Abstract
Pattern classification of brain imaging data can enable the automatic detection of differences in cognitive processes of specific groups of interest. Furthermore, it can also give neuroanatomical information related to the regions of the brain that are most relevant to detect these differences by means of feature selection procedures, which are also well-suited to deal with the high dimensionality of brain imaging data. This work proposes the application of recursive feature elimination using a machine learning algorithm based on composite kernels to the classification of healthy controls and patients with schizophrenia. This framework, which evaluates nonlinear relationships between voxels, analyzes whole-brain fMRI data from an auditory task experiment that is segmented into anatomical regions and recursively eliminates the uninformative ones based on their relevance estimates, thus yielding the set of most discriminative brain areas for group classification. The collected data was processed using two analysis methods: the general linear model (GLM) and independent component analysis (ICA). GLM spatial maps as well as ICA temporal lobe and default mode component maps were then input to the classifier. A mean classification accuracy of up to 95% estimated with a leave-two-out cross-validation procedure was achieved by doing multi-source data classification. In addition, it is shown that the classification accuracy rate obtained by using multi-source data surpasses that reached by using single-source data, hence showing that this algorithm takes advantage of the complimentary nature of GLM and ICA.
Keywords: fMRI, pattern classification, composite kernels, feature selection, recursive feature elimination, independent component analysis, support vector machines, schizophrenia
1. Introduction
Functional magnetic resonance imaging (fMRI) is a non-invasive technique that has been extensively used to better understand the dynamics of brain function. In order to understand the cognitive processes associated to certain activities, fMRI experimental designs usually present subjects both active and control tasks and collect several scans periodically in time from thousands of locations of the brain. One way of characterizing fMRI data is through standard statistical techniques, which fit a general linear model (GLM) to each voxel’s time series to see how correlated each of them is with the experimental task. Such methods emphasize task-related activity in each voxel separately. Another way of analyzing fMRI data is to use data-driven methods such as independent component analysis (ICA) that search for functional connectivity in the brain, i.e., they detect different components of voxels that have temporally coherent neural activity. GLM and ICA approaches are complementary to each other. For this reason, it would be sensible to devise a method that could gain more insight of the underlying processes of brain activity by combining data from both approaches. Pattern recognition techniques have been applied successfully to fMRI to detect different subject conditions. In this work, a pattern recognition system that combines GLM and ICA data to better characterize a subject’s condition is presented.
ICA has been extensively applied to fMRI data to identify differences among healthy controls and schizophrenia patients (Kim et al., 2008; Demirci et al., 2009; Calhoun et al., 2006). Thus, Calhoun et al. (2008) showed that the temporal lobe and the default mode components (networks) could reliably be used together to identify patients with bipolar disorder and schizophrenia from each other and from healthy controls. Furthermore, Garrity et al. (2007) demonstrated that the default mode component showed abnormal activation and connectivity patterns in schizophrenia patients. Therefore, there is evidence that suggest that the default mode and temporal lobe components are disturbed in schizophrenia. Based on the reported importance of the temporal lobe in the characterization of schizophrenia we used data from an auditory oddball discrimination (AOD) task, which provides a consistent activation of this part of the brain. Three sources were extracted from fMRI data using two analysis methods: model-based information via the GLM and functional connectivity information retrieved by ICA. The first source is a set of β-maps generated by the GLM. The other two sources come from an ICA analysis and include a temporal lobe component and a default mode network component.
Several works have applied pattern recognition to fMRI data for schizophrenia detection. Ford et al. (2003) projected fMRI statistical spatial maps to a lower dimensional space using principal component analysis (PCA) and then applied Fisher’s linear discriminant to differentiate between controls and patients with schizophrenia, Alzheimer’s disease and mild traumatic brain injury. On another approach, Shinkareva et al. (2006) used whole brain fMRI time series and identified voxels which had highly dissimilar time courses among groups employing the RV-coefficient. Once those voxels were detected, their fMRI time series data were used for subject classification. Finally, Demirci et al. (2008) applied a projection pursuit algorithm to reduce the dimensionality of fMRI data acquired during an AOD task and to classify schizophrenia patients from healthy controls. There have been a number of papers published on the topic of pattern recognition applied to fMRI which are not related to schizophrenia characterization. D.D. Cox and R.L. Savoy (2003) applied linear discriminant analysis and a linear support vector machine (SVM) to classify among 10-class visual patterns; LaConte et al. (2003, 2005) presented a linear SVM for left and right motor activation; Wang et al. (2004) used an SVM to distinguish between brain cognitive states; Kamitani and Tong 2005) and (Haynes and Rees (2005) detected different visual stimuli; Martínez-Ramón et al. (2006a) introduced an approach which combined SVMs and boosting for 4-class interleaved classification; more recently, Bayesian networks have been used to detect between various brain states (Friston et al., 2008); in addition, a review of pattern recognition works for fMRI was presented by Decharms (2007). All these papers used kernel-based learning methods as base classifiers.
One of the main difficulties of using pattern recognition in fMRI is that each collected volume contains tens of thousands of voxels, i.e., the dimensionality of each volume is very high when compared with the number of volumes collected in an experiment, whose order of magnitude is in the order of tens or hundreds of images. The huge difference between the data dimensionality and the number of available observations affects the generalization performance of the estimator (classifier or regression machine) or even precludes its use due to the low average information per dimension present in the data. Thus, it is desirable to reduce the data dimensionality with an algorithm that loses the least amount of information possible with an affordable computational burden.
Two approaches to solve this problem are feature extraction and feature selection. Feature extraction projects the data in high-dimensional space to a space of fewer dimensions. PCA is the most representative method of feature extraction and was used by Mourão-Miranda et al. (2005) for whole-brain classification of fMRI attention experiments. The second approach is feature selection, which determines a subset of features that optimizes the performance of the classifier. The latter approach is suitable for fMRI under the assumption that information in the brain is sparse, i.e., informative brain activity is concentrated in a few areas, making the rest of them irrelevant for the classification task. In addition, feature selection can improve the prediction performance of a classifier as well as provide a better understanding of the underlying process that generated the data. Feature selection methods can be divided into three categories: filters, wrappers and embedded methods (Guyon and Elisseeff, 2003). Filters select a subset of features as a preprocessing step to classification. On the other hand, wrappers and embedded methods use the classifier itself to find the optimal feature set. The difference between them is that while wrappers make use of the learning machine to select the feature set that increases its prediction accuracy, embedded methods incorporate feature selection as part of the training phase of the learning machine. The work presented in Mourão-Miranda et al. (2006) is an example of a filter approach; in this paper temporal compression and space selection were applied to fMRI data on a visual experiment. Haynes and Rees (2005) also applied filter feature selection by selecting the top 100 voxels that had the strongest activation in two different visual stimuli. The aforementioned methods apply univariate strategies to perform variable selection, thus not accounting for the (potentially nonlinear) multivariate relationships between voxels. De Martino et al. (2008) used a hybrid filter/wrapper approach by applying univariate voxel selection strategies prior to using recursive feature elimination SVM (RFE-SVM) (Guyon et al., 2002) on both simulated and real data. Despite its robustness, RFE-SVM is a computational intensive method since it has been designed to eliminate features one by one at each iteration, requiring the SVM to be retrained M times, where M is the data dimensionality. While it is possible to remove several features at a time, this could come at the expense of classification performance degradation (Guyon et al., 2002). Moreover, this would add an extra parameter to be tuned, which would be the fraction of features to be eliminated at each iteration that degrades the classification accuracy the least. An alternative approach is the use of embedded feature selection methods such as the one presented by Ryali et al. (2010), which has a smaller execution time since it does not require to be repeatedly retrained. The disadvantage of this method relies on the fact that it achieves just average classification accuracy when applied to real fMRI data. Multivariate, nonlinear feature selection is computationally intensive, so usually only linear methods are applied to do feature selection in fMRI due to its high dimensionality. Thus, models assume that there is an intrinsic linear relationship between voxels. In fact, all of the previously cited feature selection methods make use of linear methods. Models that assume nonlinear relationships between voxels may lead to an unaffordable computational burden. A convenient tradeoff consists on assuming that there are nonlinear relationships between voxels that are close to each other and that are part of the same anatomical brain region, and that voxels in different brain regions are linearly related. This region-based approach resembles the spherical multivariate searchlight technique (Kriegeskorte et al., 2006), which moves a sphere through the brain image and measures how well the multivariate signal in the local spherical neighborhood differentiates experimental conditions. However, our approach works with fixed regions and assumes that long range interactions between these are linear. Another characteristic shared by feature selection methods applied to fMRI is that they focus on performing voxel-wise feature selection. We propose a nonlinear method based on composite kernels that achieves a reasonable classification rate in real fMRI data, specifically in the differentiation of groups of healthy controls and schizophrenia patients. In this approach, RFE is implemented by performing a ranking of anatomically defined brain regions instead of doing it for voxels. By doing so we not only reduce the number of iterations of our approach and thus its execution time compared to other RFE-based approaches such as RFE-SVM, but we are also capable of reporting the relevance of those brain regions in detecting group differences. The measurement of the relevance of each region indicates the magnitude of differential activity between groups of interest. The proposed methodology also presents two important advantages. Firstly, it allows the use of a nonlinear kernel within a RFE procedure in a reasonable computational time, which cannot be achieved by using conventional SVM implementations. Secondly, the detection of the most relevant brain regions for a given task is developed by including all of the voxels present in the brain, without the need to apply data compression in these regions. Moreover, such an approach can lead to a more robust understanding of cognitive processes compared to voxel-wise analyses since reporting the relevance of anatomical brain areas is potentially more meaningful than reporting the relevance of isolated voxels.
