Abstract
Background
Bla g 2, one of the major cockroach allergens, induces a strong IgE response against conformational epitopes, and on reexposure, sensitized individuals often display symptoms of allergic rhinitis and asthma. The aim of the current study was to perform a test of the efficacy of a modified phage display screening, characterization of selected phages and an automated algorithm, EpiSearch, in locating an important conformational epitope.
Methods
The monoclonal antibody 7C11, which partially inhibits the binding of patient IgE antibodies to Bla g 2, was used to screen a random peptide phage library. After 3 rounds of panning, 32 phage clones were isolated and the amino acid sequences of their peptides were determined. The relative affinity and specificity of the binding of these peptides to 7C11 were tested in ELISAs. The amino acid composition of these peptides was then matched with clusters of residues on the surface of the 3-dimensional (3D) structure of Bla g 2, using our EpiSearch algorithm.
Results
The amino acid sequences of the peptides on selected phages differed at only one position, occupied by 1 of 2 negatively charged residues. The two 12-mer sequences bound to 7C11 with similar avidity and specificity. There was good concordance between the residues in the 3D clusters identified from our phage display/computational method with the co-crystal structural analysis.
Conclusion
Conformational epitopes may be mapped through screening of clones from random peptide phage display libraries and EpiSearch.
Key Words: Bla g 2, Cockroach allergen, Conformational epitope, EpiSearch, Monoclonal antibody, Random peptide phage display library
Introduction
Hypersensitivity to aeroallergens is a major cause of allergic diseases (e.g. asthma, allergic rhinitis and allergic conjunctivitis) that affect up to 30% of some populations [1]. Symptoms of rhinitis and asthma due to aeroallergens are a major cause of morbidity, lost productivity and increased health care costs [2,3]. The clinical efficacy of current pharmaco- and immunotherapy for allergic diseases is limited [4].
Previously, most IgE epitopes were thought to be dependent, at least in part, on the secondary or tertiary structures of unique regions of the allergen (conformational epitopes) [5,6,7]. The majority of IgE epitopes have been defined in terms of linear sequences of amino acids that can bind IgE in vitro. Recent findings that only about 10% of antibodies raised against intact proteins react with any peptide fragment derived from the parent protein suggest that the remaining antibodies are directed against discontinuous or conformational epitopes [6,8]. Previous studies using site-directed mutagenesis suggest that the loss of even one epitope on an allergen may be sufficient to prevent an allergen from activating basophils/mast cells [9]. Such genetically modified allergens have also been shown to bind less IgE from patient sera and to be less active in acute hypersensitivity skin tests than their wild-type counterparts [10]. However, the process of designing such mutants is currently largely empirical.
Immune complexes that induce basophil/mast cell degranulation consist of at least five molecules on the outer surface of the cells: at least two IgE molecules, usually of different antibody specificity, bound to two different α-chains of Fc∊ receptors I, and an allergen with at least two epitopes recognized by the IgE molecules. While the crystal structures of complexes containing the interacting portions of the Fc∊ receptor I and IgE molecules have been resolved [11,12], much less is known about the structural requirements for specific allergens to participate in the formation of activating complexes. The 3-dimensional (3D) structures of approximately 70 allergens have been determined by X-ray crystallography or nuclear magnetic resonance techniques, and are listed in the Structural Database of Allergenic Proteins [13], whereas more than 1,000 allergens are listed in the World Health Organization International Union of Immunological Societies Allergen Nomenclature (www.allergen.org) [14]. However, the locations of IgE-binding sites have only been established for a few allergens of known structure. Establishing the repertoire of a large number of IgE epitopes may enhance our understanding of the structural requirements for allergens to complete the pentameric complexes necessary to activate basophils and mast cells.
The most definitive approach to elucidating the structure of conformational epitope(s) of an allergen is by crystallographic analysis of allergen-antibody complexes [15,16,17,18,19,20]. This approach is not always available to investigators and can be laborious and time consuming. Another approach is to use phage display of random peptides and IgE antibodies from allergic donors, in combination with computational approaches, to determine important conformational epitopes [21,22,23]. However, the phage display approach has not been validated by comparison with the co-crystal structure in a blind test. We have recently modified the method for isolating and confirming the reactivity of phage-expressing random peptides and coupled this with a fully automated computational method, EpiSearch (http://curie.utmb.edu/episearch.html) [24]. The algorithm uses the peptide sequences obtained from the phage display experiment as input and ranks all surface-exposed patches on the antigen according to the frequency distribution of similar residues. The program allows mapping of a single peptide or a set of peptides onto the antigen structure, and the results of the calculation can be visualized on an interactive web server.
We describe here a validation of the combination of these two approaches by mapping a conformational epitope on Bla g 2, using the monoclonal antibody (mAb) 7C11 as a surrogate for the human IgE antibody. The results from these experiments were validated by comparing the individual amino acids of Bla g 2 we identified with those residues that were shown by X-ray co-crystallography to be involved in the interaction of the same mAb with Bla g 2 [19].
