Abstract
We evaluated the in vivo efficacy of structurally flexible, cationic PAMAM dendrimers as a small interfering RNA (siRNA) delivery system in a Rag2-/-γc-/- (RAG-hu) humanized mouse model for HIV-1 infection. HIV-infected humanized Rag2-/-γc-/- mice (RAG-hu) were injected intravenously (i.v.) with dendrimer-siRNA nanoparticles consisting of a cocktail of dicer substrate siRNAs (dsiRNAs) targeting both viral and cellular transcripts. We report in this study that the dendrimer-dsiRNA treatment suppressed HIV-1 infection by several orders of magnitude and protected against viral induced CD4+ T-cell depletion. We also demonstrated that follow-up injections of the dendrimer-cocktailed dsiRNAs following viral rebound resulted in complete inhibition of HIV-1 titers. Biodistribution studies demonstrate that the dendrimer-dsiRNAs preferentially accumulate in peripheral blood mononuclear cells (PBMCs) and liver and do not exhibit any discernable toxicity. These data demonstrate for the first time efficacious combinatorial delivery of anti-host and -viral siRNAs for HIV-1 treatment in vivo. The dendrimer delivery approach therefore represents a promising method for systemic delivery of combinations of siRNAs for treatment of HIV-1 infection.
Introduction
Small interfering RNAs (siRNAs) are 21–22 base long RNAs that guide the sequence-specific degradation of target mRNAs.1,2 Given the ability to direct sequence-specific silencing of target mRNAs, siRNAs have the potential to function as biodrugs for the treatment of a wide range of human maladies.3,4,5,6 Several recent studies have demonstrated the use of siRNAs in functionally downregulating expression of HIV-1 and host mRNAs against viral and cellular targets.5,7,8,9 In similarity with the use of combinatorial antiviral drugs [such as Highly Active Antiretroviral Therapy (HAART)], siRNA-based therapeutics that target combinations of distinct genomic regions of HIV-1 or target HIV-1 host dependency factors can be administered in a tailored fashion to minimize the possibility of viral escape mutants.10,11
Efficient systemic delivery of siRNA in vivo remains a principal challenge to achieve the desired RNA interference (RNAi) potency for successful clinical application.12,13 Most HIV-susceptible cells (such as, CD4+ lymphocytes and monocytes) are exceedingly difficult to transfect with nonviral agents such as cationic lipids. Several delivery strategies such as carbon nanotubes,14 carbosilane dendrimers,15 and immunoliposomes16 have been shown to deliver siRNAs into cultured human T cells and primary peripheral blood mononuclear cells (PBMCs). However, the complicated formulations (nanotube functionalization and nanotube–siRNA conjugation) and high concentrations of siRNA (>300 nmol/l in the case of carbosilane dendrimers and 50-µg siRNA per injection in the case of immunoliposomes) make them less favorable for in vivo anti-HIV applications. Other strategies, such as the use of cell-penetrating peptide-dsRNA-binding fusion proteins,17 Fab antibody fragment–protamine fusions18 and a CD7-antibody-polyarginine conjugate19 have been used to functionally deliver siRNAs and induce RNAi responses in vivo.
To date the use of synthetic nanoparticles for delivery of combinatorial dicer substrate siRNAs (dsiRNAs) in vivo has not been described. In the present study, we utilized a humanized mouse model20,21,22,23,24 to evaluate the in vivo antiviral efficacy of a cationic poly(amidoamine) (PAMAM) dendrimer-mediated siRNA delivery system. PAMAM dendrimers are a class of highly branched, structurally well-defined chemical polymers which bear cationic primary amine groups on their spherical surface and are able to form stable and uniform nanoscale complexes with negatively charged nucleic acids via electrostatic interactions. Once formed, the complexes can protect nucleic acids from ribonuclease degradation and promote cell uptake via endocytosis.25,26 These dendrimers possess numerous tertiary amines in the interior, which are protonated in the acidic endosomes leading to endosomal disruption and the release of nucleic acids from dendrimer/nucleic acid complexes. We have previously developed a series of structurally flexible PAMAM dendrimers bearing a triethanolamine core (Figure 1) as effective vectors for RNA interaction27 and siRNA delivery in adherent cells.28 The flexible structure imparts these dendrimers with stronger interactions with nucleic acids via a mutually induced fit process. At the same time, it promotes efficient dissociation of nucleic acids in endosomes via a proton sponge effect. Recently, these triethanolamine-core PAMAM dendrimers have been used to deliver an Hsp27 siRNA into human prostate cancer (PC-3) cells, resulting in specific silencing of the hsp27 gene and inhibition of cell proliferation.29 Thus, these dendrimers represent an attractive and relatively simple vehicle for siRNA delivery.
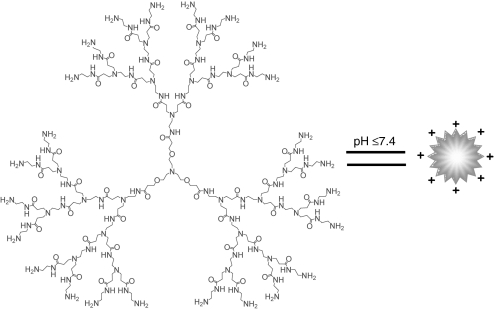
Figure 1.
Structure of a flexible poly(amidoamine) (PAMAM) dendrimer with a triethanolamine core. At pH ≤7.4, the terminal amino groups (NH2) possess positive charges via protonation. For clarity, the generation 3 (G3) dendrimer is shown as an example.
We describe here the use of PAMAM dendrimers for delivering anti-HIV-1 dsiRNAs in a combinatorial fashion for targeting both viral and cellular transcripts. Our initial in vitro studies demonstrated that these dendrimers are capable of delivering functional anti-HIV-1 dsiRNAs into cultured human T-cells and primary PBMCs. We also demonstrate that the dendrimer-dsiRNA nanoparticles functionally deliver anti-HIV-1 siRNAs to HIV-1 infected, viremic humanized Rag2-/-γc-/- mice (RAG-hu). Post-treatment analyses showed sequence-specific knockdown of the targeted viral and cellular transcripts, resulting in several logs of inhibition of HIV-1 viral load and protection of CD4+ T-cells from HIV-1 induced depletion. These results provide the first demonstration of efficacious in vivo delivery and of a combination of anti-host and -viral dsiRNA for the treatment of HIV-1 infection.
Results
Dendrimer-dsiRNAs form stable nanoscale complexes which protect the siRNAs from degradation and facilitate siRNA delivery into human T-cells in vitro
As shown in Supplementary Figure S1a, the triethanolamine-core PAMAM dendrimer of generation 5 (G5) form stable complexes with the 27-mer Dicer substrate tat/rev siRNA (dsiRNA) when the N/P (total terminal amines in the cationic dendrimer/phosphates in the siRNA) ratio ≥2. Transmission electron microscopy30 of the dendrimer G5-dsiRNA complexes at an N/P ratio of five reveals condensed spherical particles with diameters of ~100 nm (Supplementary Figure S1b). The stable G5-siRNA nanoparticles at an N/P ratio of five protected the dsiRNA from RNase degradation, even after 90-minute exposure to RNase (Supplementary Figure S1c).
The cellular uptake of the G5–dsiRNA complexes was analyzed using fluorescent microscopy. Cy3-labeled dsiRNAs were complexed with the G5 dendrimer and the complex was added to human T-lymphoblast CCRF-CEM cells (a suspension cell line). For comparison purposes, the same cell lines were transfected with the Cy3-labeled dsiRNAs using a commercial lipid-based transfection agent (Trans IT-TKO). Fluorescence microscopic imaging (Supplementary Figure S1d) revealed efficient delivery of the Cy3-dsiRNA to the suspension CEM T-cells cells, with an efficiency comparable to a commercial lipid-based reagent. No uptake was observed when uncomplexed Cy3-dsiRNAs were tested.
