Abstract
The University of California, Santa Cruz Genome Browser (http://genome.ucsc.edu) offers online access to a database of genomic sequence and annotation data for a wide variety of organisms. The Browser also has many tools for visualizing, comparing and analyzing both publicly available and user-generated genomic data sets, aligning sequences and uploading user data. Among the features released this year are a gene search tool and annotation track drag-reorder functionality as well as support for BAM and BigWig/BigBed file formats. New display enhancements include overlay of multiple wiggle tracks through use of transparent coloring, options for displaying transformed wiggle data, a ‘mean+whiskers’ windowing function for display of wiggle data at high zoom levels, and more color schemes for microarray data. New data highlights include seven new genome assemblies, a Neandertal genome data portal, phenotype and disease association data, a human RNA editing track, and a zebrafish Conservation track. We also describe updates to existing tracks.
INTRODUCTION
The University of California, Santa Cruz (UCSC) Genome Browser provides online access to sequence and annotation data for the human genome and those of several other species (1,2). The level of annotation differs among species, with recent assemblies of the human genome being the most richly annotated. The Genome Browser contains mapping and sequencing annotation tracks describing assembly, gap and GC percent details for all assemblies. Most organisms also have tracks containing alignments of RefSeq genes (3,4), mRNAs and ESTs from GenBank (5) as well as gene and gene prediction tracks such as Ensembl Genes (6). UCSC Genes, a gene prediction track generated at UCSC that is based on data from RefSeq, GenBank, CCDS and UniProt (2,7,8), is present for the most recent human and mouse assemblies. Most organisms also have comparative genomic tracks showing pairwise genomic alignments between assemblies. Roughly half the organisms hosted in the browser have a multiple sequence alignment track (multiz) (9). Expression, regulation, variation and phenotype tracks are available for many organisms. Track descriptions can be accessed by clicking on a track item, the track title or the vertical bar to the left of the track in the image. Links to the corresponding locations on the NCBI Map Viewer (10) and Ensembl (6) genome browsers are also provided.
UCSC hosts the Data Coordination Center for the Encyclopedia of DNA Elements (ENCODE) project, using the Genome Browser website as its primary data portal (11–13). Genome-wide production phase data were initially published on the hg18 (NCBI build 36) human assembly and are currently being migrated to the hg19 (Genome Reference Consortium GRCh37) browser. (For more detail see Raney et al. in this issue.)
The Genome Browser includes many tools for visualizing and analyzing genomic data. Sequence data can be retrieved via the ‘Get DNA’ utility, the Table Browser (14) or direct download (see below). The Table Browser also serves as a tool for retrieving and exploring Genome Browser data through filtering, intersecting and correlating the underlying database tables. Output from the Table Browser can also be sent to other tools such as Galaxy (15) or GREAT (see ‘New features’ section) for subsequent analysis. The BLAT (16) and in silico PCR tools align sequences to genomes available in the browser. The LiftOver utility, available as both a web interface (at http://genome.ucsc.edu/cgi-bin/hgLiftOver) and a command-line executable program (from http://hgdownload.cse.ucsc.ed/admin/exe/) translates genomic coordinates between assemblies. The Gene Sorter (17) allows users to explore the relationships between genes by comparing expression profiles, protein homology and other useful metrics of similarity. The Proteome Browser displays protein properties and sequence data as tracks and histograms (18). VisiGene (19) is a searchable database of Xenopus and mouse in situ images showing cytology and expression patterns. Users can upload and view their data in the context of the other browser tracks using the custom tracks tool (8). Once uploaded, custom track data can be manipulated using any of the standard Genome Browser functionalities including the Table Browser. Track display configurations can also be saved and shared using the Sessions tool (8). Finally, Genome Graphs displays hosted tracks and user-generated custom tracks in the context of a genome-wide view.
Bulk downloads of sequence and annotation data and Genome Browser source code can be found at http://hgdownload.cse.ucsc.edu/. The source includes the browser bioinformatic command-line utilities (http://genomewiki.ucsc.edu/index.php/Kent_source_utilities). Instructions for setting up a mirror are available at http://genome.ucsc.edu/admin/mirror.html. Assemblies and data of interest can also be mirrored selectively (see http://genomewiki.ucsc.edu/index.php/Minimal_Browser_Installation).