Materials and Methods
Biopanning of a 12-mer Phage Display Library
The Ph.D.-12-mer random peptide library (NEB, Ipswich, Mass., USA) was used in this study. This library displays 2.7 × 109 unique 12-amino acid peptides fused to the pIII minor coat protein of the M13 filamentous phage. Three successive rounds of biopannings were performed with anti-Bla g 2 mAb 7C11 [19] as the capture reagent on protein G beads (NEB), as described in the manufacturer's manual with modifications. In brief, protein G magnetic beads were washed with 0.1% Tween-Tris-buffered saline (T-TBS; 50 mM Tris-HCl, pH 7.5, 150 mM NaCl) and incubated with blocking buffer [0.1 M NaHCO3, pH 8.6, 5 mg/ml bovine serum albumin (BSA), 0.02% NaN3] for 1 h at 4°C to reduce nonspecific binding sites. In the first round of biopanning, an aliquot of 1.5 × 1011 plaque-forming units (PFUs) of phage were precleared by incubation for 20 min at room temperature with mouse IgG mAb 6G-2 of unrelated specificity [25] bound to protein G beads. Unbound phages were then incubated with 300 ng of purified mAb 7C11 for 20 min at room temperature. This mixture was incubated for 15 min at room temperature with the BSA-blocked and washed protein G beads. The phages that did not bind to the antibody-coated beads were washed away with T-TBS. The phages weakly bound to mAb 7C11 were eluted with 0.2 M glycine-HCl, pH 4.0, containing 1 mg/ml BSA. The phages that remained bound to the 7C11-coated beads were eluted with 0.2 M glycine-HCl, pH 2.2, containing 1 mg/ml BSA by rocking gently for 10 min. Eluted fractions were neutralized immediately with 1 M Tris-HCl, pH 9.1 to pH 7.0, and cultured on Luria-Bertani plates. The PFUs in the eluted fractions were titered with 1 μl of the eluate on Luria-Bertani plates containing 20 μg/ml tetracycline. The phage in the pH 2.2 eluate was amplified in Escherichia coli (E. coli) ER2738 at 37°C for 4.5 h. Three rounds of this panning procedure were carried out, applying the same PFUs of total phage in each round. The mAb concentration in the subsequent panning rounds was decreased 10-fold from that used in the previous round to select for the phage that bound to 7C11 with the highest affinity.
Amplification and Purification of the Selected Phage
Thirty-two individual plaques from the pH 2.2 elution fraction from the third round of panning were randomly picked and amplified by infecting a log-phase culture of E. coli ER2738 by shaking at 37°C for 5 h. The cultures were centrifuged and the phage-containing supernatants were collected and precipitated by incubation with 20% (wt/vol) polyethylene glycol 8,000 (Sigma, St. Louis, Mo., USA) and 2.5 M NaCl overnight, according to the NEB protocol. The supernatant was removed by centrifugation and the phage pellet was suspended in 1 ml TBS and stored at 4°C.
Nucleotide Sequencing
Phage single-strand DNA was extracted as recommended in the NEB manual. The purified phage DNA was sequenced with −96g III primer 5′-HOCCC TCA TAG TTA GCG TAA CG-3′ (NEB) by 3100 Capillary Automated DNA Sequencers (Applied Biosystems, Carlsbad, Calif., USA) at the Biomolecular Research Facility, University of Texas Medical Branch. The amino acid sequences of the peptides were deduced from the nucleotide sequence.
Phage ELISA for Assessing the Relative Affinity of Binding of Phage Clones to mAb 7C11
After 3 rounds of selection, the enrichment and specificity of the phage clones were evaluated by ELISAs. Briefly, 3 dilutions (107, 109 and 1011 in 100 μl) of phage particles in T-TBS were added to microtiter wells, coated with 10 μg/ml purified anti-Bla g 2 mAb 7C11 and incubated overnight at 4°C. The wells were washed, and bound phage were detected with horseradish peroxidase-labeled mouse anti-M13 phage antibody (Amersham, Piscataway, N.J., USA), followed by incubation with o-phenylenediamine dihydrochloride. The absorbance was determined using a SoftmaxPro 4.0 plate spectrophometer (Molecular Devices, Sunnyvale, Calif., USA). A nonspecific streptavidin-binding phage clone was used as a negative control.