Live-cell confocal microscopy using HeLa cells and a G5 dendrimer-Alexa 488 labeled dsiRNA complex demonstrated that the complex is effectively internalized (Supplementary Figure S2a). To further evaluate the mechanism of dendrimer-mediated dsiRNA delivery, we tested different endocytic inhibitors (Supplementary Figure S2b). No significant effects on uptake were observed when Genistein (a caveolae-mediated endocytosis inhibitor) or Chlorpromazine (a Clathrin-mediated endocytosis inhibitor) were used. However, an increasing concentration of cytochalasin D (a macropinocytosis inhibitor) reduced the cellular uptake of the dendrimer–dsiRNA complex, suggesting a cell uptake mechanism involving macropinocytosis. It is known that actin depolymerization is a hallmark of macropinocytosis. Therefore, we also asked whether the uptake of the G5 dendrimer–dsiRNA complex depolymerized actin. The results obtained, show G5 dendrimer-dsiRNA mediated actin depolarization (Supplementary Figure S2c), providing further evidence for macropinocytosis as the dominant G5 dendrimer-dsiRNA cellular uptake mechanism.
Dendrimer-dsiRNA nanoparticles mediate specific gene silencing via the RNAi pathway and inhibit HIV 1 infection in human T-lymphoblast CEM cells and primary PBMCs
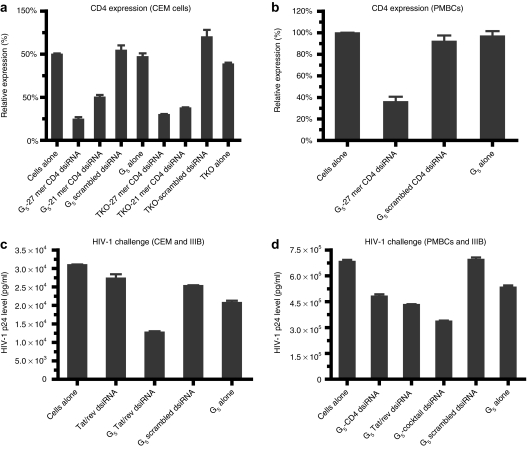
TNPO3 (Transportin-3) and CD4 are HIV-1 host dependency factors that are potential therapeutic targets.31,32 CD4 is the primary receptor for HIV-1, and transient knocking down of this receptor blocks HIV-1 infection.10,33 TNPO3 is a cellular factor that is involved in facilitating cytoplasmic—nuclear trafficking of the HIV-1 preintegration complex and was previously shown by an siRNA screen to block HIV-1 infection at the afferent stage.32,34,35 We therefore asked whether the G5 dendrimer delivered dsiRNAs targeting transcripts encoding these proteins effectively mediated gene silencing in human T-lymphoblast CEM cells and primary human PBMCs. The siRNA mediated knockdowns of the target transcripts were evaluated via quantitative real-time PCR (qRT-PCR) (Figure 2a,b) and flow cytometry (Supplementary Figure S3a). The G5 dendrimer delivered anti-CD4 dsiRNA complex resulted in a 75% decrease in CD4 mRNA levels 48 hours post-transfection (Figure 2a), with an efficiency comparable to a commercial lipid-based reagent TKO. The dendrimer G5 delivered dsiRNA gave better target knockdown than the corresponding conventional 119+2 mer siRNA.36 In similarity to the CD4 knockdown results, the G5 dendrimer delivered dsiRNA targeting TNPO3 resulted in better target knockdown than the 119+2 siRNA (Supplementary Figure S3b). Furthermore, in the primary PBMCs the G5 dendrimer delivered dsiRNA resulted in ~60% knockdown of the levels of the CD4 mRNA at the tested concentration of 50 nmol/l (Figure 2b). In contrast, no inhibition was observed with G5 dendrimer alone or a G5 dendrimer delivered scrambled dsiRNA. The commercial transfection agents were ineffective for delivery to primary blood mononuclear cells (PBMCs) (data not shown).
Figure 2.
Dendrimer-dicer substrate siRNAs (dsiRNA) nanoparticles mediate specific gene silencing in vitro. Inhibition of CD4 expression by dendrimer G5-mediated delivery of 21-mer or 27 mer anti-CD4 siRNAs (at an N/P ratio of 5) was assayed using quantitative real-time (qRT)-PCR 2 days post-treatment in culture (a) CEM T-cells and (b) human peripheral blood mononuclear cells (PBMCs). Dendrimer–small interfering RNA (siRNA) complexes inhibit HIV-1 infection in (c) CEM T-cells and (d) human PBMCs previously infected with HIV-1. HIV-1 infected cells were incubated at 37 °C with the G5-27-mer anti-tat/rev dsiRNA complexes or G5-cocktail siRNAs (combination of 27-mer anti-tat/rev dsiRNA and anti-CD4 dsiRNA) complexes. The culture supernatants were collected at 3 days following addition of the complexes for HIV p24 antigen analyses. Data represent the average of triplicate measurements of p24 and the average of three replicates.
Since the dsiRNAs are Dicer cleavage substrates, we also asked whether the dendrimer G5 delivered dsiRNAs were processed into 21–23 base siRNAs in the cells. Northern blotting analyses (Supplementary Figure S3c,d) demonstrated Dicer processed 21–22 mer siRNAs along with unprocessed 27 mer dsiRNAs in both cultured and primary cells. Illumina deep sequencing analyses of the transfected dsiRNAs confirmed the Dicer processed products (data not shown). To further validate that the G5 dendrimer delivered dsiRNAs were functioning through the RNAi mechanism, we carried out a modified 5′-RACE (Rapid amplification of cDNA ends) PCR assay on total RNAs isolated from the control and G5 dendrimer delivered anti-TNPO3 dsiRNAs in CEM cells. For these analyses we used the Ago2 cleavage specificity which takes place between bases 10 and 11 relative to the 5′ end of the guide strand siRNA.37,38 RACE PCR product sequence analyses should reveal a linker addition on the siRNA complementary target at the base 10 nucleotides downstream from the 5′ end of the antisense siRNA strand. We obtained PCR bands of the predicted length for a Dicer processed siRNA directed cleavage product following nested PCR reactions (Supplementary Figure S3e). No appropriately sized products were generated from total RNAs prepared from cells treated with RNA from G5 dendrimer scrambled dsiRNA-treated cells. The PCR amplified, gel-purified bands of the predicted lengths were cloned and the individual clones characterized by DNA sequencing. The results obtained show that the cleavage reaction took place at the predicted position for the Dicer processed dsiRNA between positions 10 and 11 from the 5′ end of the predicted dicer processed siRNA antisense strand. Based upon the position of the cleavage site these results also reconfirm Dicer processing of the dsiRNA to 21–23 base siRNAs (as shown in Supplementary Figure S3f).
Having established that the G5 dendrimer delivered anti-HIV dsiRNAs effectively downregulate the cellular CD4 and TNPO3 targets, we next assessed the anti-HIV-1 activities of the G5 dendrimer-delivered dsiRNAs. In these assays, the G5 dendrimer dsiRNA nanoparticles were incubated with cells that had been previously infected with HIV-1 IIIB followed by assays for the HIV-1 encoded p24 gag peptide release as previously reported.35,39 Results presented in Figure 2c,d show the HIV-1 p24 antigen levels following treatment with the G5 dendrimer–dsiRNA complexes. The individual as well as the combination of dsiRNAs all provided inhibition of HIV-1 replication in these stringent assays which reflect spread of viral infection. To validate the RNAi efficacy in these experiments, we evaluated the downregulation of HIV-1 tat/rev gene expression following treatment of CEM cells with the G5 dendrimer anti-tat/rev dsiRNA complex. The expression levels of tat/rev encoding RNAs were determined by quantitative RT-PCR analysis 3 days post-treatment of cells with the complex. Substantial down regulation of the tat/rev mRNA was observed, a direct consequence of the G5 dendrimer-anti-tat/rev dsiRNA delivery. In contrast, treatment of infected cells with the empty dendrimer or dsiRNA alone had no affect on HIV tat/rev levels (Supplementary Figure S3g).