NEW FEATURES
Gene search
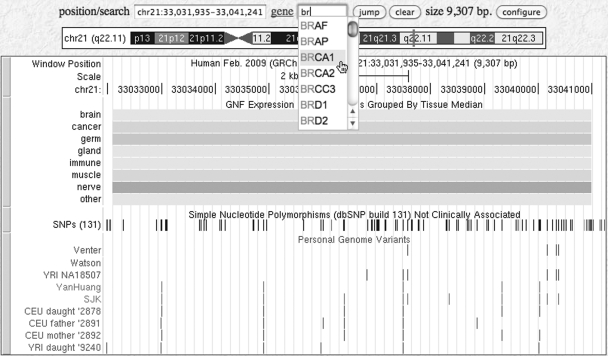
The gene search box takes a user directly to the UCSC/Known Genes or RefSeq record associated with a gene of interest, bypassing the default search of the entire database (see Figure 1). After two or more characters are entered, the search box suggests gene names, and upon selection of a particular gene, the gene’s coordinates appear in the position/search bar. In cases where the gene has several isoforms, the gene region is immediately displayed rather than requiring the user to first select a particular isoform, thus eliminating an extra navigation step.
Figure 1.
Genome Browser display on the hg19 human assembly showing the gene search box in use. After two or more characters are typed, the software suggests possible matching gene names. Tracks shown in this image, from top to bottom: base position, GNF Atlas 2 (with the red/blue on yellow background microarray color option enabled), SNPs (131) and Genome Variants (which includes five new datasets).
Drag-reorder
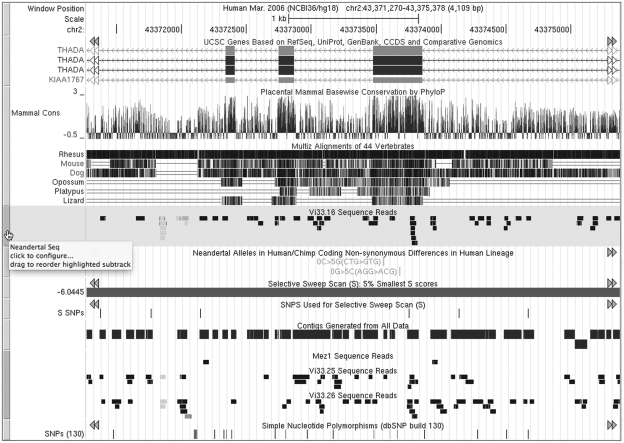
Tracks within the browser image can now be reordered more easily by clicking on the label or vertical bar to the left of the track and dragging it to a new position within the image. If the track is a member of a composite track, hovering over the bar will cause the bars of all related subtracks to turn blue, making it easier to distinguish the reordered subtracks that belong to a single composite track. Tracks can be restored to their default order via a button below the track image (see Figure 2).
Figure 2.
Genome Browser image on the hg18 human assembly showing the UCSC Genes, Conservation and Neandertal tracks (Human-Chimp coding differences, regions with the 5% lowest S, SNPs used to calculate S and alignments of Neandertal sequence reads). The Vi33.6 sequence read subtrack highlighted in green is being vertically dragged to a new position. Note that hovering the mouse over any component subtrack causes the vertical bars to the left of all related subtracks to turn blue.
BigBed/BigWig and BAM file support
In late 2009 we introduced two new file formats for very large data sets, BigBed and BigWig (20), and have continued to add support for the display of these files as built-in tracks and custom tracks. BigWig and BigBed files are compressed binary indexed files containing data at several resolutions that allow the high-performance display of next-generation sequencing experiment results in the Genome Browser. A big advantage of these file formats is that only the portions of the files needed to display a particular region are transferred to UCSC, enabling fast remote access to large distributed data sets.
We have also introduced support for the Binary Alignment/Map (BAM) file format in custom tracks and in multi-view composite tracks. BAM is the compressed binary version of the Sequence Alignment/Map (SAM) format (21), a compact and indexable representation of nucleotide sequence alignments. BAM file format employs an architecture similar to BigWig/BigBed files and thus segments of the BAM file are transmitted as needed to display the current browser view, unlike PSL and other human-readable alignments formats. This makes it possible to load very large BAM files as custom tracks in situations where the file size would preclude upload in other file formats. BAM custom tracks enable the display of high-coverage sequencing read alignments from the 1000 Genomes Project (http://www.1000genomes.org/), other sequencing projects, and the underlying data from which SNPs and CNVs were called. (See the Neandertal Sequence Reads track in Figure 2 for an example of the BAM track display.)