Assessing the Specificity of Binding of Phage Clones to mAb 7C11
Inhibition ELISAs were performed to determine the degree to which cloned phage inhibited the binding of recombinant Bla g 2 (rBla g 2) to solid-phase mAb 7C11. Briefly, ELISA plates were coated with 10 μg/ml mAb 7C11 in 0.125 M borate buffer, pH 8.5. After washing the wells, selected phage clones bearing the peptides were added to duplicate wells at concentrations of 1 × 108–1015 PFU/ml in T-TBS and incubated overnight at 4°C. Unlabelled rBla g 2 protein was used as a positive control, and a streptavidin-specific phage was used as a negative control. Wells were washed and incubated with 20 ng/ml biotin-labeled rBla g 2 on ice. Bound rBla g 2 was detected by incubation with horseradish peroxidase-conjugated streptavidin, followed by o-phenylenediamine dihydrochloride.
Sequence Alignment
Sequences from the clones were aligned with the Bla g 2 sequence using ClustalW multiple sequence alignment.
Mapping of the Conformational Epitope
The EpiSearch [24] program was used to map the conformational epitope recognized by mAb 7C11, based on the peptide sequences from the selected phage clones. The algorithm uses the peptide sequences from the phage display experiment as input and matches these with surface-exposed patches on the 3D structure of the allergens that possess similar physical-chemical properties to those of the input peptides. EpiSearch uses 5 quantitative descriptors of 20 amino acids, as previously derived by a multidimensional scaling of 237 physical-chemical properties [26]. The 5 descriptors correlate well with the hydrophobicity, size and preferences of amino acids in α-helices, the number of degenerate triplet codons and the frequency of amino acids in β-strands. Those 5 quantitative descriptors are used to calculate the physicochemical distance between residues in the input sequences and in the surface patches, as described elsewhere [24]. The X-ray crystal structure of the Bla g 2 monomer [Protein Data Bank (PDB) ID: 1YG9] was used to define surface patches with a radius of 12 Å. Only amino acids of Bla g 2 that have a surface-exposed area ≥10 Å2 were considered constituents of these patches [27].
Comparing the Residues within the Selected Patches on Bla g 2 with those that Interact with mAb 7C11 in the Co-Crystal Structure
The amino acids within the high-scoring patches were compared with the Bla g 2 residues that interacted with the antibody in the co-crystal structure of Bla g 2 complexed with mAb 7C11 (PDB ID: 2NR6 [19]).
Statistical Analysis
Groups with statistically significant differences were identified by the Student's t test. Differences at p < 0.05 were defined as statistically significant.
Results
Enrichment of the Selected Phage from the Library
The results of ELISAs for enrichment of phage during each of the 3 consecutive rounds of panning indicated that the PFUs of phage bound to the mAb 7C11-coated beads increased approximately three-fold between the products of the first to third rounds of panning (data not shown).
Peptide Sequences
The implied amino acid sequences of the peptides displayed on 32 randomly picked clones from the third round of panning expressed 1 of the only 2 peptides selected, and these two differed from each other at only one position, as follows: peptide 1, Ser Met Met Lys Ala Asp Phe Asp Glu Glu Pro Arg, and peptide 2, Ser Met Met Lys Ala Asp Phe Glu Glu Glu Pro Arg. Twenty-four out of 32 clones expressed peptide 1 and the other 8 clones expressed peptide 2. Interestingly, the residues that varied between the two sequences were both negatively charged (underlined).
Relative Affinity and Specificity of Phage Binding to mAb 7C11
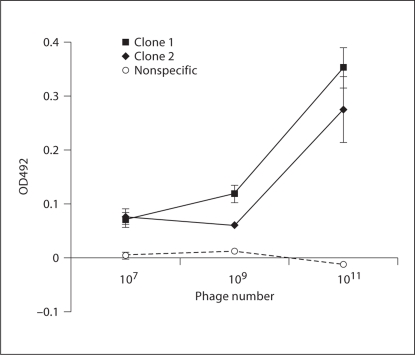
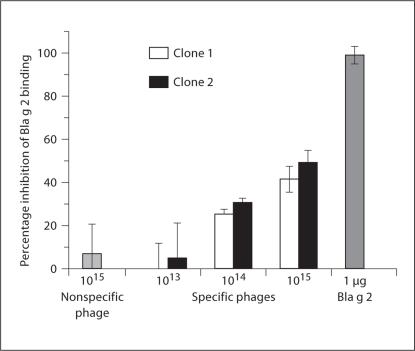
The binding of 2 phage clones to mAb 7C11 in ELISA suggested that binding of both clones required similarly high concentrations of phage, though the binding for clone 1 (from the dominant clones) was relatively stronger than that for clone 2 (fig. 1; p >0.05 for clone 1 vs. clone 2 the two high concentrations tested). The ability of both phage clones to inhibit the binding of rBla g 2 to 7C11 (fig. 2) was similar (42% inhibition for clone 1 and 49% for clone 2 at a concentration of 1 × 1015 PFU/ml; p > 0.05 for clone 1 vs. clone 2 at all concentrations tested), confirming that these two peptides bind to mAb 7C11 with similar specificity and affinities.
Fig. 1.