As a final test for nonspecific inhibitory activity, the G5 dendrimer delivered siRNAs were monitored for induction of type I interferon responses (IFN). The levels of two different type I IFN-stimulated gene expression (mRNAs) were quantified by quantitative RT-PCR (Supplementary Figure S4a,b). From these data, we concluded that the G5 dendrimer-dsiRNA nanoparticles did not activate the type I IFN pathways and did not trigger any apparent cellular toxicity (Supplementary Figure S4c,d) under the experimental conditions employed.
Systemic administration of the G5 dendrimer-dsiRNA nanoparticles suppress HIV-1 loads in viremic RAG-hu mice
Given the promising results of the G5 dendrimer-mediated dsiRNA delivery in CD4+ T cells, we next sought to determine whether the dendrimer–siRNA complexes could functionally deliver dsiRNAs in vivo and provide protection and/or inhibition of HIV-1 infection in humanized mice. RAG-hu mice were first infected with HIV-1 NL4-3 by intraperitoneal injection until they became viremic, usually 3 weeks postinfection. The viral loads in these animals averaged 105 viral RNA copies/ml. These viremic mice were subsequently given one intravenous (i.v.) injection/week of dendrimer G5–dsiRNA complexes as described in the Supplementary Materials and Methods section. As control groups, animals were administered a G5 dendrimer-mutant tat/rev dsiRNA complex, dsiRNA alone or G5 dendrimer alone.
Plasma RNA viral levels were monitored by qPCR on a weekly basis. There were six animals in each treatment group. The siRNA treatment groups consisted of dendrimer complexed with 0.25 nmol (0.15 mg/kg) of the anti-tat/rev dsiRNA or 0.25 nmol total of a combination of the anti-tat/rev, anti-CD4 and anti-TNPO3 dsiRNAs (Figure 3a). We observed a general pattern of decreased viral loads in the majority of the G5 dendrimer-treated mice when the functional dsiRNAs were applied (weeks 8–12 as shown). The suppression of viral loads was on average three logs relative to the controls and these reductions reached statistical significance with a rank sum P = 0.0002 (Figure 3a and Supplementary Table S1 and Supplementary Figure S5a with SD). Even 3 weeks following the termination of the treatment, viral loads were still suppressed in the majority of treated animals (weeks 13–15), indicating a prolonged antiviral effect in these animals. Importantly, there was no viral suppression in the animals treated with the control G5 dendrimer delivered mutant anti tat/rev dsiRNA complex (Figure 3a and Supplementary Table S1 and Supplementary Figure S5a with SD) or the G5 dendrimer alone (Supplementary Figure S5c and Supplementary Table S4), validating the activities of the functional siRNAs. While viral suppression is significant in all the treated animals, variations seen between mice could be due to inherent differences imparted by different human donor cells used to reconstitute the animals.
Figure 3.
Dendrimer–dicer substrate siRNAs (dsiRNA) complexes suppress viral loads in HIV-1 infected RAG-hu mice. HIV-1 viral loads at different weeks postinfection and treatment are indicated. The treatment period is indicated by the yellow framed in region. Viral loads at each indicated week pre- and post-treatment. Weeks postinjection and the time point of treatment start and end are indicated. (a) The first-treatment included five weekly injections: The viral loads of uninfected mice (n = 2), nontreated mice (n = 7), naked cocktail dsiRNA-treated mice (n = 5), G5-mutant-tat/rev dsiRNA complex-treated mice (n = 3), G5-tat/rev dsiRNA complexes treated mice (n = 6) and G5-cocktail small interfering RNA (siRNA) complex treated mice (n = 6) are indicated. (b) The retreatment including twice weekly injections with G5-cocktail siRNA complexes: The viral loads of uninfected mice (n = 2), nontreated mice (n = 4), G5-tat/rev dsiRNA complex treated mice (n = 5) and G5-cocktail dsiRNA complex treated mice (n = 1) are indicated. P values for both experiments were determined as described in Materials and Methods section. The viral RNA was detected through quantitative real-time (qRT)-PCR as described in Materials and Methods section. If there was no detectable viral RNA we established this as a value of 1 (100) to allow for the use of logarithmic values on the y-axis.
Once the dendrimer-siRNA treatments were stopped, steady increases in viral loads were observed in the majority of the animals. We therefore asked if retreatment with the G5 dendrimer–dsiRNA complexes could restore suppression of viral levels to those seen with the first set of treatments. Experimental animals in which the viral loads had become elevated were retreated with two separate injections of the G5 dendrimer cocktail of dsiRNAs at 3 months following the last administration from the previous treatment period. The retreatments were conducted at weeks 24.5 and 25.5 postinfection as shown (Figure 3b). These retreatments resulted in a statistically significant (P = 0.0492) reinhibition of HIV-1 levels (Figure 3b and Supplementary Table S2 and Supplementary Figure S5b with SD). Complete suppression persisted for 3 weeks beyond the retreatment period in the G5 dendrimer-cocktail dsiRNA treated mice.
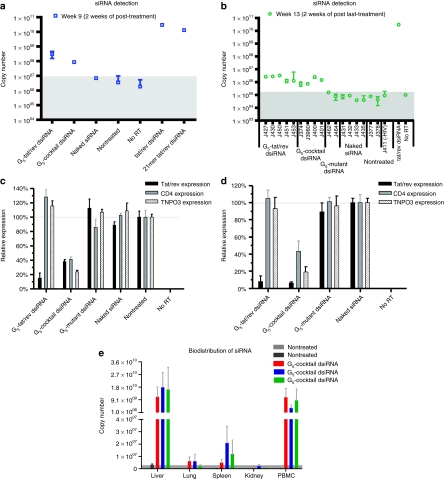
Taqman qPCR assays were carried out to quantify the tat/rev dsiRNA levels in cells collected from peripheral blood at various times during and after treatment. The results presented in Figure 4a reveal that the tat/rev dsiRNA was quantifiable in all of the G5 dendrimer dsiRNA treated mice at 2 weeks into the treatment period (week 9). At 2 weeks following the last injection (week 13), the dsiRNAs were still detectable in cells prepared from all the dendrimer-dsiRNA-treated mice (Figure 4b). Importantly, no dsiRNAs were detected in the cellular fractions from mice treated with naked dsiRNAs.
Figure 4.
The detection and function of small interfering RNAs (siRNAs) in blood cells of HIV-1 infected RAG-hu mice and in vivo biodistribution. (a,b) Detection of the tat/rev small interfering RNA (siRNA) sequences at weeks 9 and 13 postinfection usin naked siRNAs versus G5–dicer substrate siRNAs (dsiRNA) complex treated animals from Figure 3a. The background copy number of siRNA is <107 (gray). Error bars indicate SD (n = 4). (c,d) Expression levels of targeted tat/rev, CD4, and TNPO3 gene transcripts at weeks 8 and 9 postinfection are shown relative to HIV-1 infected, (c) nontreated animal samples and (d) naked siRNA-treated animal samples, respectively. (e) In vivo biodistribution analyses. The delivery of the anti-tat/rev siRNA was monitored using Taqman quantitative real-time (qRT)-PCR on RNAs isolated from various tissues and peripheral blood mononuclear cells (PBMCs) following systemic administration of the G5 dendrimer-dsiRNA nanoparticles in RAG-hu mice. The background copy number of siRNA is <5 × 106 (gray). Error bars indicate SD (n = 3).
Reductions in the levels of the three targeted genes (HIV tat/rev, CD4 and TNPO3) in PBMCs of treated mice were evaluated by quantitative real-time PCR. The G5 dendrimer-dsiRNA-treated mice had reduced levels of the corresponding targeted mRNA levels relative to the controls following the first and second treatments with the G5 dendrimer-dsiRNA complexes (weeks 8 and 9) (Figure 4c,d), confirming the effective sequence-specific gene silencing.