Display enhancements
We have augmented the Genome Browser’s wiggle and microarray track display functionalities. Log-transformed wiggle data values may now be viewed in the browser, and we have also added a new windowing function for viewing wiggle data at zoomed-out levels. When a zoom-level is too large to show individual data values, the values must be combined to produce a plot point. With the ‘mean + whiskers’ function, it is possible to simultaneously view the mean data value overlaid with measures of its central tendency. The mean appears in a dark shade, 1 standard deviation around the mean in a medium shade, and the maximum/minimum in a light shade. Another new display feature is the transparent, multicolored overlay of multiple wiggles for some tracks (for an example, see Raney et al. in this issue). Standard and custom microarray tracks can be viewed in one of five combinations of red, green, blue, and yellow by selecting a scheme on the track details page (Figure 1) or by specifying an ‘expColor’ value in the custom track’s settings (see http://genomewiki.cse.ucsc.edu/index.php/Microarray_track#Microarray_Custom_Tracks).
Table Browser support for external software
This year the Table Browser benefited from an addition that allows users to send genomic region data to the Genomic Regions Enrichment of Annotations Tool (GREAT) (22). Given a set of genomic regions, such as segments of DNA selected through ChIP-Seq experiments, GREAT analyzes the cis-regulatory patterns in these regions and assesses their functional significance. GREAT users can also create UCSC custom tracks from these term-enriched subsets of genomic regions.
NEW DATA
We constantly add new annotation tracks and update existing tracks in the Genome Browser. Tracks that were released this year as a part of the ENCODE project are described in a separate publication. (See Raney et al. in this issue for a more information.)
Neandertal data portal
In May 2010 we released a group of tracks on the hg18 human browser and the panTro2 chimpanzee browser to accompany the initial publication of the Neandertal genome (23) (see Figure 2). Both the human and chimpanzee browsers display alignments of Neandertal sequence reads and assembled contig sequences, and the human browser also offers human-chimp coding differences, a selective sweep scan (S) score, regions with the 5% lowest S score, SNPs used to calculate S score, and Neandertal mitochondrial sequence from a prior publication (24). These tracks can be viewed in the human and chimpanzee browsers or accessed through the Neandertal portal page (http://genome.ucsc.edu/Neandertal/), which also provides links to download the associated tables and data files.
Phenotype and disease association data
In the past year we released two new human phenotype and disease association tracks. The first is based on DECIPHER, a database of submicroscopic chromosomal imbalances based on clinical information about chromosomal microdeletions/duplications/insertions, translocations and inversions (25). This track shows genomic regions of reported cases and their associated phenotype information. The second track displays SNPs from the Catalog of Published Genome-Wide Association Studies (http://www.genome.gov/gwastudies), a curated and regularly updated collection of SNPs identified by published studies attempting to assay at least 100 000 SNPs (26).
Human RNA editing
We have added an RNA editing track on the human (hg18) assembly based on the DARNED database (27), a catalog of RNA sequences that are edited after transcription, along with their corresponding genomic coordinates. Only post-transcriptional editing that results in small changes to the identity of a nucleic acid are included in this track; it does not include other RNA processing such as splicing or methylation. The data were obtained from several research papers on RNA editing and were mapped to the human reference genome.
New assemblies
Since September 2009 we have updated the genome assemblies for marmoset, tetraodon, zebrafish and cat. We have also added new browsers for pig, European rabbit, giant panda, African savannah elephant and California sea hare. Each browser contains the baseline set of tracks and an additional complement of comparative genomic and other annotation tracks.
New tracks in other organisms
The chicken browser now features a track displaying the alignment of California condor (Gymnogyps californianus) transcripts (sequenced using 454 high-throughput DNA sequencing) to the galGal3 chicken genome. The condor read sequences were obtained from the NCBI Trace Archives (28). We have also released a Conservation track for the danRer6 zebrafish assembly showing multiple alignments of six vertebrate species and measurements of evolutionary conservation using phastCons from the PHAST package. Conserved elements identified by phastCons are displayed in the companion ‘Most Conserved’ track.