Specificity and affinity for phage binding to mAb 7C11. The binding of clones obtained after the third panning to mAb 7C11 was assessed by ELISA. The two clones bound to 7C11 with similar affinity, while the control phage did not bind. Mean and standard deviation of duplicated data were shown.
Fig. 2.
Phage peptide inhibition of Bla g 2 binding to mAb 7C11. The specificity of the binding of phage clones to 7C11 was confirmed by ELISA. The phage clones bearing the two different peptides were added to an mAb 7C11-coated plate. Unlabelled rBla g 2 protein and a streptavidin phage clone served as positive and negative controls, respectively. Bound phages were detected with biotin-labeled rBla g 2 and HRP-conjugated streptavidin. % inhibition = 100 × [1 – OD(preincubation)/OD(no preincubation)]. Mean and standard deviation of triplicate data were shown.
Sequence Alignment
No consensus linear sequence was observed in a ClustalW multiple sequence alignment between the peptides expressed by the selected phage and the Bla g 2 sequence.
Mapping of Peptide Sequences on the Bla g 2 Structure
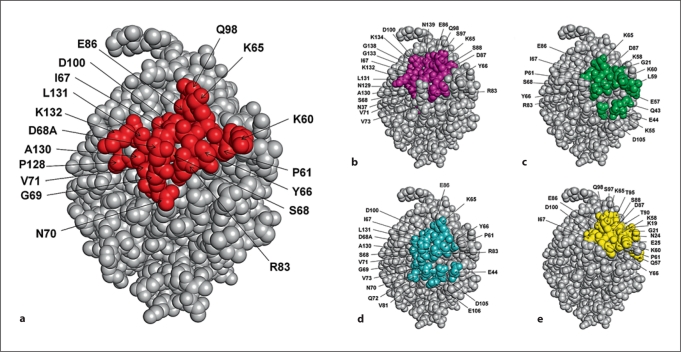
The two peptide sequences obtained from phage analysis were mapped on the crystal structure of Bla g 2 using EpiSearch. The EpiSearch program uses the 3D structure of an antigenic protein and the peptide sequences selected from a phage display experiment as input and ranks all surface-exposed patches according to the distribution of similar amino acids in the peptides and in the patch. Each patch predicted on the antigen surface represents a cluster of amino acids that have similar physical-chemical properties to those observed in the peptides expressed on the phage display clones [16]. Using default values of parameters in EpiSearch, the analysis was repeated using each of the peptide sequences individually and then both peptide sequences together as inputs. The analysis using peptide 1 identified 2 high-scoring patches centered at residue Asp100 (score = 1.0) and Asp87 (score = 0.87), respectively. Analysis with peptide 2 identified 1 high-scoring patch, centered at Pro61 (score = 1.0), and 1 low-scoring patch, centered at Arg83 (score = 0.67). The same 4 patches, centered at Asp100, Pro61, Asp87 and Arg83 (fig. 3), were also identified using the sequences of peptides 1 and 2 simultaneously. The performance of EpiSearch in mapping all 4 patches is shown in table 1. All 4 of these surface patches on Bla g 2 overlap with each other and share 4 amino acids, namely Lys65, Tyr66, Ile67 and Glu86, that were present in the genuine epitope-binding site on the Bla g 2 surface, as shown in figure 3. The residues within the overlapping regions of these 4 patches correspond to a linear ‘core’ of amino acids, while the other residues of the putative 7C11 epitope were derived from 3 discontinuous regions of the Bla g 2 sequence.
Fig. 3.
Comparison between the genuine epitope on Bla g 2 surface, obtained from X-ray crystallography, with the epitope site predicted by the EpiSearch method. (a) The amino acids in the genuine epitope on Bla g 2 surface are highlighted. The amino acids predicted in the high scoring patches centered at residue D100 (patch 1) (b), at residue P61 (patch 2) (c), at residue R83 (patch 3) (d) and at residue D87 (patch 4) (e) are shown.
Table 1.
The performance of the EpiSearch method using two peptide sequences selected with the phage display method
| Patch No. | Patch center residue | Interface residues, n | ColA1 | Overlap with peptides2 | Peptide coverage3 | ColB4 | Overlap with epitope5 | Epitope coverage6 |
|---|---|---|---|---|---|---|---|---|
| 1 | 100 | 18 | 22/12 | 0.54 | 0.66 | 22/12 | 0.54 | 0.66 |
| 2 | 61 | 18 | 17/12 | 0.70 | 0.66 | 17/8 | 0.47 | 0.44 |
| 3 | 83 | 18 | 20/11 | 0.57 | 0.61 | 20/14 | 0.70 | 0.77 |
| 4 | 87 | 18 | 19/11 | 0.57 | 0.61 | 19/8 | 0.42 | 0.44 |
The score profiles of the top 4 patches predicted in EpiSearch analysis are listed. The peptide sequences share 8 residues in common with the Bla g 2 epitope binding site. Not all amino acids in the peptide sequences selected from the phage library are present in the genuine binding site.