G5 dendrimer delivered dsiRNAs distribute primarily to PBMCs and the liver
To evaluate the biodistribution of G5 dendrimer delivered dsiRNAs in vivo, the RAG-hu mice were systemically injected with a dendrimer G5—cocktail of dsiRNAs as described above. The tat/rev dsiRNA levels isolated from various tissues and PBMCs were quantified by Taqman qRT-PCR. As shown in Figure 4e, tat/rev dsiRNAs were primarily detected in the human PBMCs and mouse liver 1 week postinjection, but not in lung and kidney. We also utilized a C57BL/6 mouse to image the in vivo biodistribution of dendrimer–dsiRNA complex. The results from these analyses demonstrated that the G5 dendrimer-Cy5-labeled siRNA nanoparticles mainly distributed to and accumulated in the mouse liver (Supplementary Figure S6). In contrast, naked siRNA was quickly cleared through the liver within several hours following the injection (Supplementary Figure S6a). The marked accumulation of the dsiRNAs in PBMCs rationally accounts for the marked inhibition of HIV-1 replication. Further evaluation of dsiRNA accumulation in different human hematopoietic cell subsets such as CD4+ T cells and macrophages was hampered by limited sample volumes from these mice. The liver accumulation is characteristic of many delivery vehicles, and perhaps could be taken advantage of for treatment of liver-specific diseases. Although the concentration of dsiRNAs available to the HIV-1 susceptible cells might be reduced due to the accumulation in the liver, the dendrimer-dsiRNA nanoparticles administered by systemic delivery showed effective suppression of HIV-1 loads in viremic RAG-hu mice in our experimental setup. These results suggest that local delivery to HIV sanctuaries such as the CNS might prove to be a rational use of the dendrimer delivery.
The G5 dendrimer-dsiRNA treatment protects against HIV-1-mediated T-cell depletion
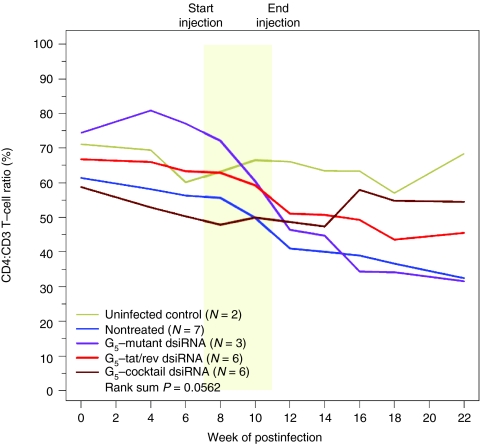
HIV-1 infection characteristically results in a progressive loss of helper CD4+ T cells. During the course of infection, CD4+ T cell levels fall transiently during the acute stage of infection followed by a return to a set point for several months/years with an eventual depletion leading to AIDS.40,41 The levels of CD4+ T-cells show similar declines in the HIV-1 infected humanized mice.42 Considering the siRNA mediated suppression of HIV-1 viral replication in the G5 dendrimer—dsiRNA treated animals, we sought to determine whether the reduced viral loads impacted on the T-cell levels of the treated animals by monitoring the levels of CD4+ T-cells in peripheral blood collected at different time points post-treatment (Figure 5). In uninfected mice, the levels of CD4+ T cells remained stable (within a 10% variation range) throughout the course of the experiments. For those animals treated with the tat/rev or a combination dsiRNAs, the protection against T-cell depletion reached near statistical significance (P = 0.0562) (Figure 5 and Supplementary Table S3 and Supplementary Figure S7a with SD). In contrast, in the animals that were HIV-1 infected but not receiving treatment, the viral infection resulted in a rapid decline in CD4+ T cells beginning at week 4 postinfection, falling to <40% of the starting levels at 20 weeks postinfection. There was no protection in the mice treated with the G5 dendrimer-mutant tat/rev dsiRNA complex (Figure 5 and Supplementary Table S3) or the empty G5 dendrimer (Supplementary Figure S7b and Supplementary Table S5). In animals treated with the G5 dendrimer-cocktail of dsiRNAs the levels of CD4+ T cells remained stable well beyond the last treatment.
Figure 5.
Dendrimer–dicer substrate siRNAs (dsiRNA) complexes protect RAG-hu mice from CD4+ T-cell loss. CD4+ T-cell levels were assessed by fluorescence-activated cell sorting (FACS) at each indicated week pre- and post-small interfering RNA (siRNA) treatment. Start and end of treatments are indicated by the yellow framed in region. Mice from the first treatment depicted in Figure 3a. Uninfected mice (n = 2), nontreated mice (n = 7), G5-mutant-tat/rev dsiRNA complex treated mice (n = 3), G5-tat/rev dsiRNA complex-treated mice (n = 6) and G5-cocktail dsiRNA complex-treated mice (n = 6) are indicated. P values for the experiment are indicated and were determined as described in Materials and Methods section. BL (base line): each individual mouse was bled two times before HIV-1 infection and the CD4:CD3 levels were averaged within treatment groups to establish a baseline CD4:CD3 level.
The G5 dendrimer-mediated dsiRNA delivery does not trigger type I IFN responses and does not show apparent toxicities in vivo
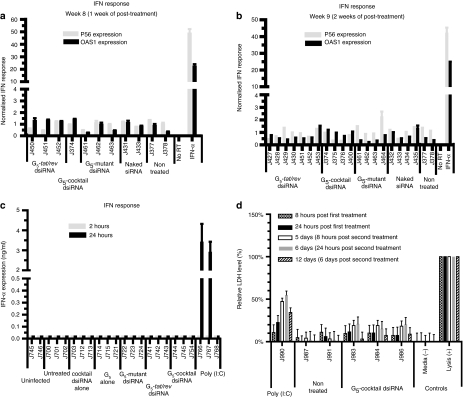
It has been previously reported that synthetic siRNAs delivered by liposomes or other polymers can trigger innate immune responses, such as Toll-like receptor mediated induction of type I IFN, tumor necrosis factor-α (TNF-α), and interleukin-6.43,44,45 We therefore assessed whether or not treatment of the animals with the G5 dendrimer-dsiRNAs resulted in induction of type I IFN–regulated gene expression. To assay for this we used quantitative RT-PCR expression assays on RNAs derived from PBMCs isolated from the G5 dendrimer-dsiRNA-treated animals as well as controls. IFN-α treated PBMCs was used as a positive control. No significant differences in IFN responsive gene expression were discernable in samples from either the treated or control animals at different times post-treatment (Figure 6a,b). Additionally, the acute responses were evaluated within a short time frame (2–24 hours post-treatment). Using an enzyme-linked immunosorbent assay, we directly monitored IFN-α levels in treated mice at 2 hours and 24 hours postinjection of the experimental RNAs and found no significant elevation of IFN-α (Figure 6c). To monitor for possible liver toxicity triggered by the dendrimer delivered dsiRNAs we performed lactate dehydrogenase (LDH) assays. No apparent toxicities were observed in samples from animals injected with the G5 dendrimer-dsiRNAs nanoparticles (Figure 6d). Histopathological analyses of hematoxylin and eosin-stained tissue sections were used to evaluate potential toxicity of the dendrimer–dsiRNA complexes (Supplementary Table S6). The anatomic histopathologic data revealed no specific pathologic alterations in the livers of all treated animals. Livers from mice J983 and J984 showed a focal mild to minimal area of mineralization, which is considered a background lesion. The other organs showed no significant histological lesions.
Figure 6.
In vivo administration of dendrimer–dicer substrate siRNAs (dsiRNA) complexes do not induce interferon or toxicities. (a,b) The expression of type I interferon response genes (P56 and OAS1) at (a) week 8 and (b) 9 postinfection after treatment with siRNAs and G5–dsiRNA complexes. Interferon (IFN)-α treated, HIV-1-infected human peripheral blood mononuclear cells (PBMCs) were used as a positive control. Gene expression was normalized to the gapdh mRNA. Error bars indicate SD (n = 4). (c) The expression levels of IFN-α at 2 hours or 24 hours after G5-dsiRNA nanoparticle injections as measured by an enzyme-linked immunosorbent assay (ELISA) are shown. Poly (I:C) treated infected Rag-hu mice were used as a positive control. Error bars indicate SD (n = 3). (d) The levels of LDH (lactate dehydragenase) after G5-dsiRNA nanoparticles injections are shown. Cellular lysate-treated human PBMCs were used as a positive control. The data are cumulative from three mice per experimental group.