Composite kernels were first applied to multiple kernel learning methods that were intended to iteratively select the best among various kernels applied to the same data through the optimization of a linear combination of them (Bach and Lanckriet, 2004; Sonnenburg et al., 2006). Composite kernels can also be generated by applying kernels to different subspaces of the data input space (segments) that are linearly recombined in a higher dimensional space, thus assuming a linear relationship between segments. Such an approach was followed by Martínez-Ramón et al. (2006b) and Camps-Valls et al. (2008). As a result, the data from each segment is analyzed separately, permitting an independent analysis of the relevance of each of the segments in the classification task. Specifically, in this work a segment represents an anatomical brain region while activity levels in voxels are the features. Composite kernels can be used to estimate the relevance of each area by computing the squared norm of the weight vector projection onto the subspace given by each kernel. Therefore, RFE can be applied to this nonlinear kernel-based method to discard uninformative regions. The advantage of this approach, which is referred to as recursive composite kernels (RCK), is based on the fact that it does not need to use a set of regions of interest (ROIs) to run the classification algorithm; instead, it can take whole-brain data segmented into anatomical brain regions and by applying RFE, it can automatically detect the regions which are the most relevant ones for the classification task. In the present approach we hypothesized that nonlinear relationships exist between voxels in an anatomical brain region and that relationships between brain regions are linear, even between regions from different sources. This specific set of assumptions is used to balance computational complexity and also incorporate nonlinear relationships.
Once the sources are extracted, volumes from both the GLM and ICA sources are segmented into anatomical regions. Each of these areas is mapped into a different space using composite kernels. Then, a single classifier (an SVM) is used to detect controls and patients. By analyzing the classifier parameters related to each area separately, composite kernels are able to assess their relevance in the classification task. Hence, RFE is applied to composite kernels to remove uninformative areas, discarding the least informative region at each iteration. An optimal set of regions is obtained by the proposed approach and it is composed by those regions that yield the best validated performance across the iterations of the recursive analysis. In all cases, the performance of the classifier is estimated using a leave-two-out cross-validation procedure, using the left out (test) observations only to assess the classifier accuracy rate and not including them for training purposes. The same applies to model selection, such as parameter tuning and the criteria to select the most relevant regions for classification purposes.
2. Materials and Methods
2.1. Participants
Data were collected at the Olin Neuropsychiatric Research Center (Hart-ford, CT) from healthy controls and patients with schizophrenia. All subjects gave written, informed, Hartford hospital IRB approved consent. Schizophrenia was diagnosed according to DSM-IV-TR criteria (American Psychiatric Association, 2000) on the basis of both a structured clinical interview (SCID) (First et al., 1995) administered by a research nurse and the review of the medical file. All patients were on stable medication prior to the scan session. Healthy participants were screened to ensure they were free from DSM-IV Axis I or Axis II psychopathology using the SCID for non-patients (Spitzer et al., 1996) and were also interviewed to determine that there was no history of psychosis in any first-degree relatives. All participants had normal hearing, and were able to perform the AOD task (see Section 2.2) successfully during practice prior to the scanning session.
Data from 106 right-handed subjects were used, 54 controls aged 17 to 82 years (mean=37.1, SD=16.0) and 52 patients aged 19 to 59 years (mean=36.7, SD=12.0). A two-sample t-test on age yielded t = 0.13 (p = 0.90). There were 29 male controls (M:F ratio=1.16) and 32 male patients (M:F ratio=1.60). A Pearson’s chi-square test yielded χ2 = 0.67 (p = 0.41).
2.2. Experimental Design
The AOD task involved subjects that were presented with three frequencies of sounds: target (1200 Hz with probability, p = 0.09), novel (computer generated complex tones, p = 0.09), and standard (1000 Hz, p = 0.82) presented through a computer system via sound insulated, MR-compatible earphones. Stimuli were presented sequentially in pseudorandom order for 200 ms each with inter-stimulus interval varying randomly from 500 to 2050 ms. Subjects were asked to make a quick button-press response with their right index finger upon each presentation of each target stimulus; no response was required for the other two stimuli. There were two runs, each comprising 90 stimuli (3.2 minutes) (Kiehl and Liddle, 2001).
2.3. Image Acquisition
Scans were acquired at the Institute of Living, Hartford, CT on a 3T dedicated head scanner (Siemens Allegra) equipped with 40mT/m gradients and a standard quadrature head coil. The functional scans were acquired using gradient-echo echo planar imaging (EPI) with the following parameters: repeat time (TR) = 1.5 sec, echo time (TE) = 27 ms, field of view = 24 cm, acquisition matrix = 64 × 64, flip angle = 70°, voxel size = 3.75 × 3.75 × 4 mm3, slice thickness = 4 mm, gap = 1 mm, number of slices = 29; ascending acquisition. Six dummy scans were carried out at the beginning to allow for longitudinal equilibrium, after which the paradigm was automatically triggered to start by the scanner.
2.4. Preprocessing
fMRI data were preprocessed using the SPM5 software package (http://www.fil.ion.ucl.ac.uk/spm/software/spm5/). Images were realigned using INRIalign, a motion correction algorithm unbiased by local signal changes (Freire et al., 2002). Data were spatially normalized into the standard Montreal Neurological Institute (MNI) space (Friston et al., 1995), spatially smoothed with a 9×9×9–mm3 full width at half-maximum Gaussian kernel. The data (originally acquired at 3.75 × 3.75 × 4 mm3) were slightly upsampled to 3 × 3 × 3 mm3, resulting in 53 × 63 × 46 voxels.
2.5. Creation of Spatial Maps
The GLM analysis performs a univariate multiple regression of each voxel’s timecourse with an experimental design matrix, which is generated by doing the convolution of pulse train functions (built based on the task onset times of the fMRI experiment) with the hemodynamic response function (Friston et al., 2000). This results in a set of β-weight maps (or β-maps) associated with each parametric regressor. The β-maps associated with the target versus standard contrast were used in our analysis. The final target versus standard contrast images were averaged over two runs.