Total number of residues in the predicted patch/number of similar residues present in input peptides.
Number of similar residues present in input peptides/total number of residues in the predicted patch.
Number of similar residues present in input peptides/total number of residues in the epitope binding site.
Total number of residues in the predicted patch/number of correctly predicted residues in the epitope binding site.
Number of correctly predicted residues in the epitope binding site/total number of residues in the predicted patch.
Number of correctly predicted residues in the epitope binding site/total number of residues in the epitope binding site.
Comparison of the Phage Library/EpiSearch Solution with the Co-Crystallization Results
The mAb 7C11 recognizes a large conformational epitope on the surface of Bla g 2 [19] consisting of 18 surface-exposed residues (table 2). The binding of mAb 7C11 reduces the surface-exposed area on Bla g 2 by approximately 920 Å2 (table 2). A residue is considered to be part of a conformational epitope if its surface area changes by more than 10 Å2 upon formation of a complex with the antibody. The amino acids Ser68 and Asp68A, present in the linear part of the epitope, also formed contacts with the heavy chain. After antibody binding, amino acids Lys60, Lys65, Ile67, Asp68 and Arg83 lost more than 50 Å2 of their surface area, while Try66 and Lys132 lost 100 Å2 or more of their surface area (table 2).
Table 2.
Amino acids present on the Bla g 2 surface in contact with mAb 7C11
| Patch No. | Residue No. | Amino acid | Surface area in monomeric form, Å2 | Surface area upon complex formation with mAb 7C11, Å2 | Difference in surface area, Å2 |
|---|---|---|---|---|---|
| 2, 4 | 60 | K | 181.3 | 116.2 | 65.1 |
| 2, 3, 4 | 61 | P | 31.2 | 8.4 | 22.8 |
| 1, 2, 3, 4 | 65 | K | 118.0 | 32.2 | 85.8 |
| 1, 2, 3, 4 | 66 | Y | 110.0 | 9.1 | 100.9 |
| 1, 2, 3, 4 | 67 | I | 68.7 | 1.3 | 67.4 |
| 1, 2, 3 | 68 | S | 35.0 | 9.1 | 25.9 |
| 3 | 68A | D | 152.4 | 20.5 | 131.9 |
| 3 | 69 | G | 37.5 | 3.8 | 33.7 |
| 3 | 70 | N | 88.6 | 56.5 | 32.1 |
| 1, 3 | 71 | V | 32.6 | 14.8 | 17.8 |
| 1, 2, 3 | 83 | R | 85.0 | 20.6 | 64.4 |
| 1, 2, 3, 4 | 86 | E | 26.1 | 9.6 | 16.5 |
| 1, 4 | 98 | Q | 110.2 | 89.1 | 21.1 |
| 1, 3, 4 | 100 | D | 22.1 | 4.0 | 18.1 |
| 128 | P | 118.4 | 98.5 | 19.9 | |
| 1, 3 | 130 | A | 33.5 | 19.5 | 14.0 |
| 1, 3 | 131 | L | 23.2 | 4.4 | 18.8 |
| 1 | 132 | K | 183.3 | 19.3 | 164.0 |
The surface area was calculated using GetArea (http://curie.utmb.edu/getarea.html).
Discussion
IgE antibody responses to conformational epitopes often dominate the allergic response to aeroallergens [28]. Defining the structure of the epitopes from the allergens may provide insights into the structural basis of allergenicity, but current techniques often require resources that only a few laboratories possess.
The availability of the crystal structure of Bla g 2 [29] (PDB ID: 1YG9) prior to the publication of the co-crystal structure of Bla g 2 and mAb 7C11 [19] allowed us to perform a blind evaluation of our phage display/computational algorithm for identifying and mapping conformational epitopes. This made it possible to predict which residues or 3D regions within the identified epitope were most likely to be involved in the interaction with an mAb that interfered with the binding of IgE antibody.
A comparison of the amino acid composition of the two peptides expressed on the phage that bound with the highest affinity and specificity to mAb 7C11 indicated that they had 11 residues in common. No consensus linear sequence was observed in a ClustalW multiple sequence alignment between the peptides expressed by the selected phage and the Bla g 2 sequence. EpiSearch was used to find the best matches between the amino acid composition of the two peptides expressed by the selected phage and surface-exposed patches on the monomeric crystal structure of Bla g 2. The amino acid residues identified in these surface patches were compared with the Bla g 2 residues that formed the interface with mAb 7C11 in the co-crystal structure (table 1). Out of 18 residues present in the true epitope, the phage display/EpiSearch method identified 12 residues in a patch centered at Asp100, 14 residues in patch Arg83 and 8 residues in patches centered at Pro61 and Asp87, respectively. These 4 patches revealed a total of 17 out of 18 residues. All 4 patches overlapped with each other, indicating the ability of the modified phage display/EpiSearch approach to identify not only the probable location of the epitope but also specific residues of that epitope represented by individual phage peptides. While both peptides that were identified bound with a similar affinity and specificity to mAb 7C11, it is possible that human IgE antibodies will bind differentially to these peptides. This could be of value in developing mutants that reduce the allergenicity of Bla g 2.