Updates to existing tracks and assemblies
Many Genome Browser tracks are updated regularly. The Database of Genomic Variants (DGV) (29,30) tracks, which detail genomic variants found among healthy human individuals, were updated to version 9 in the hg17 and hg18 assemblies and added to the hg19 assembly browser. The ORFeome Clones tracks, which show alignments of clones from the ORFeome Collaboration (31), were also updated for human assemblies. The human Genome Variants tracks were augmented to include Korean (SJK) (32) and 1000 Genomes high-coverage pilot individuals (NA12878, NA12891, NA12892 and NA19240) (Figure 1).
A number of annotations present on the human assembly hg18 were added to the new hg19 assembly, most notably tracks showing UCSC Genes and conservation. New in hg19 is the SNP track based on dbSNP build 131 (33) (Figure 1). The UCSC Genes track is a moderately conservative set of gene predictions based on data from RefSeq, Genbank, CCDS and UniProt. The Conservation track shows multiple alignments of 46 vertebrate species and measurements of evolutionary conservation using two methods (phastCons and phyloP) from the PHAST package for all vertebrate species, as well as primate and placental mammal subsets. The SNP track contains over 26 million mappings of more than 23 million reference SNPs that have been mapped to the reference genome by dbSNP. This represents a significant increase from the provisional hg19 mappings of build 130 (33). As we continue to migrate the bulk of our hg18 annotation tracks to the hg19 assembly, we encourage our contributors to submit hg19-based data sets for inclusion in this effort.
Tracks that are regularly updated on the mouse browsers include the International Gene Trap Consortium (IGTC) tracks (34) (updated monthly), the Mouse Genome Informatics MGI tracks (35), which show quantitative trait loci, phenotypes and alleles, and the IKMC Genes tracks (36), which show the genes targeted by the International Knockout Mouse Consortium for generating mouse embryonic stem cells containing a null mutation in every gene in the mouse genome.
Some of our regularly updated tracks appear on multiple browsers. These include the Consensus Coding Sequence (CCDS) (37) tracks, which were updated on the human and mouse genomes, the Mammalian Gene Collection (MGC) tracks (38) on the human, mouse, rat, cow and frog genomes, and the Ensembl Genes tracks (6), available on approximately 25 different organisms. RefSeq and mRNA tracks, which display aligned sequences from all organisms in GenBank (5), are updated nightly, and EST tracks are updated weekly.
FUTURE DIRECTIONS
We plan to incorporate several new features as well as exciting new variation and medical genomics data over the next year. We will also continue to add new and updated vertebrate and other selected model organism assemblies that have been deposited into GenBank. (Only assemblies registered and deposited at NCBI will be considered for hosting at UCSC, as stipulated in the Browser Genome Release Agreement instituted by NCBI, Ensembl, and UCSC.)
By late 2010 we plan to release a utility that enables users to quickly search track names and descriptions. This tool will provide both simple and advanced search interfaces, with the advanced interface allowing users to further refine their search criteria and search the metadata associated with ENCODE tracks (e.g. cell line, transcription factor, stage, etc.). Also by the end of 2010, users will be able to quickly access configuration and navigation shortcuts on the Genome Browser image by right-clicking on the vertical bar to the left of a track.
We are developing data hub support that will make it possible to view user-supplied data (such as BigWig, BigBed and BAM files) with the more sophisticated track display options currently used on other UCSC tracks such as composite tracks. We are also working on several improvements to the display of BAM files such as filtering by flag and density-wiggle view. We plan to enable data extraction from BAM file-based tracks via the Table Browser.
We anticipate adding a number of new variation tracks, including data from the 1000 Genomes project as well as from dbVar, a new structural variation database at NCBI. We are currently working on browser display support for data stored in Variant Call Format (VCF; http://1000genomes.org/wiki/doku.php?id=1000_genomes:analysis:vcf4.0), a format developed by the 1000 Genomes Project to represent variant data. Additionally we are discussing strategies for distinguishing SNPs annotated as ‘clinically associated’ by dbSNP in our SNP annotations, and looking into other stratifications of these data such as singly mapped versus multiply mapped.