In addition, group spatial ICA (Calhoun et al., 2001) was used to decompose all the data into 20 components using the GIFT software (http://icatb.sourceforge.net/) as follows. Dimension estimation, which was used to determine the number of components, was performed using the minimum description length criteria, modified to account for spatial correlation (Li et al., 2007). Data from all subjects were then concatenated and this aggregate data set reduced to 20 temporal dimensions using PCA, followed by an independent component estimation using the infomax algorithm (Bell and Sejnowski, 1995). Individual subject components were back-reconstructed from the group ICA analysis to generate their associated spatial maps (ICA maps). Component maps from the two runs were averaged together resulting in a single spatial map of each ICA component for each subject. It is important to mention that this averaging was performed after the spatial ICA components were estimated. The two components of interest (temporal lobe and default mode) were identified in a fully automated manner using different approaches. The temporal lobe component was detected by temporally sorting the components in GIFT based on their similarity with the SPM design regressors and retrieving the component whose ICA timecourse had the best fit (Kim et al., 2009). By contrast, the default mode network was identified by spatially sorting the components in GIFT using a mask derived from the Wake Forest University pick atlas (WFU-PickAtlas) (Lancaster et al., 1997, 2000; Maldjian et al., 2003), (http://www.fmri.wfubmc.edu/download.htm). For the default mode mask we used precuneus, posterior cingulate, and Brodmann areas 7, 10, and 39 (Correa et al., 2007; Franco et al., 2009). A spatial multiple regression of this mask with each of the networks was performed, and the network which had the best fit was automatically selected as the default mode component.
2.6. Data Segmentation and Normalization
The spatial maps obtained from the three available sources were segmented into 116 regions according to the automated anatomical labeling (AAL) brain parcellation (Tzourio-Mazoyer et al., 2002) using the WFU-PickAtlas. In addition, the spatial maps were normalized by subtracting from each voxel its mean value across subjects and dividing it by its standard deviation. Multiple kernel learning methods such as composite kernels and RCK further required each kernel matrix to be scaled such that the variance of the training vectors in its associated feature space were equal to 1. This procedure is explained in more detail in the next section.
2.7. Composite Kernels Method
2.7.1. Structure of the learning machine based on composite kernels
Each area from observation i is placed in a vector xi;l where i, 1 ≤ i ≤ N is the observation index and l, 1 ≤ l ≤ L is the area index. An observation is defined as either a single-source spatial map or the combination of multiple sources spatial maps of a specific subject. In the particular case of our study N = 106. For single-source analysis, composite kernels map each observation i into L = 116 vectors xi;l; for two-source analysis, composite kernels map each observation into L = 2 × 116 = 232 vectors xi;l, and so on. Then, each vector is mapped through a nonlinear transformation ϕl(·). These transformations produce vectors in a higher (usually infinite) dimension Hilbert space
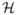 provided with a kernel inner product < ϕl(xi;l), ϕl(xj;l) > = kl(xi;l, xj;l), where < · > is the inner product operator and kl(·, ·) is a Mercer’s kernel. In this work, kernels kl(·, ·) are defined to be Gaussian kernels with the same parameter σ (see Appendix 1 for details about kernels).
provided with a kernel inner product < ϕl(xi;l), ϕl(xj;l) > = kl(xi;l, xj;l), where < · > is the inner product operator and kl(·, ·) is a Mercer’s kernel. In this work, kernels kl(·, ·) are defined to be Gaussian kernels with the same parameter σ (see Appendix 1 for details about kernels).
When the kernel function kl(·, ·) is applied to the training vectors in the dataset, matrix Kl is generated. Component i, j of this matrix is computed as Kl(i, j) = kl(xi;l, xj;l). In order for training vectors transformed by ϕl(·) to have unit variance in this Hilbert space, its matrix kernel is applied the following transformation (Kloft et al., 2011)
| (1) |
where the denominator of Eq.1 is the variance of the observations in the feature space.
All areas of the observation (example) can be stacked in a single vector
| (2) |
where T is the transpose operator.
The output of the learning machine can be expressed (see Appendix 2) as a sum of learning machines
| (3) |
where wl is the vector of parameters of the learning machine inside each Hilbert space and x* is a given test pattern.
Assuming that the set of parameters is a linear combination of the data, the classifier can be expressed as
| (4) |
where αi are the machine parameters that have to be optimized using a simple least squares approach or SVMs. In this work, SVMs are used by means of the LIBSVM software package (Chang and Lin, 2001) (http://www.csie.ntu.edu.tw/~cjlin/libsvm). Note that the output is a linear combination of kernels, which is called composite kernel. This specific kind of composite kernel is called summation kernel (see Appendix 2).
2.7.2. Brain areas discriminative weights estimation
As it is explained in Appendix 2, if a given area l contains information relevant for the classification, its corresponding set of parameters wl will have a high quadratic norm; otherwise the norm will be low. Usually vectors wl are not accessible, but their quadratic norms can be computed using the equation
| (5) |
where Kl is a matrix containing the kernel inner products between training vectors corresponding to area l. For each of the sources, a map can be drawn in which each of their correspondent brain areas l is colored proportionally to ||wl||2. These coefficients will be referred to as discriminative weights.
2.7.3. Recursive algorithm
Once the data from each observation is split into different areas, each of them is mapped to high dimensional spaces by means of composite kernels, as it has been explained in Section 2.7.1. Since composite kernels are capable of estimating the discriminative weights of each of these areas, RFE procedures can be applied to them; the application of RFE to composite kernels yields the RCK algorithm. This recursive algorithm trains an SVM with the training set of observations and estimates the discriminative weights from all the areas at its first iteration, after which it removes the area with smallest associated weight from the analyzed area set (backward elimination). At the next iteration, the SVM is trained with the data from all the areas but the previously removed one and their discriminative weights are recalculated, eliminating the area with current minimum weight. This procedure is applied repeatedly until a single area remains in the analyzed area set, with the optimal area set being the one that achieved the best validation accuracy rate across the iterations of the recursive algorithm.
2.7.4. Parameter selection, optimal area set selection and prediction accuracy estimation
The recursive algorithm presented in Section 2.7.3 is run for both single-source and multi-source data. There are two parameters that need to be tuned in order to achieve the best performance of the learning machine. These parameters are the SVM error penalty parameter C (Burges, 1998) and the Gaussian kernel parameter σ. Based on preliminary experimentation, it was discovered that the problem under study was rather insensitive to the value of C, so it was fixed to C = 100. In order to select σ, a set of 10 logarithmically spaced values between 1 and 100 were provided to the classifier.
The validation procedure consists of finding the optimal parameter pair {σ, Iareas}, where Iareas specifies a subset of the areas indexes. If a brute-force approach were used, then the validation errors obtained for all possible values of σ and all combinations of areas would need to be calculated.
The previously mentioned approach is computationally intensive. For this reason, we propose a recursive algorithm based on the calculation of discriminative weights (please refer to previous sections). Based on this method, a grid search can be performed by calculating the validation error and the training discriminative weights for each value of σ and each remaining subset of areas at each iteration of the recursive algorithm. The algorithm starts with all brain regions, calculate the discriminative weights for each value of σ and eliminates at each iteration the regions with least discriminative weight in the area sets associated to each σ value. After executing the whole grid search, the pair {σ, Iareas} that yielded the minimum validation error rate would be selected.
The aforementioned method can be further simplified by calculating only the training discriminative weights associated to the optimal value of σ at each iteration of the recursive algorithm. This procedure is suboptimal compared to the previous one, but it reduces its computational time. The following paragraphs provide more details of the previously discussed validation procedure and the test accuracy rate calculation.