EpiSearch has been previously validated using 6 published phage display data sets [24]. In these 6 test cases, the conformational epitopes were found within the highest-scoring patches from EpiSearch. In the present paper, we demonstrated the validity of the combined approach of collecting phage display data and performing EpiSearch calculations in practice. For other cases, it might be necessary to perform a cluster analysis of the phage display sequences before submitting the sequences to EpiSearch. For example, if the input peptide sequences have very low sequence similarities, then it is difficult to obtain a consensus sequence pattern, and EpiSearch might predict many surface patches with low score values. We recommend that EpiSearch users select only surface patches with score values of at least 0.7 in the prediction of conformational epitopes.
The experimental results from the phage display panning suggested that very few 12-mer sequences expressed in the phage library bound strongly to mAb 7C11 under the conditions that were used. The fact that the two sequences that were selected by the panning differed by 1 residue, and that these residues in both phages (Asp and Glu) were negatively charged, strongly suggests that the convergence from 109 unique sequences to only 2 was due to the stringent structural requirements for binding to mAb 7C11 in the panning protocol. This finding also indicated the success of the protocol in reducing background binding of phage to mouse IgG1, protein G and the matrix of the magnetic beads. The progressive increase in the binding of the phage from consecutive rounds of panning to mAb 7C11-coated beads also suggests a high specificity and avidity of the binding of the selected phage to this antibody.
The encouraging results we describe here may have resulted from the refinements of the phage panning protocol that selects phages which bind between pH 4 and pH 2.2 or the high degree of specificity and affinity of mAb 7C11 for a highly structured region of Bla g 2. For this study, we used a 12-mer library instead of 7-mer or cis-conformation-restricted peptides, because it provided more diversity of peptide sequences and conformations. However, the restricted clonal diversity of the selected phage may be due to the relatively low complexity of the library that we used (2.7 × 109), compared to the larger number (4.1 × 1015) of all possible peptide sequences (considering that each of the 12 positions could have any of the 20 amino acids). Nonetheless, our approach allowed us to propose an epitopic region that was subsequently proven by comparison with the co-crystal analysis.
In the absence of a larger number of dissimilar phage peptides, we found that submitting the sequences from two selected peptides individually and together to EpiSearch gave an expanded group of solutions (patches). This allowed a frequency analysis, based on the hypothesis that Bla g 2 residues that interact well with mAb 7C11 will recur within the region of intense overlap between the selected patches.
It has been suggested that conformational epitopes dominate the allergic reactions to aeroallergens, whereas linear epitopes are the major causes of allergy to food allergens [30]. A growing body of evidence supports the importance of conformational epitopes, even for class 1 food allergens [23,31,32,33]. The detection of IgE epitopes in most previous studies has been restricted by methodology defining linear sequences that can bind IgE. Systems such as EpiSearch that can rapidly identify conformational epitopes and assign them to structural regions of proteins would enhance our understanding of the structural requirements for allergenicity [24]. Using phage display/EpiSearch analysis to characterize the interactions between allergens and IgE antibodies may provide a better understanding of the structural features of allergens that are most likely to elicit IgE sensitization. An additional application of the phage display technology could be the development of Bla g 2 mimotopes based on phage peptides, which might allow the development of peptide-based diagnostics or immunotherapeutics.