Discussion
Recent studies have demonstrated that applications of siRNAs targeting viral or cellular transcripts provide inhibition of HIV-1 replication in vitro and in vivo.14,15,17,18,19 Since negatively charged nucleic acids do not readily traverse cellular membranes and are vulnerable to degradation without some protective covering, such direct administration ultimately results in poor pharmacokinetics and lack of gene silencing.
We demonstrate in this study the first results of delivery of combinatorial siRNAs via a synthetic dendrimer nanoparticle. We have utilized a PAMAM generation 5 dendrimer-dsiRNA delivery system for the treatment of HIV-1 infection both in cell culture (human T-lymphoblast cells and primary PBMCs) and in a humanized mouse model. Dendrimers are a class of highly branched and well-defined synthetic polymers. The G5 dendrimer used in this study contains 96 terminal amine groups on the surface and a triethanolamine core with a flexible structure. This dendrimer is able to form stable and uniformly sized complexes with nucleic acids via electrostatic interactions, and the flexible structure further enhances the interaction with nucleic acids through mutually induced fit. By simply mixing the G5 dendrimer with dsiRNAs, stable nanometer-scale (ca 100 nmol/l in diameter) particles were formed. Following internalization in cells (which we show is primarily via macropinocytosis), the G5 dendrimer–dsiRNA complexes release the dsiRNAs, which are subsequently processed by Dicer into 21–23 base siRNAs that trigger sequence specific gene silencing of the targeted mRNAs.
Development of viral resistance is a common setback for HIV therapies due to the generation of viral escape mutants. One of the strategies for achieving the desired long-term RNAi efficacy is to mitigate viral escape from RNAi through multiple RNAi effectors, a strategy akin to the HAART approach. In addition to targeting conserved viral targets, transient knockdown of the levels of HIV-1 host-dependency factors (such as CD4, TNPO3, CCR5) that are essential for viral replication may reduce the emergence of viral escape mutants. We have demonstrated in this study that a cocktail of three dsiRNAs targeting HIV-1 tat/rev and two cellular targets, CD4 and TNPO3 resulted in downregulation of all three targeted transcripts in vivo. We initially chose to transiently inhibit CD4 expression since it is absolutely required for HIV-1 infection, and would therefore affect both M-tropic and T-tropic viral infection. Future experiments will explore the effectiveness and potential toxicities of transiently knocking down the expression of other host dependency factors in combination with conserved viral sequences. The inhibition of these targets resulted in a reduction of the viral RNA load and protection of CD4+ T cells from HIV-1-mediated depletion. Furthermore, the results of this study demonstrate that a combination of dsiRNAs can be functionally delivered to T-lymphocytes in vivo in the absence of any apparent or measured toxicity. Furthermore we show that 3 months after cessation of the treatments (allowing viral loads to become elevated again), animals retreated with the dendrimer-cocktail dsiRNA once again showed potent knockdown of viral loads. This suppression persisted for an additional 3 weeks beyond the retreatment period. Collectively, our results demonstrate the capacity of the G5 dendrimer delivered dsiRNAs to achieve statistically significant viral suppression in vivo, resulting in protection of CD4+ T cells from HIV-1-mediated depletion. Given the importance and challenge of siRNA delivery, it is of significance that these results provide the first demonstration of dendrimer-mediated delivery of siRNAs to hematopoietic cells in vivo, and provide the impetus for further clinical development of this strategy. Our results demonstrate a lack of discernable toxicity and a significant level of efficacy following systemic dendrimer delivery of dsiRNAs. The data presented strongly support the combinatorial use of dsiRNAs targeting both cellular and viral transcripts for treating HIV-1 infection as a stand-alone therapy or as an adjuvant to the HAART currently in clinical use for the treatment of HIV-1 infection.
Materials and Methods
Materials. Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich (St Louis, MO), all restriction enzymes were obtained from New England BioLabs (Ipswich, MA) and all cell culture products were purchased from GIBCO (Gibco BRL/Life Technologies, Carlsbad, CA, a division of Invitrogen, Carlsbad, CA). Reverse transcriptase III, random primers, and oligo(dT)20 primers (Invitrogen); Lipofectamine 2000 (Invitrogen, CA); Trans IT-TKO (Mirus, Madison, WI); (Invitrogen); DNase I (Ambion, Austin, TX); CCRF-CEM (ATCC, Manassas, VA); the HIV-1 NL4.3 and HIV-1 IIIB viruses were obtained from the AIDS Research and Reference Reagent Program.
siRNAs. siRNA and Cy3-labeled single strand RNA were purchased from Integrated DNA Technologies, Coralville, IA.
Site I (tat/rev) 27 mer. Sense: 5′-GCG GAG ACA GCG ACG AAG AGC UCA UCA-3′ antisense: 5′-UGA UGA GCU CUU CGU CGC UGU CUC CGC dTdT-3′.
Mutated site I (tat/rev) 27 mer. Sense: 5′-GCG CUA ACA GCG UGU AAG AGC GAC UCA-3′ antisense: 5′-UGA GUC GCU CUU ACA CGC UGU UAG CGC UU-3′. (The mutated sequences were underlined.)
Anti-CD4 21 mer. Sense: 5′-GAU CAA GAG ACU CCU CAG U dGdA-3′ antisense: 5′-ACU GAG GAG UCU CUU GAU C dTdG-3′.
Anti-CD4 27 mer. Sense: 5′-GAU CAA GAG ACU CCU CAG UGA GAA G-3′ antisense: 5′-CUU CUC ACU GAG GAG UCU CUU GAU CUG-3′ (2′-OMe modified U was underlined.)
Anti-TNPO3 21 mer. Sense: 5′-CGA CAU UGC AGC UCG UGU AUU-3′ antisense: 5′-UAC ACG AGC UGC AAU GUC GUU-3′.
Anti-TNPO3 27 mer. Sense: 5′-CGA CAU UGC AGC UCG UGU ACC AG dGdC-3′ antisense: 5′-GCC UGG UAC ACG AGC UGC AAU GUC GUU-3′.
Dendrimer synthesis and characterization. The PAMAM dendrimers were synthesized as previously described and characterized by IR, NMR, MS, and HPLC.27,28
Cell culture. CCRF-CEM cells were purchased from ATCC and cultured in RPMI 1640 supplemented with 10% fetal bovine serum. Cells were cultured in a humidified 5% CO2 incubator at 37 °C.
PBMCs. Peripheral blood mononuclear samples were obtained from healthy donors from the City of hope National Medical Center. PBMCs were isolated from whole blood by centrifugation through a Ficoll–Hypaque solution (Histopaque-1077; Sigma, St Louis, MO). CD8 cells (T-cytotoxic/suppressor cells) were depleted from the PBMCs by CD8 Dynabeads (Invitrogen, CA) according to the manufacturer's instructions. CD8+ T cell-depleted PBMCs were washed twice in phosphate-buffered saline and resuspended in culture media (RPMI 1640 with 10% fetal bovine serum, 1× PenStrep and 100 U/ml interleukin-2). Cells were cultured in a humidified 5% CO2 incubator at 37 °C.
Transfection. 2 × 105 CEM cells or PBMCs per well were seeded in 24-well tissue culture plates in 300 µl fresh complete medium containing 10% fetal bovine serum. Before transfection, complexes of dsiRNA/dendrimer reagents were prepared. The desired amount of dsiRNA and dendrimer reagent was diluted in 50 µl of serum-free medium (Opti-MED), respectively. The dendrimer solution was mixed gently and incubated for 10 minutes at room temperature. After the 10-minute incubation, the diluted dsiRNA and the dendrimer reagent was mixed gently and incubated for 30 minutes at room temperature. The 100 µl of dsiRNA/dendrimer complex was added to each well containing cells and medium and mixed gently by rocking the plate back and forth. The cells were incubated at 37 °C in a CO2 incubator for 24–48 hours for the further assay. A commercial transfection reagent TransIT-TKO (Mirus) was used as control according to the manufacturer's instructions.