First of all, a pair of observations (one from a patient and one from a control) is set aside to be used for test purposes and not included in the validation procedure. The remaining data, which is called TrainValSet in algorithm 1, is further divided into training and validation sets, the latter one being composed by another control/patient pair of observations, as shown in algorithm 2.
Algorithm 1.
Estimate optimal sigma and optimal area set
| 1: | Inputs: TrainV alSet |
| 2: | Outputs: SigmaOpt, Iopt |
| 3: | Define I(1): indexes for all areas |
| 4: | Define P: number of areas |
| 5: | for p = 1 to P − 1 do |
| 6: | Validate sigma, one RCK iteration (TrainV alSet, I(p)) ⇒ Sigma(p) and E(p) |
| 7: | Train with TrainV alSet, Sigma(p) and I(p) |
| 8: | Compute discriminative weights |
| 9: | Remove area with lowest weight |
| 10: | Store indexes of remaining areas ⇒ I (p + 1) |
| 11: | end for |
| 12: | Find p that minimizes E(p) ⇒ pmin |
| 13: | Sigma(pmin) ⇒ SigmaOpt, I (pmin) ⇒ I opt |
Algorithm 2.
Validate sigma, one RCK iteration
| 1: | Inputs: TrainV alSet and I(p) |
| 2: | Outputs: Sigma(p) and E(p) |
| 3: | Define N: number of subject pairs in TrainV alSet |
| 4: | Define L: Number of possible values for sigma |
| 5: | for j = 1 to N do |
| 6: | Extract Train(j) from TrainV alSet |
| 7: | Extract Val(j) from TrainV alSet |
| 8: | for k = 1 to L do |
| 9: | Train with Train(j), sigma(k) and I(p) ⇒ SV Mparameters |
| 10: | Test with Val(j), sigma(k), I(p) and SV Mparameters |
| 11: | Store error ⇒ e(j, k) |
| 12: | end for |
| 13: | end for |
| 14: | Average e(j, k) over j ⇒ e(k) |
| 15: | Find k that minimizes e(k) ⇒ E(p) |
| 16: | sigma(k) ⇒ Sigma(p) |
The classifier is trained by using all the brain regions and all possible σ values and the validation error rates are estimated as shown in Algorithms 1 and 2. The above process is repeated for all control/patient pairs. Next, the value of σ that yields the minimum validation error is selected and this error is stored. Then, the algorithm is retrained with this value of σ and the discriminative weights are estimated, eliminating the area with minimum associated value. This procedure is then repeated until a single brain region remains.
Afterwards, the pair {σ, Iareas} that achieves minimum validation error is selected. Then, another control/patient pair is selected as the new test set and the entire procedure is repeated for each of these test set pairs.
In the next step, the areas selection frequency scores across TrainValSet datasets are estimated by using the information in their associated Iareas parameters. The ones that achieve a score higher than or equal to 0.5 define the overall optimal area set.
The test error rate is then estimated by training a model with each TrainValSet dataset with the previously defined area set and the optimal value of σ associated to each of them and testing it using the reserved test set. Finally, the test accuracy rate is estimated by averaging the accuracy rates achieved by each test set.
2.7.5. Comparison of composite kernels and RCK with other methods
The composite kernels algorithm allows the analysis of non-linear relationships between voxels within a brain region and captures linear relationships between those regions. We compare the performance of the proposed algorithm for single-source and multi-source analyses with both a linear SVM, which assumes linear relationships between voxels, and a Gaussian SVM, which analyzes all possible non-linear relationships between voxels. The data from each area, which is extracted by the segmentation process (please refer to Section 2.6), is input to the aforementioned conventional kernel-based methods after been concatenated.
Besides analyzing the classification accuracy rate obtained by our proposed feature selection approach (RCK) compared to the previously mentioned algorithms, we are interested in evaluating the performance of RCK by comparing it against another RFE-based procedure: RFE-SVM applied to linear SVMs (which will be hereafter referred to as RFE-SVM).
Parameter selection for the aforementioned algorithms is performed as follows. As stated before, the problem under study is rather insensitive to the value of C. Therefore, its value is fixed to 100 for linear SVM, Gaussian SVM and RFE-SVM. In addition, the Gaussian kernel parameter σ values are retrieved from a set of 100 logarithmically spaced values between 1 and 1000.
3. Results
3.1. RCK Applied to Single Sources
This section presents the sets of most relevant areas and the test results of RCK applied to each source.
The mean test accuracy achieved by using ICA default-mode component data is 90%. The list of overall 40 brain regions that were selected by RCK for the ICA default mode component data are listed in Table 1, alongside the statistics of their discriminative weights. These regions are grouped in macro regions to better identify their location in the brain. Furthermore, the rate of training sets that selected each region (selection frequency) is also specified.
Table 1.
Optimal area set and associated discriminative weights for RCK analysis applied to ICA default mode data. The most informative anatomical regions retrieved by RCK when applied to ICA default mode data are grouped in macro brain regions to give a better idea of their location in the brain. The mean and the standard deviation of the discriminative weights of each area are listed in this table. In addition the rate of training sets in the cross-validation procedure that selected each area (selection frequency) is also reported in order to measure the validity of the inclusion of each region in the optimal area set.
| Source | Areas and Discriminative Weights
|
||||
|---|---|---|---|---|---|
| Macro Regions | Regions | Discriminative Weights
|
|||
| Mean | Std. Dev. | Sel. Freq. | |||
| ICA default mode | Central Region | Right Precentral Gyrus | 2.32 | 0.06 | 1.00 |
| Left Precentral Gyrus | 2.31 | 0.04 | 1.00 | ||
| Left Postcentral Gyrus | 2.22 | 0.03 | 1.00 | ||
| Right Postcentral Gyrus | 2.21 | 0.02 | 1.00 | ||
|
| |||||
| Frontal lobe | Right Paracentral Lobule | 3.44 | 0.16 | 1.00 | |
| Left Superior Frontal Gyrus, Medial | 2.97 | 0.15 | 1.00 | ||
| Left Middle Frontal Gyrus, Orbital Part 1 | 2.52 | 0.15 | 1.00 | ||
| Right Superior Frontal Gyrus, Medial | 2.51 | 0.10 | 1.00 | ||
| Left Superior Frontal Gyrus | 2.28 | 0.09 | 1.00 | ||
| Right Superior Frontal Gyrus | 2.27 | 0.06 | 1.00 | ||
| Left Inferior Frontal Gyrus, Triangular Part | 2.24 | 0.04 | 1.00 | ||
| Right Middle Frontal Gyrus | 2.21 | 0.04 | 0.94 | ||
| Right Inferior Frontal Gyrus, Opercular Part | 2.19 | 0.08 | 0.79 | ||
| Left Inferior Frontal Gyrus, Orbital Part | 2.16 | 0.08 | 0.55 | ||
| Right Gyrus Rectus | 2.38 | 0.21 | 0.94 | ||
|
| |||||
| Temporal lobe | Left Middle Temporal Gyrus | 2.27 | 0.03 | 1.00 | |
| Right Middle Temporal Gyrus | 2.22 | 0.05 | 1.00 | ||
|
| |||||
| Parietal lobe | Left Angular Gyrus | 2.72 | 0.11 | 1.00 | |
| Left Supramarginal Gyrus | 2.45 | 0.11 | 1.00 | ||
| Right Cuneus | 2.72 | 0.08 | 1.00 | ||
| Right Superior Parietal Gyrus | 2.31 | 0.06 | 1.00 | ||
| Left Superior Parietal Gyrus | 2.25 | 0.08 | 0.96 | ||
|
| |||||
| Occipital lobe | Right Superior Occipital Gyrus | 2.94 | 0.13 | 1.00 | |
| Left Superior Occipital Gyrus | 2.88 | 0.09 | 1.00 | ||
| Left Middle Occipital Gyrus | 2.58 | 0.07 | 1.00 | ||
| Right Inferior Occipital Gyrus | 2.50 | 0.14 | 1.00 | ||
| Left Cuneus | 2.38 | 0.07 | 1.00 | ||
| Left Fusiform Gyrus | 2.31 | 0.05 | 1.00 | ||
|
| |||||
| Limbic lobe | Left Anterior Cingulate Gyrus | 3.33 | 0.10 | 1.00 | |
| Right Anterior Cingulate Gyrus | 2.71 | 0.09 | 1.00 | ||
| Right Middle Cingulate Gyrus | 2.46 | 0.06 | 1.00 | ||
| Left Middle Cingulate Gyrus | 2.41 | 0.06 | 1.00 | ||
| Left Temporal Pole: Middle Temporal Gyrus | 2.40 | 0.13 | 1.00 | ||
| Right Temporal Pole: Superior Temporal Gyrus | 2.36 | 0.10 | 0.96 | ||
| Left Parahippocampal Gyrus | 2.27 | 0.11 | 0.87 | ||
|
| |||||
| Insula | Right Insular Cortex | 2.25 | 0.07 | 0.98 | |
|
| |||||
| Sub cortical gray cortex | Left Thalamus | 2.53 | 0.12 | 1.00 | |
|
| |||||
| Cerebellum | Right Inferior Posterior Lobe of Cerebellum | 3.83 | 0.19 | 1.00 | |
| Left Anterior Lobe of Cerebellum 48 | 2.35 | 0.07 | 1.00 | ||
| Left Superior Posterior Lobe of Cerebellum | 2.32 | 0.07 | 1.00 | ||
When RCK is applied ICA temporal lobe component data, it achieves a mean test accuracy rate of 85%. The optimal area set obtained by using ICA temporal lobe data is reported in Table 2.