Acknowledgements
We thank Monique R. Ferguson, PhD, for suggestions for modifying the panning protocol. This work was supported by grants R01AI052428 (R.M.G.), R01AI064913 (W.B.), R01AI077653 (A.P. and M.D.C.) and K08AI055792 (T.M.-H.) from the National Institute of Allergy And Infectious Diseases, ES06676 (R.M.G. and T.M.-H.) from the National Institute of Environmental Health Sciences’ Center for Environmental Science and RD833137 (W.B.) from the US Environmental Protection Agency. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
References
- 1.Akinbami LJ, Moorman JE, Liu X. Asthma prevalence, health care use, and mortality: United States, 2005–2009. Natl Health Stat Report. 2011;(32):1–14. [PubMed] [Google Scholar]
- 2.Reed SD, Lee TA, McCrory DC. The economic burden of allergic rhinitis: a critical evaluation of the literature. Pharmacoeconomics. 2004;22:345–361. doi: 10.2165/00019053-200422060-00002. [DOI] [PubMed] [Google Scholar]
- 3.Bukstein D. The Obama dilemma: allergic rhinitis (animal dander allergy) – the great burden of illness. Allergy Asthma Proc. 2009;30:567–572. doi: 10.2500/aap.2009.30.3297. [DOI] [PubMed] [Google Scholar]
- 4.Meltzer EO, Blaiss MS, Derebery MJ, Mahr TA, Gordon BR, Sheth KK, Simmons AL, Wingertzahn MA, Boyle JM. Burden of allergic rhinitis: results from the Pediatric Allergies in America survey. J Allergy Clin Immunol. 2009;124:S43–S70. doi: 10.1016/j.jaci.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Rihs HP, Chen Z, Rueff F, Petersen A, Rozynek P, Heimann H, Baur X. IgE binding of the recombinant allergen soybean profilin (rGly m 3) is mediated by conformational epitopes. J Allergy Clin Immunol. 1999;104:1293–1301. doi: 10.1016/s0091-6749(99)70027-8. [DOI] [PubMed] [Google Scholar]
- 6.Van Regenmortel MHV. Mapping epitope structure and activity: from one-dimensional prediction to four-dimensional description of antigenic specificity. Methods. 1996;9:465–472. doi: 10.1006/meth.1996.0054. [DOI] [PubMed] [Google Scholar]
- 7.Aalberse RC. Structural biology of allergens. J Allergy Clin Immunol. 2000;106:228–238. doi: 10.1067/mai.2000.108434. [DOI] [PubMed] [Google Scholar]
- 8.Barlow DJ, Edwards MS, Thornton JM. Continuous and discontinuous protein antigenic determinants. Nature. 1986;322:747–748. doi: 10.1038/322747a0. [DOI] [PubMed] [Google Scholar]
- 9.Satoh R, Koyano S, Takagi K, Nakamura R, Teshima R. Identification of an IgE-binding epitope of a major buckwheat allergen, BWp16, by SPOTs assay and mimotope screening. Int Arch Allergy Immunol. 2010;153:133–140. doi: 10.1159/000312630. [DOI] [PubMed] [Google Scholar]
- 10.Smith AM, Chapman MD, Taketomi EA, Platts-Mills TA, Sung SS. Recombinant allergens for immunotherapy: a Der p 2 variant with reduced IgE reactivity retains T-cell epitopes. J Allergy Clin Immunol. 1998;101:423–425. doi: 10.1016/S0091-6749(98)70259-3. [DOI] [PubMed] [Google Scholar]
- 11.Garman SC, Kinet JP, Jardetzky TS. Crystal structure of the human high-affinity IgE receptor. Cell. 1998;95:951–961. doi: 10.1016/s0092-8674(00)81719-5. [DOI] [PubMed] [Google Scholar]
- 12.Wan T, Beavil RL, Fabiane SM, Beavil AJ, Sohi MK, Keown M, Young RJ, Henry AJ, Owens RJ, Gould HJ, Sutton BJ. The crystal structure of IgE Fc reveals an asymmetrically bent conformation. Nat Immunol. 2002;3:681–686. doi: 10.1038/ni811. [DOI] [PubMed] [Google Scholar]
- 13.Ivanciuc O, Mathura V, Midoro-Horiuti T, Braun W, Goldblum RM, Schein CH. Detecting potential IgE-reactive sites on food proteins using a sequence and structure database, SDAP-Food. J Agric Food Chem. 2003;51:4830–4837. doi: 10.1021/jf034218r. [DOI] [PubMed] [Google Scholar]
- 14.Chapman MD, Pomés A, Breiteneder H, Ferreira F. Nomenclature and structural biology of allergens. J Allergy Clin Immunol. 2007;119:414–420. doi: 10.1016/j.jaci.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Niemi M, Jylha S, Laukkanen ML, Soderlund H, Makinen-Kiljunen S, Kallio JM, Hakulinen N, Haahtela T, Takkinen K, Rouvinen J. Molecular interactions between a recombinant IgE antibody and the beta-lactoglobulin allergen. Structure. 2007;15:1413–1421. doi: 10.1016/j.str.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 16.Padavattan S, Schirmer T, Schmidt M, Akdis C, Valenta R, Mittermann I, Soldatova L, Slater J, Mueller U, Markovic-Housley Z. Identification of a B-cell epitope of hyaluronidase, a major bee venom allergen, from its crystal structure in complex with a specific Fab. J Mol Biol. 2007;368:742–752. doi: 10.1016/j.jmb.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 17.Mirza O, Henriksen A, Ipsen H, Larsen JN, Wissenbach M, Spangfort MD, Gajhede M. Dominant epitopes and allergic cross-reactivity: complex formation between a Fab fragment of a monoclonal murine IgG antibody and the major allergen from birch pollen Bet v 1. J Immunol. 2000;165:331–338. doi: 10.4049/jimmunol.165.1.331. [DOI] [PubMed] [Google Scholar]