Determination of CD4 or TNPO3 gene silencing (qRT-PCR analysis). For CD4 expression, cells were transfected with 50 nmol/l of 21-mer or 27-mer anti-CD4 siRNA or anti-TNPO3 siRNA and dsiRNA using dendrimers as described above. Forty-eight hours post-transfection, the total RNAs were isolated with STAT-60 (TEL-TEST “B”, Friendswood, TX). Expression of the target genes was analyzed by quantitative real-time-PCR using 2× iQ SyberGreen Mastermix (Bio-Rad, Hercules, CA) and specific primer sets at a final concentration of 400 nmol/l. Primers were as follows: CD4 forward primer: 5′-GCT GGA ATC CAA CAT CAA GG-3′ CD4 reverse primer: 5′-CTT CTG AAA CCG GTG AGG AC-3′ TNPO3 forward primer: 5′-CCT GGA AGG GAT GTG TGC-3′ TNPO3 reverse primer: 5′-AAA AAG GCA AAG AAG TCA CAT CA-3′ GAPDH forward primer: 5′-CAT TGA CCT CAA CTA CAT G-3′ GAPDH reverse primer: 5′-TCT CCA TGG TGG TGA AGA C-3′.
RNA-Stat60 was used to extract total RNA according to the manufacturer's instruction (Tel-Test). Residual DNA was digested using the DNA-free kit per the manufacturer's instructions (Ambion). cDNA was produced using 2 µg of total RNA, Moloney murine leukemia virus reverse transcriptase and random primers in a 15 µl reaction according to the manufacturer's instructions (Invitrogen). GAPDH expression was used for normalization of the qPCR data.
HIV-1 challenges and p24 antigen assay. The CEM cells or PBMCs were infected with HIV IIIB for 5 days (multiplicity of infection: 0.001). Before transfection, the infected cells were gently washed with phosphate-buffered saline three times to remove free virus. 1.0 × 105 infected cells and 1.0 × 105 uninfected cells were mixed and transfected with 50 nmol/l 27-mer tat/rev siRNA using dendrimers in 24-well plates as previously described. The culture supernatants were collected at the 3rd day. The p24 antigen analyses were performed using a Coulter HIV-1 p24 Antigen Assay (Beckman Coulter, Brea, CA) according to the manufacturer's instructions.
Determination of tat/rev mRNA silencing (qRT-PCR analysis). For detection of the tat/rev mRNA levels, cells were infected with HIV-1 IIIB for 5 days (multiplicity of infection: 0.001). Before the assays the infected cells were washed gently three times to eliminate free virus. 1.0 × 105 infected cells and 1.0 × 105 uninfected cells (total 2 × 105/well) were mixed and transfected with 50 nmol/l Tat-rev siRNA using dendrimers in 24-well plates as described above. Seventy-two hours post-transfection, total RNAs were isolated and expression of the tat/rev coding RNAs was analyzed by qRT-PCR as previously described. Primers were as follows: tat/rev forward primer: 5′-GGC GTT ACT CGA CAG AGG AG-3′ tat/rev reverse primer: 5′-TGC TTT GAT AGA GAA GCT TGA TG-3′ GAPDH expression was used for normalization of the qPCR data.
Generation and HIV-1 infection of humanized Rag2-/-γc-/- mice (RAG-hu mice). Humanized BALB/c-Rag2-/-γc-/- mice were prepared as previously described using human fetal liver-derived CD34+ cells. Briefly, neonatal mice were conditioned by irradiating at 350 rads and then injected intrahepatically with 0.5–1×106 human CD34+ cells. Approximately 12 weeks after reconstitution, mice were screened for human cell engraftment. Blood was collected by tail bleeds, and red blood cells were lysed using the Whole Blood Erythrocyte Lysing Kit (R&D Systems, Minneapolis, MN). The white blood cell fraction was stained with antibodies against the human pan-leukocyte marker CD45 (Caltag) and fluorescence-activated cell sorting analyzed as described. To infect human cell reconstituted RAG-hu mice, HIV-1 NL4-3 (1.2 × 105 i.u.) in a 100 µl volume was injected intraperitoneally at least 12 weeks after cell engraftment. Viral loads were examined weekly and viremia was established in all the mice by 4 weeks.
Treatment with dendrimer–siRNA complexes. G5 dendrimers were diluted to an appropriate concentration in phosphate-buffered saline buffer (pH 7.4) and the experimental siRNAs were diluted with H2O, with all solutions stored at -20 °C. Both solutions were mixed at N/P (=[total end amines in cationic dendrimer]/[phosphates in siRNA]) ratio 5 and incubated at 37 °C for 30 minutes in phosphate-buffered saline buffer. Treatment was done by i.v. injection on the last day of week 4 with 0.25 nmol experimental RNAs (4.6 µg cocktailed siRNAs including equal amount of three siRNAs: tat/rev siRNA, TNPO3 siRNA, and CD4 siRNA, or preparations of G5 dendrimer–siRNAs conjugates with equal amount of three siRNA portions) in a 40 µl volume, followed by another the next day. Later, the injections were continued on a weekly basis for 4 weeks. In the second in vivo treatment experiment, 0.25 nmol conjugates in a 40 µl volume were administered at 12.5 and 13.5 weeks after last-infection like above.
Measurement of viral loads in plasma. To quantify cell-free HIV-1 by qRT-PCR, RNA was extracted from 25 to 50 µl of EDTA-treated plasma using the QIAamp Viral RNA kit (Qiagen). cDNAs were produced with Superscript III reverse transcriptase (Invitrogen) using a primer set specific for the HIV-1 LTR sequence, and qPCR was performed with the same primer set and a LTR-specific probe using Supermix UDG (Invitrogen) as described.
Flow cytometry. Whole blood was collected and red blood cells were lysed as reported previously. Peripheral blood cells were stained for hCD3-PE and hCD4-PECy5 (Caltag) markers and analyzed using a Coulter EPICS XL-MCL fluorescence-activated cell sorting analyzer (Beckman Coulter). CD4+ T-cell levels were calculated as a ratio of the entire CD3 population (CD4+CD3+:CD4-CD3+). To establish baseline CD4+ T-cell ratios, all mice were analyzed before infection.
Determination of tat/rev siRNA. At 1, 3, and 9 weeks postinjection, blood samples were collected and small RNAs were isolated with MirVana miRNA isolation kit (Applied Biosystems, Carlsbad, CA) according to the manufacturer's instruction. The siRNA quantification was performed using TaqMan MicroRNA Assay according to manufacturer's recommended protocol (Applied Biosystems). Ten nanograms of small RNA, 0.2 µmol/l stem-loop RT primer, RT buffer, 0.25 mmol/l dNTPs, 3.33 U/ml MultiScribe reverse transcriptase (RT) and 0.25 U/ml RNase inhibitor were used in 15 µl RT reactions for 30 minutes at 16 °C, 30 minutes at 42 °C, and 5 minutes at 85 °C, using the TaqMan MicroRNA reverse transcription Kit (Applied Biosystems). For real-time PCR, 1.33 µl of cDNA, 0.2 mmol/l TaqMan Probe, 1.5 mmol/l forward primer, 0.7 mmol/l reverse primer, and TaqMan Universal PCR Master Mix were added in 20 µl reactions for 10 minutes at 95 °C and 40 cycles of 15 seconds at 95 °C and 1 minute at 60 °C. All real-time PCR experiments were done using an iCycler iQ system (Bio-Rad). Primers were as follows: Site I looped RT primer: 5′-GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACA CAG CG-3′ site I forward primer: 5′-GCT GAT GAG CTC TTC GTC G-3′ site I reverse primer: 5′-GTG CAG GGT CCG AGG T-3′ site I probe primer: 5′-6-FAM-TCG CAC TGG ATA CGA CAC AGC GAC GA–BHQ1-3′. In this case, a synthetic 27mer duplex RNA was used as positive control.
