analysis, lysates of fresh frozen injected striata were evaluated for YAC18 and BACHD Htt levels by immunoblotting with a human-specific HTT antibody, 2168 (1:1000 Millipore, Billerica, MA) and normalization to calnexin (1:2500 Sigma, St. Louis, MO).
SUPPLEMENTARY MATERIAL Figure S1. Flowchart diagramming the allele-specific targeting pipeline, with indicated number of target SNPs and ASO designs at each step. Table S1. Phased allele frequencies in 234 human HD patients. Table S2. Summary of the distribution of different classes of SNPs in the HTT coding region. Table S3. SNP sequences at described sites in primary human fibroblasts (Corriell). Materials and Methods.
Acknowledgments
William Yang kindly provided the BACHD mice. We thank C. Ross and C. Carter for their assistance with genotyping, as well as L. Lau, M. Wang, and N. Bissada for support with animal work. This work was supported by grants from the Canadian Institute for Health Research and Michael Smith Foundation (J.B.C. and S.C.W.). M.R.H. is a Killam University Professor and holds a Canada Research Chair. S.G., G.H., S.M.F., and C.F.B. are employees of, with a financial interest in, Isis Pharmaceuticals. Isis pharmaceuticals synthesized and provided the ASOs used in this study, and supported work in the laboratory of M.R.H.
Supplementary Material
Flowchart diagramming the allele-specific targeting pipeline, with indicated number of target SNPs and ASO designs at each step.
Phased allele frequencies in 234 human HD patients.
Summary of the distribution of different classes of SNPs in the HTT coding region.
SNP sequences at described sites in primary human fibroblasts (Corriell).
REFERENCES
- The Huntington Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- The Huntington Study Group Tetrabenazine as antichorea therapy in Huntington disease: a randomized controlled trial. Neurology. 2006;66:366–372. doi: 10.1212/01.wnl.0000198586.85250.13. [DOI] [PubMed] [Google Scholar]
- De Marchi N, Daniele F., and, Ragone MA. Fluoxetine in the treatment of Huntington's disease. Psychopharmacology (Berl) 2001;153:264–266. doi: 10.1007/s002130000575. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Valenza M., and, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington's disease. Physiol Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- Graham RK, Slow EJ, Deng Y, Bissada N, Lu G, Pearson J.et al. (2006Levels of mutant huntingtin influence the phenotypic severity of Huntington disease in YAC128 mouse models Neurobiol Dis 21444–455. [DOI] [PubMed] [Google Scholar]
- Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q.et al. (2005RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model Proc Natl Acad Sci USA 1025820–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Lucas JJ., and, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lebron E, Denovan-Wright EM, Nash K, Lewin AS., and, Mandel RJ. Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington's disease transgenic mice. Mol Ther. 2005;12:618–633. doi: 10.1016/j.ymthe.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouet V, Perrin V, Hassig R, Dufour N, Auregan G, Alves S.et al. (2009Sustained effects of nonallele-specific Huntingtin silencing Ann Neurol 65276–285. [DOI] [PubMed] [Google Scholar]
- Boudreau RL, McBride JL, Martins I, Shen S, Xing Y, Carter BJ.et al. (2009Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington's disease mice Mol Ther 171053–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Liu W, Wada E, Murata M, Wada K., and, Kanazawa I. Clinico-pathological rescue of a model mouse of Huntington's disease by siRNA. Neurosci Res. 2005;53:241–249. doi: 10.1016/j.neures.2005.06.021. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, Sass M.et al. (2007Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits Proc Natl Acad Sci USA 10417204–17209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I.et al. (2008Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi Proc Natl Acad Sci USA 1055868–5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL.et al. (2007Small molecules enhance autophagy and reduce toxicity in Huntington's disease models Nat Chem Biol 3331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov AS, Miller J, Arrasate M, Wong JS, Pleiss MA., and, Finkbeiner S. A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc Natl Acad Sci USA. 2010;107:16982–16987. doi: 10.1073/pnas.1004498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir J, Floresco SB, O'Kusky JR, Diewert VM, Richman JM, Zeisler J.et al. (1995Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes Cell 81811–823. [DOI] [PubMed] [Google Scholar]
- White JK, Auerbach W, Duyao MP, Vonsattel JP, Gusella JF, Joyner AL.et al. (1997Huntingtin is required for neurogenesis and is not impaired by the Huntington's disease CAG expansion Nat Genet 17404–410. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Leavitt BR, van Raamsdonk JM, Dragatsis I, Goldowitz D, MacDonald ME.et al. (2006Huntingtin inhibits caspase-3 activation EMBO J 255896–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt BR, van Raamsdonk JM, Shehadeh J, Fernandes H, Murphy Z, Graham RK.et al. (2006Wild-type huntingtin protects neurons from excitotoxicity J Neurochem 961121–1129. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L.et al. (2001Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease Science 293493–498. [DOI] [PubMed] [Google Scholar]
- Leavitt BR, Guttman JA, Hodgson JG, Kimel GH, Singaraja R, Vogl AW.et al. (2001Wild-type huntingtin reduces the cellular toxicity of mutant huntingtin in vivo Am J Hum Genet 68313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigamonti D, Sipione S, Goffredo D, Zuccato C, Fossale E., and, Cattaneo E. Huntingtin's neuroprotective activity occurs via inhibition of procaspase-9 processing. J Biol Chem. 2001;276:14545–14548. doi: 10.1074/jbc.C100044200. [DOI] [PubMed] [Google Scholar]