Table 2.
Optimal area set and associated discriminative weights for RCK analysis applied to ICA temporal lobe data. The most informative anatomical regions retrieved by RCK when applied to ICA temporal lobe data are grouped in macro brain regions to give a better idea of their location in the brain. The mean and the standard deviation of the discriminative weights of each area are listed in this table. In addition the rate of training sets in the cross-validation procedure that selected each area (selection frequency) is also reported in order to measure the validity of the inclusion of each region in the optimal area set.
| Source | Areas and Discriminative Weights
|
||||
|---|---|---|---|---|---|
| Macro Regions | Regions | Discriminative Weights
|
|||
| Mean | Std. Dev. | Sel. Freq. | |||
| ICA temporal lobe | Central region | Right Rolandic Operculum | 8.63 | 0.25 | 1.00 |
| Left Precentral Gyrus | 7.70 | 0.09 | 1.00 | ||
|
| |||||
| Frontal lobe | Left Inferior Frontal Gyrus, Orbital Part | 7.79 | 0.21 | 1.00 | |
| Right Superior Frontal Gyrus, Medial | 7.58 | 0.10 | 0.96 | ||
| Right Superior Frontal Gyrus | 7.56 | 0.05 | 1.00 | ||
|
| |||||
| Temporal lobe | Right Middle Temporal Gyrus | 7.39 | 0.04 | 0.81 | |
|
| |||||
| Occipital lobe | Right Middle Occipital Gyrus | 7.97 | 0.09 | 1.00 | |
| Left Middle Occipital Gyrus | 7.67 | 0.15 | 1.00 | ||
| Right Fusiform Gyrus | 7.57 | 0.12 | 0.98 | ||
| Right Calcarine Fissure | 7.46 | 0.11 | 0.83 | ||
|
| |||||
| Limbic lobe | Left Middle Cingulate Gyrus | 7.67 | 0.11 | 1.00 | |
|
| |||||
| Insula | Left Insular Cortex | 7.64 | 0.12 | 1.00 | |
|
| |||||
| Cerebellum | Right Inferior Posterior Lobe of Cerebellum | 7.36 | 0.25 | 0.52 | |
Finally, RCK achieves a mean test accuracy rate of 86% when it is applied to GLM data. The list of areas selected by RCK in this case is displayed in Table 3.
Table 3.
Optimal area set and associated discriminative weights for RCK analysis applied to GLM data. The most informative anatomical regions retrieved by RCK when applied to GLM data are grouped in macro brain regions to give a better idea of their location in the brain. The mean and the standard deviation of the discriminative weights of each area are listed in this table. In addition the rate of training sets in the cross-validation procedure that selected each area (selection frequency) is also reported in order to measure the validity of the inclusion of each region in the optimal area set.
| Source | Areas and Discriminative Weights
|
||||
|---|---|---|---|---|---|
| Macro Regions | Regions | Discriminative Weights
|
|||
| Mean | Std. Dev. | Sel. Freq. | |||
| GLM | Central region | Left Postcentral Gyrus | 3.12 | 0.16 | 1.00 |
| Right Precentral Gyrus | 2.78 | 0.12 | 1.00 | ||
| Left Precentral Gyrus | 2.67 | 0.09 | 1.00 | ||
| Right Postcentral Gyrus | 2.64 | 0.12 | 1.00 | ||
|
| |||||
| Frontal lobe | Left Superior Frontal Gyrus | 4.12 | 0.12 | 1.00 | |
| Right Middle Frontal Gyrus | 4.02 | 0.14 | 1.00 | ||
| Left Inferior Frontal Gyrus, Triangular Part | 3.64 | 0.19 | 1.00 | ||
| Left Middle Frontal Gyrus | 3.45 | 0.12 | 1.00 | ||
| Left Middle Frontal Gyrus, Orbital Part 2 | 3.15 | 0.17 | 1.00 | ||
| Right Superior Frontal Gyrus | 2.71 | 0.10 | 1.00 | ||
| Left Middle Frontal Gyrus, Orbital Part 1 | 2.59 | 0.17 | 1.00 | ||
| Left Supplementary Motor Area | 2.48 | 0.12 | 1.00 | ||
| Left Superior Frontal Gyrus, Medial | 2.43 | 0.10 | 1.00 | ||
| Right Inferior Frontal Gyrus, Orbital Part | 2.31 | 0.16 | 0.96 | ||
| Right Superior Frontal Gyrus, Medial | 2.23 | 0.11 | 1.00 | ||
| Left Inferior Frontal Gyrus, Opercular Part | 2.15 | 0.12 | 0.98 | ||
| Left Inferior Frontal Gyrus, Orbital Part | 2.10 | 0.11 | 0.92 | ||
| Right Paracentral Lobule | 2.07 | 0.16 | 0.83 | ||
|
| |||||
| Temporal lobe | Right Middle Temporal Gyrus | 3.87 | 0.13 | 1.00 | |
| Left Superior Temporal Gyrus | 2.79 | 0.15 | 1.00 | ||
| Right Superior Temporal Gyrus | 2.37 | 0.12 | 1.00 | ||
| Left Middle Temporal Gyrus | 2.30 | 0.07 | 1.00 | ||
| Left Inferior Temporal Gyrus | 2.28 | 0.14 | 1.00 | ||
| Right Inferior Temporal Gyrus | 2.14 | 0.08 | 0.98 | ||
|
| |||||
| Parietal lobe | Right Precuneus | 2.35 | 0.10 | 1.00 | |
| Left Inferior Parietal Gyrus | 2.18 | 0.17 | 0.96 | ||
|
| |||||
| Occipital lobe | Left Calcarine Fissure | 3.00 | 0.19 | 1.00 | |
| Right Fusiform Gyrus | 2.55 | 0.13 | 1.00 | ||
| Right Middle Occipital Gyrus | 2.50 | 0.11 | 1.00 | ||
|
| |||||
| Limbic lobe | Right Hippocampus | 2.27 | 0.12 | 1.00 | |
| Right Middle Cingulate Gyrus | 2.24 | 0.08 | 1.00 | ||
| Right Anterior Cingulate Gyrus | 2.21 | 0.12 | 0.98 | ||
|
| |||||
| Insula | Left Insular Cortex | 1.96 | 0.07 | 0.52 | |
|
| |||||
| Sub cortical gray nuclei | Right Caudate Nucleus | 2.30 | 0.14 | 1.00 | |
| Right Amygdala | 2.26 | 0.15 | 0.98 | ||
|
| |||||
| Cerebellum | Anterior Lobe of Vermis | 2.83 | 0.21 | 1.00 | |
| Posterior Lobe of Vermis | 2.67 | 0.22 | 1.00 | ||
| Right Inferior Posterior Lobe of Cerebellum | 2.30 | 0.16 | 0.98 | ||
3.2. RCK Applied to Multiple Sources
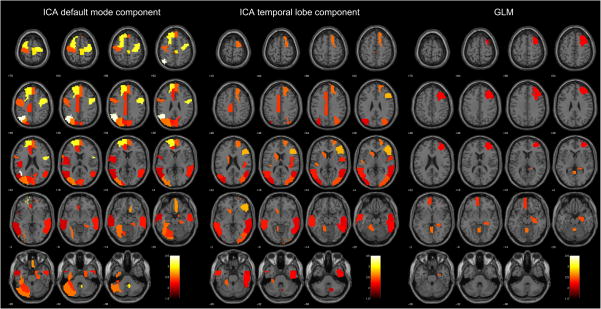
All possible combinations of data sources were analyzed by RCK, and we report the obtained results for each of them (please refer to Table 6). It can be seen that RCK achieves its peak performance when it is applied to all of the provided sources (95%). Due to this fact, we think that special attention should be given to the areas retrieved by this multi-source analysis and its characterization by means of their discriminative weights. Therefore, we present Table 4, which displays this information. In addition, a graphical representation of the coefficients associated to those areas is presented in Fig. 1, which overlay colored regions on top of a structural brain map for each of the three analyzed sources.