We plan to import additional personal genome variant tracks from the Pennsylvania State University Bioinformatics Genome Browser (http://main.genome-browser.bx.psu.edu/), including updated 1000 Genomes high-coverage trio variants as well as variants from five Khoisan and Bantu genomes (39) and a Paleo-Eskimo Saqqaq genome (40).
We intend to add medical genomics data from the International Standards for Cytogenomic Arrays (ISCA) consortium (41). These data should help clinicians interpret array CGH results by aggregating the results of potentially thousands of cases from clinics all over the world in one place. The data will be released to dbGaP and dbVar at NCBI and then integrated into the browser for display in the context of our other content.
Finally, we plan to offer cloud support for mirrors, providing a Genome Browser image that will enable labs to instantiate a browser for private use without the overhead of a local server (42) and supporting the simple construction of new Genome Browsers on novel genome sequences.
CONTACTING US
We have two public, moderated mailing lists for user support: genome@soe.ucsc.edu for general questions about the Genome Browser, and genome-mirror@soe.ucsc.edu for questions specific to the setup and maintenance of Genome Browser mirrors. Archives of both lists are searchable from our contacts page at http://genome.ucsc.edu/contacts.html. You may also reach us at genome-www@soe.ucsc.edu, the preferred address for inquiring about mirror site licenses and reporting server errors. Messages sent to this address are not archived in a publicly searchable location.
FUNDING
Grants from the NHGRI (P41HG002371 to G.B., H.C., M.D., P.F., A.H., F.H., D.K., V.K., W.J.K., R.K., C.L., L.M, B.R. and A.Z.; U41HG004568 to M.C., T.D., M.G., F.H., W.J.K., K.L., K.R. and B.R.); NCI (U24CA143858 to F.H. and W.J.K); subcontracts from the NIDCR (U01DE20057 to G.B. and R.K.), NHGRI (P01HG5062 to G.B., W.J.K. and B.R; U54HG004555 to M.D. and R.H.; U41HG004269 to A.H. and W.J.K.; U01HG004695 to W.J.K.); NICHD (RC2HD064525 to H.C., A.H. and R.K.); NIEHS (U01ES017154 to W.J.K). Support from HHMI to D.H. Funding for open access charges: HHMI.
Conflict of interest statement. P.A.F., B.R., A.S.Z., A.S.H., D.K., G.P.B., H.C., M.D., T.R.D., B.M.G., R.A.H., F.H., V.K., R.M.K., K.L., C.H.L., L.R.M., A.P., B.J.R., K.R.R., K.E.S., D.H. and W.J.K. receive royalties from the sale of UCSC Genome Browser source code licenses to commercial entities.
ACKNOWLEDGEMENTS
The authors would like to thank the many data contributors whose work makes the Genome Browser possible, our Scientific Advisory Board for steering our efforts, our users for their consistent support and valuable feedback, and our outstanding team of system administrators: Jorge Garcia, Erich Weiler, Victoria Lin and Alex Wolfe.
REFERENCES
- 1.Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhead B., Karolchik D., Kuhn R.M., Hinrichs A.S., Zweig A.S., Fujita P.A., Diekhans M., Smith K.E., Rosenbloom K.R., Raney B.J., et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pruitt K.D., Tatusova T., Maglott D.R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pruitt K.D., Tatusova T., Klimke W., Maglott D.R. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2010;38:D46–D51. doi: 10.1093/nar/gkp1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flicek P., Aken B.L., Ballester B., Beal K., Bragin E., Brent S., Chen Y., Clapham P., Coates G., Fairley S., et al. Ensembl's 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu F., Kent W.J., Clawson H., Kuhn R.M., Diekhans M., Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 8.Karolchik D., Kuhn R., Baertsch R., Barber G., Clawson H., Diekhans M., Giardine B., Harte R., Hinrichs A., Hsu F., et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset al.gner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sayers E.W., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 12.Thomas D.J., Rosenbloom K.R., Clawson H., Hinrichs A.S., Trumbower H., Raney B.J., Karolchik D., Barber G.P., Harte R.A., Hillman-Jackson J., et al. The ENCODE Project at UC Santa Cruz. Nucleic Acids Res. 2007;35:D663–D667. doi: 10.1093/nar/gkl1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenbloom K.R., Dreszer T.R., Pheasant M., Barber G.P., Meyer L.R., Pohl A., Raney B.J., Wang T., Hinrichs A.S., Zweig A.S., et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010;38:D620–D625. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kent W.J. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent W.J., Hsu F., Karolchik D., Kuhn R.M., Clawson H., Trumbower H., Haussler D. Exploring relationships and mining data with the UCSC Gene Sorter. Genome Res. 2005;15:737–741. doi: 10.1101/gr.3694705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu F., Pringle T.H., Kuhn R.M., Karolchik D., Diekhans M., Haussler D., Kent W.J. The UCSC Proteome Browser. Nucleic Acids Res. 2005;33:D454–D458. doi: 10.1093/nar/gki100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhn R.M., Karolchik D., Zweig A.S., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., et al. The UCSC Genome Browser Database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kent W.J., Zweig A.S., Barber G., Hinrichs A.S., Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. and 1000_Genome_Project_Data_Processing_Subgroup (2009) The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green R.E., Krause J., Briggs A.W., Maricic T., Stenzel U., Kircher M., Patterson N., Li H., Zhai W., Fritz M.H., et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green R.E., Malaspinas A.S., Krause J., Briggs A.W., Johnson P.L., Uhler C., Meyer M., Good J.M., Maricic T., Stenzel U., et al. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell. 2008;134:416–426. doi: 10.1016/j.cell.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiran A., Baranov P.V. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics. 2010;26:1772–1776. doi: 10.1093/bioinformatics/btq285. [DOI] [PubMed] [Google Scholar]
- 28.Romanov M.N., Tuttle E.M., Houck M.L., Modi W.S., Chemnick L.G., Korody M.L., Mork E.M., Otten C.A., Renner T., Jones K.C., et al. The value of avian genomics to the conservation of wildlife. BMC Genomics. 2009;10(Suppl. 2):S10. doi: 10.1186/1471-2164-10-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 30.Feuk L., Carson A.R., Scherer S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 31.Lamesch P., Li N., Milstein S., Fan C., Hao T., Szabo G., Hu Z., Venkatesan K., Bethel G., Martin P., et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahn S.M., Kim T.H., Lee S., Kim D., Ghang H., Kim D.S., Kim B.C., Kim S.Y., Kim W.Y., Kim C., et al. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res. 2009;19:1622–1629. doi: 10.1101/gr.092197.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherry S., Ward M.-H., Kholodov M., Baker J., Phan L., Smigielski E., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nord A.S., Chang P., Conklin B., Cox A., Harper C., Hicks G.G., Huang C.C., Johns S.J., Kawamoto M., Liu S., et al. The International Gene Trap Consortium Website: a portal to all publicly available gene trap cell lines in mouse. Nucleic Acids Res. 2006;34:D642–D648. doi: 10.1093/nar/gkj097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bult C.J., Eppig J.T., Kadin J.A., Richardson J.E., Blake J.A. and the Mouse Genome Database Group (2008) The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The Comprehensive Knockout Mouse Project Consortium. Austin C.P., Battey J.F., Bradley A., Bucan M., Capecchi M., Collins F.S., Dove W.F., Duyk G., Dymecki S., et al. The Knockout Mouse Project. Nat. Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pruitt K.D., Harrow J., Harte R.A., Wallin C., Diekhans M., Maglott D.R., Searle S., Farrell C.M., Loveland J.E., Ruef B.J., et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;18:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerhard D.S., Wagner L., Feingold E.A., Shenmen C.M., Grouse L.H., Schuler G., Klein S.L., Old S., Rasooly R., Good P., et al. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuster S.C., Miller W., Ratan A., Tomsho L.P., Giardine B., Kasson L.R., Harris R.S., Petersen D.C., Zhao F., Qi J., et al. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–947. doi: 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rasmussen M., Li Y., Lindgreen S., Pedersen J.S., Albrechtsen A., Moltke I., Metspalu M., Metspalu E., Kivisild T., Gupta R., et al. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463:757–762. doi: 10.1038/nature08835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller D.T., Adam M.P., Aradhya S., Biesecker L.G., Brothman A.R., Carter N.P., Church D.M., Crolla J.A., Eichler E.E., Epstein C.J., et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stein L.D. The case for cloud computing in genome informatics. Genome Biol. 2010;11:207. doi: 10.1186/gb-2010-11-5-207. [DOI] [PMC free article] [PubMed] [Google Scholar]