Determination of targeted gene expression. Human PBMCs were obtained from treated mice at 1 and 3 weeks postinjection and total RNAs were isolated with STAT-60 (TEL-TEST “B”) according to the manufacturer's instructions. Residual DNA was digested using the DNA-free kit per the manufacturer's instructions (Ambion). cDNA was made using 2 µg of total RNA. Reverse transcription was carried out using Moloney murine leukemia virus reverse transcriptase and random primers in a 15-µl reaction according to the manufacturer's instructions (Invitrogen). Expression of the tat/rev coding RNAs was analyzed by quantitative RT-PCR using 2× iQ SyberGreen Mastermix (Bio-Rad, Hercules, CA) and specific primer sets at a final concentration of 400 nmol/l. Gapdh expression was used for normalization of the qPCR data. Primers were as follows: IIIB or NL4-3 tat/rev forward primer: 5′-GGC GTT ACT CGA CAG AGG AG-3′ IIIB or NL4-3 tat/rev reverse primer: 5′-TGC TTT GAT AGA GAA GCT TGA TG-3′ CD4 forward primer: 5′-GCT GGA ATC CAA CAT CAA GG-3′ CD4 reverse primer: 5′-CTT CTG AAA CCG GTG AGG AC-3′ TNPO3 Forward primer: 5′-CCT GGA AGG GAT GTG TGC-3′ TNPO3 Reverse primer: 5′-AAA AAG GCA AAG AAG TCA CAT CA-3′ gapdh forward primer 1: 5′-CAT TGA CCT CAA CTA CAT G-3′ gapdh reverse primer 2: 5′-TCT CCA TGG TGG TGA AGA C-3′.
In vivo distribution study of G5 dendrimer–dsiRNA complexes. Liver, kidney, spleen, lungs, and PBMCs were collected 12 days post-treatment (6 day after additional second treatment) and small RNAs were isolated with the MirVana miRNA isolation Kit (Applied Biosystems) according to the manufacturer's instruction. The siRNA quantification was performed using a TaqMan MicroRNA Assay according to manufacturer's recommended protocol (Applied Biosystems) using the primers described above.
Interferon assays. Total RNA was isolated from PBMCs of treated mice using STAT-60. Expression of mRNAs encoding p56(CDKL2) and OAS1 were analyzed by quantitative RT-PCR using 2× iQ SyberGreen Mastermix (Bio-Rad) as described above and specific primer sets for these genes at final concentrations of 400 nmol/l. Primers were as follows: P56 (CDKL2) forward, 5′-TCA AGT ATG GCA AGG CTG TG-3′ P56 (CDKL2) reverse, 5′-GAG GCT CTG CTT CTG CAT CT-3′ OAS1 forward, 5′-ACC GTC TTG GAA CTG GTC AC-3′ OAS1 reverse, 5′-ATG TTC CTT GTT GGG TCA GC-3′ gapdh expression was used for normalization of the qPCR data. To measure any induced IFN-α directly, human IFN-α1 enzyme-linked immunosorbent assay Ready-SET-Go! (eBioscience, San Diego, CA) was used. Mice were injected with the G5 dendrimer mutant tat/rev dsiRNA complex, dsiRNA alone, G5 dendrimer alone, G5 dendrimer-siRNA tat/rev, or G5 dendrimer-cocktail. At 2 and 24 hours post-treatment 25–50 µl of EDTA-treated plasma was collected from three mice per treatment group and from three positive control mice which had been i.v. injected with 5 µg of poly (I:C) (Sigma) in a 50 µl volume. Plasma levels were evaluated as per instructions supplied in the kit.
In vivo LDH toxicity assay. Lactate dehydrogenase activity in serum after i.v. injection of G5 dendrimerd-siRNA nanoparticles. 0.25 nmol of a dsiRNA cocktail complexed with a G5 dendrimer was injected i.v. on day 0 and again on day 5. Blood was collected at 8 and 24 hours after each treatment and on day 12 and the level of LDH was determined from serum using the Cytotoxicity Detection Kit (LDH). For a positive control, blood was drawn and PBMCs were isolated and lysed per kit protocol.
Statistical methods. The mouse viral loads and CD4:CD3 T-cell ratios were plotted by using an Lowess smoother across values. Viral loads were first log-transformed before smoothing and then anti-transformed for plotting. Missing values were imputed with a last observation carried forward scheme. The calculations were conducted as previously described.24
SUPPLEMENTARY MATERIAL Figure S1. Stable nanoscale dendrimer–dsiRNA complexes prevent dsiRNA from degradation and promote cellular uptake in vitro. Figure S2. Internalization study of dendrimer–siRNA complexes. Figure S3. Dendrimer-siRNA nanoparticles efficiently release siRNA and mediate specific gene silencing via the RNAi pathway in vitro. Figure S4. Dendrimer-mediated dsiRNA delivery system does not trigger type I interferon responses in cultured HIV-1 infected (a) CEM cells and (b) human PBMCs. Figure S5. Dendrimer–dsiRNA complexes suppress viral loads in HIV-1 infected RAG-hu mice while dendrimer alone has no affect on viral loads. Figure S6. In vivo distribution and cryosections of G5 dendrimer-Cy5 siRNA complexes in a C57BL/6 mouse model. Figure S7. Dendrimer-dsiRNA complexes protect RAG-hu mice from CD4+ T-cell loss while dendrimer alone gave no protection. Table S1. First-treatment monitoring of HIV-1 viral loads in individual RAG-hu mice. Table S2. Retreatment HIV-1 viral loads in individual RAG-hu mice. Table S3. First-treatment CD4 T-cell levels in individual treated and control RAG-hu mice. Table S4. HIV-1 viral loads in individual RAG-hu mice. Table S5. CD4+ T-cell levels in treated and control individual RAG-hu mice. Table S6. Histopathology analysis. Materials and Methods.
Acknowledgments
We thank NIH AIDS Research and Reference Reagents Program for HIV-1-related reagents used in this work. This work was supported by NIH RO1 grants AI057066 and AI073255 to R.A., AI29329 and HL07470 to J.J.R., EU FP7 EuroNanoMed program “DENANORNA” to LP, and CNRS project CNRS-USA to LP. This work has also been facilitated by the infrastructure and resources provided by the Colorado Center for AIDS Research Grant P30 AI054907. The authors declared no confilict of interest.
Supplementary Material
Stable nanoscale dendrimer–dsiRNA complexes prevent dsiRNA from degradation and promote cellular uptake in vitro.
Internalization study of dendrimer–siRNA complexes.
Dendrimer-siRNA nanoparticles efficiently release siRNA and mediate specific gene silencing via the RNAi pathway in vitro.
Dendrimer-mediated dsiRNA delivery system does not trigger type I interferon responses in cultured HIV-1 infected (a) CEM cells and (b) human PBMCs.
Dendrimer–dsiRNA complexes suppress viral loads in HIV-1 infected RAG-hu mice while dendrimer alone has no affect on viral loads.
In vivo distribution and cryosections of G5 dendrimer-Cy5 siRNA complexes in a C57BL/6 mouse model.
Dendrimer-dsiRNA complexes protect RAG-hu mice from CD4+ T-cell loss while dendrimer alone gave no protection.
First-treatment monitoring of HIV-1 viral loads in individual RAG-hu mice.
Retreatment HIV-1 viral loads in individual RAG-hu mice.
First-treatment CD4 T-cell levels in individual treated and control RAG-hu mice.
HIV-1 viral loads in individual RAG-hu mice.
CD4+ T-cell levels in treated and control individual RAG-hu mice.
Histopathology analysis.