Table 6.
Mean classification accuracy achieved by different algorithms using multi-source data. The reported results indicate the mean classification rate attained by different algorithms provided with all possible combinations of data sources. The analysis is performed using all brain regions included in the AAL brain parcellation.
| Two Sources
|
All Sources | |||
|---|---|---|---|---|
| Default & Temporal | Default & GLM | Temporal & GLM | ||
| Composite Kernels | 0.70 | 0.70 | 0.69 | 0.79 |
| Linear SVM | 0.79 | 0.78 | 0.62 | 0.80 |
| Gaussian SVM | 0.76 | 0.77 | 0.70 | 0.80 |
| RFE-SVM | 0.92 | 0.90 | 0.84 | 0.90 |
| RCK | 0.92 | 0.93 | 0.85 | 0.95 |
Table 4.
Optimal area set and associated discriminative weights for RCK analysis applied -source data. The most informative anatomical regions retrieved by RCK when to 3 data sources are grouped in macro brain regions to give a better idea of location in the brain. The mean and the standard deviation of the discriminative of each area are listed in this table. In addition the rate of training sets in the -validation procedure that selected each area (selection frequency) is also reported in to measure the validity of the inclusion of each region in the optimal area set.
| Source | Areas and Discriminative Weights
|
||||
|---|---|---|---|---|---|
| Macro Regions | Regions | Discriminative Weights
|
|||
| Mean | Std. Dev. | Sel. Freq. | |||
| ICA default mode | Central region | Right Precentral Gyrus | 3.10 | 0.13 | 1.00 |
| Left Precentral Gyrus | 2.49 | 0.08 | 1.00 | ||
| Left Rolandic Operculum | 2.18 | 0.15 | 0.89 | ||
|
| |||||
| Frontal lobe | Left Superior Frontal Gyrus | 3.06 | 0.11 | 1.00 | |
| Left Superior Frontal Gyrus, Medial | 3.05 | 0.15 | 1.00 | ||
| Right Paracentral Lobule | 2.94 | 0.16 | 1.00 | ||
| Right Gyrus Rectus | 2.66 | 0.20 | 1.00 | ||
| Right Superior Frontal Gyrus, Medial | 2.50 | 0.10 | 1.00 | ||
|
| |||||
| Temporal lobe | Right Middle Temporal Gyrus | 2.30 | 0.08 | 1.00 | |
| Left Middle Temporal Gyrus | 2.09 | 0.11 | 0.74 | ||
|
| |||||
| Parietal lobe | Left Angular Gyrus | 3.44 | 0.22 | 1.00 | |
|
| |||||
| Occipital lobe | Left Superior Occipital Gyrus | 2.62 | 0.15 | 1.00 | |
| Left Middle Occipital Gyrus | 2.59 | 0.15 | 1.00 | ||
| Left Fusiform Gyrus | 2.55 | 0.12 | 1.00 | ||
| Right Cuneus | 2.35 | 0.14 | 0.98 | ||
| Left Cuneus | 2.30 | 0.12 | 1.00 | ||
|
| |||||
| Limbic lobe | Parahippocampal Gyrus | 2.45 | 0.14 | 0.98 | |
| Left Middle Cingulate Gyrus | 2.36 | 0.11 | 1.00 | ||
| Left Anterior Cingulate Gyrus | 2.29 | 0.11 | 1.00 | ||
|
| |||||
| Cerebellum | Right Inferior Posterior Lobe of Cerebellum | 2.93 | 0.20 | 1.00 | |
| Left Superior Posterior Lobe of Cerebellum | 2.58 | 0.13 | 1.00 | ||
| Left Anterior Lobe of Cerebellum | 2.37 | 0.14 | 0.98 | ||
|
| |||||
| ICA temporal lobe | Central region | Right Rolandic Operculum | 2.33 | 0.13 | 0.98 |
|
| |||||
| Frontal lobe | Right Inferior Frontal Gyrus, Triangular Part | 2.77 | 0.13 | 1.00 | |
| Right Superior Frontal Gyrus | 2.55 | 0.11 | 1.00 | ||
|
| |||||
| Temporal lobe | Left Heschl gyrus | 2.54 | 0.17 | 1.00 | |
| Left Middle Temporal Gyrus | 2.28 | 0.12 | 1.00 | ||
| Right Inferior Temporal Gyrus | 2.24 | 0.11 | 0.98 | ||
| Right Middle Temporal Gyrus | 2.18 | 0.09 | 0.98 | ||
|
| |||||
| Occipital lobe | Right Middle Occipital Gyrus | 2.44 | 0.11 | 1.00 | |
| Left Middle Occipital Gyrus | 2.16 | 0.11 | 0.94 | ||
|
| |||||
| Limbic lobe | Left Middle Cingulate Gyrus | 2.38 | 0.13 | 1.00 | |
|
| |||||
| Sub cortical gray nuclei | Left Caudate Nucleus | 2.52 | 0.13 | 1.00 | |
|
| |||||
| Cerebellum | Left Anterior Lobe of Cerebellum | 2.47 | 0.16 | 1.00 | |
| Right Cerebellar Tonsil | 2.25 | 0.19 | 0.98 | ||
| Right Posterior Lobe of Cerebellum | 2.08 | 0.15 | 0.58 | ||
|
| |||||
| GLM | Frontal lobe | Left Middle Frontal Gyrus, Orbital Part | 2.36 | 0.16 | 1.00 |
| Right Middle Frontal Gyrus | 2.23 | 0.13 | 0.98 | ||
|
| |||||
| Limbic lobe | Right Hippocampus | 2.44 | 0.14 | 1.00 | |
|
| |||||
| Cerebellum | Posterior Lobe of Vermis | 2.56 | 0.18 | 1.00 | |
Fig. 1.