- Dragatsis I, Levine MS., and, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet. 2000;26:300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lebron E., and, Paulson HL. Allele-specific RNA interference for neurological disease. Gene Ther. 2006;13:576–581. doi: 10.1038/sj.gt.3302702. [DOI] [PubMed] [Google Scholar]
- van Bilsen PH, Jaspers L, Lombardi MS, Odekerken JC, Burright EN., and, Kaemmerer WF. Identification and allele-specific silencing of the mutant huntingtin allele in Huntington's disease patient-derived fibroblasts. Hum Gene Ther. 2008;19:710–719. doi: 10.1089/hum.2007.116. [DOI] [PubMed] [Google Scholar]
- Zhang S, Feany MB, Saraswati S, Littleton JT., and, Perrimon N. Inactivation of Drosophila Huntingtin affects long-term adult functioning and the pathogenesis of a Huntington's disease model. Dis Model Mech. 2009;2:247–266. doi: 10.1242/dmm.000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi MS, Jaspers L, Spronkmans C, Gellera C, Taroni F, Di Maria E.et al. (2009A majority of Huntington's disease patients may be treatable by individualized allele-specific RNA interference Exp Neurol 217312–319. [DOI] [PubMed] [Google Scholar]
- Pfister EL, Kennington L, Straubhaar J, Wagh S, Liu W, DiFiglia M.et al. (2009Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients Curr Biol 19774–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Liu J., and, Corey DR. Allele-selective inhibition of huntingtin expression by switching to an miRNA-like RNAi mechanism. Chem Biol. 2010;17:1183–1188. doi: 10.1016/j.chembiol.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Matsui M, Gagnon KT, Schwartz JC, Gabillet S, Arar K.et al. (2009Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs Nat Biotechnol 27478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister EL., and, Zamore PD. Huntington's disease: silencing a brutal killer. Exp Neurol. 2009;220:226–229. doi: 10.1016/j.expneurol.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon KT, Pendergraff HM, Deleavey GF, Swayze EE, Potier P, Randolph J.et al. (2010Allele-selective inhibition of mutant huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat Biochemistry 4910166–10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warby SC, Montpetit A, Hayden AR, Carroll JB, Butland SL, Visscher H.et al. (2009CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup Am J Hum Genet 84351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CF., and, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- Monia BP, Johnston JF, Ecker DJ, Zounes MA, Lima WF., and, Freier SM. Selective inhibition of mutant Ha-ras mRNA expression by antisense oligonucleotides. J Biol Chem. 1992;267:19954–19962. [PubMed] [Google Scholar]
- Smith RA, Miller TM, Yamanaka K, Monia BP, Condon TP, Hung G.et al. (2006Antisense oligonucleotide therapy for neurodegenerative disease J Clin Invest 1162290–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima WF, Rose JB, Nichols JG, Wu H, Migawa MT, Wyrzykiewicz TK.et al. (2007Human RNase H1 discriminates between subtle variations in the structure of the heteroduplex substrate Mol Pharmacol 7183–91. [DOI] [PubMed] [Google Scholar]
- Slow EJ, van Raamsdonk J, Rogers D, Coleman SH, Graham RK, Deng Y.et al. (2003Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease Hum Mol Genet 121555–1567. [DOI] [PubMed] [Google Scholar]
- Hodgson JG, Smith DJ, McCutcheon K, Koide HB, Nishiyama K, Dinulos MB.et al. (1996Human huntingtin derived from YAC transgenes compensates for loss of murine huntingtin by rescue of the embryonic lethal phenotype Hum Mol Genet 51875–1885. [DOI] [PubMed] [Google Scholar]
- Gray M, Shirasaki DI, Cepeda C, André VM, Wilburn B, Lu XH.et al. (2008Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice J Neurosci 286182–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth PP, Siwkowski A, Allerson CR, Vasquez G, Lee S, Prakash TP.et al. (2009Short antisense oligonucleotides with novel 2'-4' conformationaly restricted nucleoside analogues show improved potency without increased toxicity in animals J Med Chem 5210–13. [DOI] [PubMed] [Google Scholar]
- Crooke ST. Antisense Drug Technology. Principles, Strategies, and Applications. CRC Press: Boca Raton, FL; 2008. [Google Scholar]
- Takahashi M, Watanabe S, Murata M, Furuya H, Kanazawa I, Wada K.et al. (2010Tailor-made RNAi knockdown against triplet repeat disease-causing alleles Proc Natl Acad Sci USA 10721731–21736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Kennington LA, Rosas HD, Hersch S, Cha JH, Zamore PD.et al. (2008Linking SNPs to CAG repeat length in Huntington's disease patients Nat Methods 5951–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangalos MN, Schechter LE., and, Hurko O. Drug development for CNS disorders: strategies for balancing risk and reducing attrition. Nat Rev Drug Discov. 2007;6:521–532. doi: 10.1038/nrd2094. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED., and, Richardson EP., Jr Neuropathological classification of Huntington's disease. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- Reiner A, Del Mar N, Meade CA, Yang H, Dragatsis I, Zeitlin S.et al. (2001Neurons lacking huntingtin differentially colonize brain and survive in chimeric mice J Neurosci 217608–7619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeron MM, Hansson O, Chen N, Wellington CL, Leavitt BR, Brundin P.et al. (2002Increased sensitivity to N-methyl-D-aspartate receptor-mediated excitotoxicity in a mouse model of Huntington's disease Neuron 33849–860. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flowchart diagramming the allele-specific targeting pipeline, with indicated number of target SNPs and ASO designs at each step.
Phased allele frequencies in 234 human HD patients.
Summary of the distribution of different classes of SNPs in the HTT coding region.
SNP sequences at described sites in primary human fibroblasts (Corriell).