REFERENCES
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE., and, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA., and, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Kim DH., and, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- de Fougerolles A, Vornlocher HP, Maraganore J., and, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi JJ. RNAi as a treatment for HIV-1 infection. BioTechniques. 2006;Suppl:25–29. doi: 10.2144/000112167. [DOI] [PubMed] [Google Scholar]
- Rossi JJ, June CH., and, Kohn DB. Genetic therapies against HIV. Nat Biotechnol. 2007;25:1444–1454. doi: 10.1038/nbt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NS., and, Rossi JJ. Control of HIV-1 replication by RNA interference. Virus Res. 2004;102:53–58. doi: 10.1016/j.virusres.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Scherer L, Rossi JJ., and, Weinberg MS. Progress and prospects: RNA-based therapies for treatment of HIV infection. Gene Ther. 2007;14:1057–1064. doi: 10.1038/sj.gt.3302977. [DOI] [PubMed] [Google Scholar]
- Martínez MA. Progress in the therapeutic applications of siRNAs against HIV-1. Methods Mol Biol. 2009;487:343–368. doi: 10.1007/978-1-60327-547-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Banerjea A., and, Akkina R. Bispecific short hairpin siRNA constructs targeted to CD4, CXCR4, and CCR5 confer HIV-1 resistance. Oligonucleotides. 2003;13:303–312. doi: 10.1089/154545703322616989. [DOI] [PubMed] [Google Scholar]
- Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R.et al. (2004Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition J Virol 782601–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D., and, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead KA, Langer R., and, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Winters M, Holodniy M., and, Dai H. siRNA delivery into human T cells and primary cells with carbon-nanotube transporters. Angew Chem Int Ed Engl. 2007;46:2023–2027. doi: 10.1002/anie.200604295. [DOI] [PubMed] [Google Scholar]
- Weber N, Ortega P, Clemente MI, Shcharbin D, Bryszewska M, de la Mata FJ.et al. (2008Characterization of carbosilane dendrimers as effective carriers of siRNA to HIV-infected lymphocytes J Control Release 13255–64. [DOI] [PubMed] [Google Scholar]
- Kim SS, Peer D, Kumar P, Subramanya S, Wu H, Asthana D.et al. (2010RNAi-mediated CCR5 silencing by LFA-1-targeted nanoparticles prevents HIV infection in BLT mice Mol Ther 18370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi A, Meade BR, Chang YC, Fredrickson CT, Willert K, Puri N.et al. (2009Efficient siRNA delivery into primary cells by a peptide transduction domain-dsRNA binding domain fusion protein Nat Biotechnol 27567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM.et al. (2005Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors Nat Biotechnol 23709–717. [DOI] [PubMed] [Google Scholar]
- Kumar P, Ban HS, Kim SS, Wu H, Pearson T, Greiner DL.et al. (2008T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice Cell 134577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berges BK, Akkina SR, Folkvord JM, Connick E., and, Akkina R. Mucosal transmission of R5 and X4 tropic HIV-1 via vaginal and rectal routes in humanized Rag2-/- gammac-/- (RAG-hu) mice. Virology. 2008;373:342–351. doi: 10.1016/j.virol.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton PW., and, Garcia JV. Novel humanized murine models for HIV research. Curr HIV/AIDS Rep. 2009;6:13–19. doi: 10.1007/s11904-009-0003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand N, Ploss A, Balling R, Becker PD, Borsotti C, Brezillon N.et al. (2009Humanized mice for modeling human infectious disease: challenges, progress, and outlook Cell Host Microbe 65–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne R, Pedati C, Guendel I, Carpio L, Kehn-Hall K, Saifuddin M.et al. (2009The utilization of humanized mouse models for the study of human retroviral infections Retrovirology 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff CP, Zhou J, Remling L, Kuruvilla J, Zhang J, Li H.et al. (2011An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice Sci Transl Med 366ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boas U., and, Heegaard PM. Dendrimers in drug research. Chem Soc Rev. 2004;33:43–63. doi: 10.1039/b309043b. [DOI] [PubMed] [Google Scholar]
- Perez AP, Romero EL., and, Morilla MJ. Ethylendiamine core PAMAM dendrimers/siRNA complexes as in vitro silencing agents. Int J Pharm. 2009;380:189–200. doi: 10.1016/j.ijpharm.2009.06.035. [DOI] [PubMed] [Google Scholar]
- Wu J, Zhou J, Qu F, Bao P, Zhang Y., and, Peng L. Polycationic dendrimers interact with RNA molecules: polyamine dendrimers inhibit the catalytic activity of Candida ribozymes. Chem Commun (Camb) 2005. pp. 313–315. [DOI] [PubMed]
- Zhou J, Wu J, Hafdi N, Behr JP, Erbacher P., and, Peng L. PAMAM dendrimers for efficient siRNA delivery and potent gene silencing. Chem Commun (Camb) 2006. pp. 2362–2364. [DOI] [PubMed]
- Liu XX, Rocchi P, Qu FQ, Zheng SQ, Liang ZC, Gleave M.et al. (2009PAMAM dendrimers mediate siRNA delivery to target Hsp27 and produce potent antiproliferative effects on prostate cancer cells ChemMedChem 41302–1310. [DOI] [PubMed] [Google Scholar]
- Shen XC, Zhou J, Liu X, Wu J, Qu F, Zhang ZL.et al. (2007Importance of size-to-charge ratio in construction of stable and uniform nanoscale RNA/dendrimer complexes Org Biomol Chem 53674–3681. [DOI] [PubMed] [Google Scholar]
- Berkhout B., and, ter Brake O. Towards a durable RNAi gene therapy for HIV-AIDS. Expert Opin Biol Ther. 2009;9:161–170. doi: 10.1517/14712590802653619. [DOI] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ.et al. (2008Identification of host proteins required for HIV infection through a functional genomic screen Science 319921–926. [DOI] [PubMed] [Google Scholar]
- Novina CD, Murray MF, Dykxhoorn DM, Beresford PJ, Riess J, Lee SK.et al. (2002siRNA-directed inhibition of HIV-1 infection Nat Med 8681–686. [DOI] [PubMed] [Google Scholar]
- Christ F, Thys W, De Rijck J, Gijsbers R, Albanese A, Arosio D.et al. (2008Transportin-SR2 imports HIV into the nucleus Curr Biol 181192–1202. [DOI] [PubMed] [Google Scholar]
- Zhou, J, Swiderski, P, Li, H, Zhang, J, Neff, CP, Akkina, R, Rossi, JJ.et al. (2009Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells Nucleic Acids Re 373094–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Behlke MA, Rose SD, Chang MS, Choi S., and, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP., and, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G., and, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Zhou J, Li H, Li S, Zaia J., and, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehandru S., and, Dandekar S. Role of the gastrointestinal tract in establishing infection in primates and humans. Curr Opin HIV AIDS. 2008;3:22–27. doi: 10.1097/COH.0b013e3282f331b0. [DOI] [PubMed] [Google Scholar]
- Paiardini M, Frank I, Pandrea I, Apetrei C., and, Silvestri G. Mucosal immune dysfunction in AIDS pathogenesis. AIDS Rev. 2008;10:36–46. [PubMed] [Google Scholar]
- Berges BK, Wheat WH, Palmer BE, Connick E., and, Akkina R. HIV-1 infection and CD4 T cell depletion in the humanized Rag2-/-gamma c-/- (RAG-hu) mouse model. Retrovirology. 2006;3:76. doi: 10.1186/1742-4690-3-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Hornung V., and, Hartmann G. siRNA and isRNA: two edges of one sword. Mol Ther. 2006;14:463–470. doi: 10.1016/j.ymthe.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Behlke MA. Progress towards in vivo use of siRNAs. Mol Ther. 2006;13:644–670. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins M, Judge A., and, MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19:89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Stable nanoscale dendrimer–dsiRNA complexes prevent dsiRNA from degradation and promote cellular uptake in vitro.
Internalization study of dendrimer–siRNA complexes.
Dendrimer-siRNA nanoparticles efficiently release siRNA and mediate specific gene silencing via the RNAi pathway in vitro.
Dendrimer-mediated dsiRNA delivery system does not trigger type I interferon responses in cultured HIV-1 infected (a) CEM cells and (b) human PBMCs.
Dendrimer–dsiRNA complexes suppress viral loads in HIV-1 infected RAG-hu mice while dendrimer alone has no affect on viral loads.
In vivo distribution and cryosections of G5 dendrimer-Cy5 siRNA complexes in a C57BL/6 mouse model.
Dendrimer-dsiRNA complexes protect RAG-hu mice from CD4+ T-cell loss while dendrimer alone gave no protection.
First-treatment monitoring of HIV-1 viral loads in individual RAG-hu mice.
Retreatment HIV-1 viral loads in individual RAG-hu mice.
First-treatment CD4 T-cell levels in individual treated and control RAG-hu mice.
HIV-1 viral loads in individual RAG-hu mice.
CD4+ T-cell levels in treated and control individual RAG-hu mice.
Histopathology analysis.