Discriminative weights brain maps for multi-source analysis. The brain maps of each of these sources highlight the brain regions associated to each of them that were present in the optimal area set for this multi-source data classification. These areas are color-coded according to their associated discriminative coefficients.
3.3. Comparison of the Performance of Composite Kernels and RCK with Other Methods
For single-source data analysis, Table 5 shows that both Gaussian SVMs and composite kernels exhibit an equivalent performance for all sources, while the classification accuracy achieved by linear SVMs for both ICA temporal lobe and GLM sources are smaller than the ones attained by the aforementioned algorithms. It can also be seen that there is a moderate difference between the classification accuracy rates obtained by RCK and RFE-SVM when they are applied to all data sources, except ICA default mode.
Table 5.
Mean classification accuracy achieved by different algorithms using single-source data. The reported results indicate the mean classification rate attained by different algorithms for each data source using the data from all the brain regions included in the AAL brain parcellation.
| Default Mode | Temporal Lobe | GLM | |
|---|---|---|---|
| Composite Kernels | 0.75 | 0.64 | 0.74 |
| Linear SVM | 0.75 | 0.54 | 0.67 |
| Gaussian SVM | 0.75 | 0.62 | 0.75 |
| RFE-SVM | 0.87 | 0.75 | 0.71 |
| RCK | 0.90 | 0.85 | 0.86 |
The results of multi-source analysis are shown in Table 6. In this case, linear SVMs and Gaussian SVMs reach a similar prediction accuracy for all multi-source analyses, except for the case when they are provided with data from ICA temporal lobe and GLM sources. While composite kernels achieve almost the same classification accuracy as linear and Gaussian SVMs when provided with three-sources data, its performance is reduced on the other multi-source analyses. The differences between classification rates for RFE-based methods are small for multi-source data analyses, with RCK achieving slightly better results in some cases.
4. Discussion
A classification algorithm based on composite kernels that is applicable to fMRI data has been introduced. This algorithm analyzes nonlinear relationships across voxels within anatomical brain regions and combines the information from these areas linearly, thus assuming underlying linear relationships between them. By using composite kernels, the regions from segmented whole-brain data can be ranked multivariately, thus capturing the spatially distributed multivariate nature of fMRI data. The fact that whole-brain data is used by the composite kernels algorithm is of special importance, since the data within each region does not require any feature extraction preprocessing procedure in order to reduce their dimensionality. The application of RFE to composite kernels enables this approach to discard the least informative brain regions and hence retrieve the brain regions that are more relevant for class discrimination for both single-source and multi-source data analyses. The discriminative coefficients of each brain region indicate the degree of differential activity between controls and patients. Despite the fact that composite kernels cannot indicate which of the analyzed groups of interest is more activated for a specific brain region like linear SVMs can potentially do, the proposed method is still capable of measuring the degree of differential activity between groups for each region. Furthermore, RCK enables the use of a nonlinear kernel within a RFE procedure, a task that can become barely tractable with conventional SVM implementations. Another advantage of RCK over other RFE-based procedures such as RFE-SVM is its faster execution time; while the former takes 12 hours to be executed, the latter takes 157 hours, achieving a 13-fold improvement. Finally, this paper shows that the proposed algorithm is capable of taking advantage of the complementarity of GLM and ICA by combining them to better characterize groups of healthy controls and schizophrenia patients; the fact that the classification accuracy achieved by using data from three sources surpasses that reached by using single-source data supports this claim.
The set of assumptions upon which the proposed approach is based are the linear relationships between brain regions, the nonlinear relationships between voxels in the same brain region and the sparsity of information in the brain. These assumptions seem to be reasonable enough to analyze the experimental data based on the obtained classification results. This does not imply that cognitive processes actually work in the same way as it is stated in our assumptions, but that the complexity assumed by our method is sensible enough to produce good results with the available data. While composite kernels achieve classification accuracy rates that are greater than or equal to those reached by both linear and Gaussian SVMs when applied to single-source whole-brain data, the same does not hold for multi-source analysis. It may be possible that composite kernels performance is precluded when it is provided with too many areas, making it prone to overfitting.
The presented results suggest that for a given amount of training data, the trade-off of our proposed algorithm between the low complexity of the linear assumption, which provides the rationale of linear SVMs, and the high complexity of the fully nonlinear approach, which motivates the application of Gaussian SVMs, is convenient. In the case of composite kernels, they assume linear relationships between brain regions but are flexible enough to analyze nonlinearities within them. Nevertheless, their results are similar to the ones of the previously mentioned approaches for single-source analysis and inferior for multi-source analysis since they do not take advantage of information sparsity in the brain, thus not significantly reducing the classifier complexity. However, the accuracy rates attained by RCK are significantly better than the ones achieved by composite kernels. These results reinforce the validity of two hypotheses: first, that indeed there are brain regions that are irrelevant for the characterization of schizophrenia (information sparsity); and second, that RCK is capable of detecting such regions, therefore being capable of finding the set of most informative regions for schizophrenia detection given a specific data source.
Table 6 shows the results achieved by different classifiers using multi-source data. It is important to notice that the results obtained by all the classifiers when all of the sources are combined are greater than those obtained by these algorithms when they are provided with data from the ICA default mode component and either the ICA temporal lobe component or GLM data. The only method for which the previous statement does not hold is RFE-SVM. This finding may seem counterintuitive as one may think that both ICA temporal lobe component and GLM data are redundant, since they are detected based on their similarity to the stimuli of the fMRI task. However, the fact that ICA and GLM characterize fMRI data in different ways (the former analyzes task-related activity, while the latter detects groups of voxels with temporally coherent activity) might provide some insight of why the combination of these two sources proves to be important together with ICA default mode data.
In addition to the accuracy improvement achieved by applying feature selection to whole-brain data classification, RCK allows us to better identify the brain regions that characterize schizophrenia. The fact that several brain regions in the ICA temporal lobe component are present in the optimal area set is consistent with the findings that highlight the importance of the temporal lobe for schizophrenia detection. It is also important to note the presence of the anterior cingulate gyrus of the ICA default mode component in the optimal area set, for it has been proposed that error-related activity in the anterior cingulate cortex is impaired in patients with schizophrenia (Carter et al., 2001). The participants of the study are subject to making errors since the AOD task is designed in such a way that subjects have to make a quick button-press response upon the presentation of target stimuli. Since attention plays an important role in this fMRI task, it is sensible to think that consistent differential activation of the dorsolateral prefrontal cortex (DLPFC) for controls and patients will be present (Ungar et al., 2010). That may be the reason why the right middle frontal gyrus of the GLM is included in the optimal area set.
Brain aging effects being more pronounced in individuals after age 60 (Fjell and Walhovd, 2010) raised a concern that our results may have been influenced by the data collected from four healthy controls who exceeded this age cutoff in our sample. Thus, we re-ran our analysis excluding these four subjects. Both the resulting classification accuracy rates and the optimal area sets were consistent with the previously found ones. These findings seem to indicate that the algorithm proposed in this paper is robust enough not to be affected by the presence of potential outliers when provided with consistent features within the groups of interest.
To summarize, this work extends previous studies (Calhoun et al., 2004, 2008; Garrity et al., 2007) by introducing new elements. First, the method allows the usage of multi-source fMRI data, making it possible to combine ICA and GLM data. And second, it can automatically identify and retrieve regions which are relevant for the classification task by using whole-brain data without the need of selecting a subset of voxels or a set of ROIs prior to classification. Based on the aforementioned capabilities of the presented method, it is reasonable to think that it can be applied not only to multi-source fMRI data, but also to data from multiple imaging modalities (such as fMRI, EEG or MEG data) for schizophrenia detection and identify the regions within each of the sources which differentiate controls and patients better. Further work includes the modification of the composite kernels formulation to include scalar coefficients associated to each kernel. By applying new improved strategies based on optimizers that provide sparse solutions to this formulation, a direct sparse selection of kernels would be attainable. Such approaches are attractive because they would enable the selection of the optimal area set without the need of using a recursive algorithm, significantly improving the execution time of the learning phase of the classifier. Moreover, it is possible to analyze nonlinear relationships between groups of brain regions by using those methods, thus providing a more general setting to characterize schizophrenia. Finally, it should be stated that even though this approach is useful in schizophrenia detection and characterization, it is not restricted to this disease detection and can be utilized to detect other mental diseases.