- 18.Padavattan S, Flicker S, Schirmer T, Madritsch C, Randow S, Reese G, Vieths S, Lupinek C, Ebner C, Valenta R, Markovic-Housley Z. High-affinity IgE recognition of a conformational epitope of the major respiratory allergen Phl p 2 as revealed by X-ray crystallography. J Immunol. 2009;182:2141–2151. doi: 10.4049/jimmunol.0803018. [DOI] [PubMed] [Google Scholar]
- 19.Li M, Gustchina A, Alexandratos J, Wlodawer A, Wunschmann S, Kepley CL, Chapman MD, Pomés A. Crystal structure of a dimerized cockroach allergen Bla g 2 complexed with a monoclonal antibody. J Biol Chem. 2008;283:22806–22814. doi: 10.1074/jbc.M800937200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M, Gustchina A, Glesner J, Wunschmann S, Vailes LD, Chapman MD, Pomés A, Wlodawer A. Carbohydrates contribute to the interactions between cockroach allergen Bla g 2 and a monoclonal antibody. J Immunol. 2011;186:333–340. doi: 10.4049/jimmunol.1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tordesillas L, Pacios LF, Palacin A, Cuesta-Herranz J, Madero M, Diaz-Perales A. Characterization of IgE epitopes of Cuc m 2, the major melon allergen, and their role in cross-reactivity with pollen profilins. Clin Exp Allergy. 2010;40:174–181. doi: 10.1111/j.1365-2222.2009.03401.x. [DOI] [PubMed] [Google Scholar]
- 22.Untersmayr E, Szalai K, Riemer AB, Hemmer W, Swoboda I, Hantusch B, Scholl I, Spitzauer S, Scheiner O, Jarisch R, Boltz-Nitulescu G, Jensen-Jarolim E. Mimotopes identify conformational epitopes on parvalbumin, the major fish allergen. Mol Immunol. 2006;43:1454–1461. doi: 10.1016/j.molimm.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 23.Pacios LF, Tordesillas L, Cuesta-Herranz J, Compes E, Sanchez-Monge R, Palacin A, Salcedo G, Diaz-Perales A. Mimotope mapping as a complementary strategy to define allergen IgE-epitopes: peach Pru p 3 allergen as a model. Mol Immunol. 2008;45:2269–2276. doi: 10.1016/j.molimm.2007.11.022. [DOI] [PubMed] [Google Scholar]
- 24.Negi SS, Braun W. Automated detection of conformational epitopes using phage display peptide sequences. Bioinform Biol Insights. 2009;3:71–81. doi: 10.4137/bbi.s2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodard CS, Splawski JB, Goldblum RM, Denney RM. Characterization of epitopes of human secretory component on free secretory component, secretory IgA, and membrane-associated secretory component. J Immunol. 1984;133:2116–2125. [PubMed] [Google Scholar]
- 26.Venkatarajan MS, Braun W. New quantitative descriptors of amino acids based on multidimensional scaling of a large number of physical-chemical properties. J Mol Model. 2001;7:445–453. [Google Scholar]
- 27.Negi SS, Schein CH, Oezguen N, Power TD, Braun W. InterProSurf: a web server for predicting interacting sites on protein surfaces. Bioinformatics. 2007;23:3397–3399. doi: 10.1093/bioinformatics/btm474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H, Jeong KY, Shin KH, Yi MH, Gantulaga D, Hong CS, Yong TS. Reactivity of German cockroach allergen, Bla g 2, peptide fragments to IgE antibodies in patients’ sera. Korean J Parasitol. 2008;46:243–246. doi: 10.3347/kjp.2008.46.4.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gustchina A, Li M, Wunschmann S, Chapman MD, Pomés A, Wlodawer A. Crystal structure of cockroach allergen Bla g 2, an unusual zinc binding aspartic protease with a novel mode of self-inhibition. J Mol Biol. 2005;348:433–444. doi: 10.1016/j.jmb.2005.02.062. [DOI] [PubMed] [Google Scholar]
- 30.Sampson HA. Update on food allergy. J Allergy Clin Immunol. 2004;113:805–819. doi: 10.1016/j.jaci.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 31.Chatchatee P, Jarvinen KM, Bardina L, Beyer K, Sampson HA. Identification of IgE- and IgG-binding epitopes on alpha(s1)-casein: differences in patients with persistent and transient cow's milk allergy. J Allergy Clin Immunol. 2001;107:379–383. doi: 10.1067/mai.2001.112372. [DOI] [PubMed] [Google Scholar]
- 32.Cooke SK, Sampson HA. Allergenic properties of ovomucoid in man. J Immunol. 1997;159:2026–2032. [PubMed] [Google Scholar]
- 33.Jarvinen KM, Beyer K, Vila L, Bardina L, Mishoe M, Sampson HA. Specificity of IgE antibodies to sequential epitopes of hen's egg ovomucoid as a marker for persistence of egg allergy. Allergy. 2007;62:758–765. doi: 10.1111/j.1398-9995.2007.01332.x. [DOI] [PubMed] [Google Scholar]