Research Highlights.
Complementary sources (GLM, ICA) are combined to better characterize schizophrenia.
RCK has a lower computing load than other recursive feature elimination algorithms.
RCK provides a general setting by analyzing nonlinear relationships between voxels.
Brain regions of segmented whole-brain data are analyzed and ranked multivariately.
RCK finds the set of most discriminative brain areas for group classification.
Acknowledgments
We would like to thank the Olin Neuropsychiatry Research Center for providing the data that was used by the approach proposed in this paper. This work has been supported by NIH Grant NIBIB 2 RO1 EB000840 and Spanish government grant TEC2008-02473.
Appendix 1: Definition of Mercer’s Kernel
A theorem provided by Mercer (Aizerman et al., 1964) in the early 1900’s is of extreme relevance because it extends the principle of linear learning machines to the nonlinear case. The basic idea is that vectors x in a finite dimension space (called input space) can be mapped to a higher (possibly infinite) dimension in Hilbert space
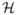 provided with a inner product, through a nonlinear transformation ϕ(·). A linear machine can be constructed in a higher dimensional space (Vapnik, 1998; Burges, 1998) (often called the feature space) which will be nonlinear from the point of view of the input space.
provided with a inner product, through a nonlinear transformation ϕ(·). A linear machine can be constructed in a higher dimensional space (Vapnik, 1998; Burges, 1998) (often called the feature space) which will be nonlinear from the point of view of the input space.
The Mercer’s theorem shows that there exists a function ϕ: ℝn →
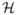 and a inner product
and a inner product
| (6) |
if and only if k(·, ·) is a positive integral operator on a Hilbert space, i.e, if and only if for any function g(x) for which
| (7) |
the inequality
| (8) |
holds. Hilbert spaces provided with kernel inner products are often called Reproducing Kernel Hilbert Spaces (RKHS). The most widely used kernel is the Gaussian. Its expression is
| (9) |
It is straightforward to show that its Hilbert space has infinite dimension.
A linear learning machine applied to these transformed data will have nonlinear properties from the point of view of the input data x. The linear learning machine can be expressed as
| (10) |
If the algorithm to optimize parameters w is linear, then they can be expressed as a linear combination of the training data
| (11) |
This expression, together with (10), give the result
| (12) |
This is, the machine can be expressed as a linear combination of inner products between the test and training data. Also, any linear algorithm to optimize w in (10) can be transformed using the same technique, leading to a linear algorithm to equivalently optimize parameters αi of expression (12). This technique is the so-called kernel trick.
Appendix 2: Composite Kernels
Summation Kernel
Vectors in different Hilbert spaces can be combined to a higher dimension Hilbert space. The most straightforward combination is the so-called direct sum of Hilbert spaces (Reed and Simon, 1980). In order to construct a direct sum of Hilbert spaces, let us assume that several nonlinear transformations ϕl(·) to Hilbert spaces and the corresponding kernel inner products kl(·, ·) are available.
Assume without loss of generality that a column vector in a finite dimension space constructed as the concatenation of several vectors as is piecewise mapped using the nonlinear transformations
| (13) |
The resulting vector is simply the concatenation of the transformations. The inner product between vectors in this space is
| (14) |
The resulting kernel is also called summation kernel.
The learning machine (12) using the kernel (14) will have the expression
| (15) |
The technique to use a learning machine based on composite kernels consists simply on computing the kernel inner products as in (14) and then proceed to train it as a regular kernel learning machine with a given optimization algorithm.
Mapping with composite kernels
Usually there is no inverse transformation to the nonlinear transformations ϕ(·). Then, the spatial information that vector w may have cannot be retrieved. But by using composite kernels each Hilbert space will hold all the properties of its particular region of the input space. That way, a straightforward analysis can provide information about that region. If a particular region of the input space contains no information relevant for the classification or regression task, then vector w will tend to be orthogonal to these space. If there is relevant information, then the vector will tend to be parallel to the space.
Then, it may be useful to compute the projection of w to all spaces. But this parameter vector is not accessible, so we need to make use of the kernel trick. Combining equations (11) and (13), the expression of the parameter vector is
| (16) |
From this, one can see that the projection of w over space l is simply , and its quadratic norm will be
| (17) |
which can be expressed in matrix version as ||wl||2 = αTKlα, where α is a vector containing all parameters αi and Kl is a matrix containing all kernel inner products kl(xi;l, xj;l).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aizerman MA, Braverman EM, Rozoner L. Theoretical foundations of the potential function method in pattern recognition learning. Automation and remote Control. 1964;25:821–837. [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders DSM-IV-TR. 4. American Psychiatric Publishing, Inc; Jun, 2000. (Text Revision) [Google Scholar]
- Bach FR, Lanckriet GRG. Multiple kernel learning, conic duality, and the smo algorithm. Proceedings of the 21st International Conference on Machine Learning (ICML). ICML ‘04; 2004. pp. 41–48. [Google Scholar]
- Bell AJ, Sejnowski TJ. An information-maximization approach to blind separation and blind deconvolution. Neural Computation. 1995;7 (6):1129–1159. doi: 10.1162/neco.1995.7.6.1129. [DOI] [PubMed] [Google Scholar]
- Burges C. A Tutorial on Support Vector Machines for Pattern Recognition. Data Mining and Knowledge Discovery. 1998;2 (2):1–32. [Google Scholar]
- Calhoun V, Adali T, Pearlson G, Pekar J. A method for making group inferences from functional mri data using independent component analysis. Human Brain Mapping. 2001;14 (3):140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Kiehl KA, Astur R, Pekar JJ, Pearlson GD. A method for multitask fmri data fusion applied to schizophrenia. Human Brain Mapping. 2006;27 (7):598–610. doi: 10.1002/hbm.20204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Kiehl KA, Liddle PF, Pearlson GD. Aberrant localization of synchronous hemodynamic activity in auditory cortex reliably characterizes schizophrenia. Biological Psychiatry. 2004;55:842–849. doi: 10.1016/j.biopsych.2004.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Pearlson GD, Maciejewski P, Kiehl KA. Temporal lobe and ‘default’ hemodynamic brain modes discriminate between schizophrenia and bipolar disorder. Hum Brain Map. 2008;29:1265–1275. doi: 10.1002/hbm.20463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps-Valls G, Gomez-Chova L, noz Mari JM, Rojo-Alvarez J, Martinez-Ramon M. Kernel-based framework for multitemporal and multisource remote sensing data classification and change detection. IEEE Transactions on Geoscience and Remote Sensing. 2008 Jun;46 (6):1822–1835. [Google Scholar]
- Carter CS, MacDonald Angus WI, Ross LL, Stenger VA. Anterior Cingulate Cortex Activity and Impaired Self-Monitoring of Performance in Patients With Schizophrenia: An Event-Related fMRI Study. Am J Psychiatry. 2001;158(9) doi: 10.1176/appi.ajp.158.9.1423. [DOI] [PubMed] [Google Scholar]
- Chang C-C, Lin C-J. LIBSVM: a library for support vector machines. 2001 Software available at http://www.csie.ntu.edu.tw/~cjlin/libsvm.
- Correa N, Adali T, Calhoun VD. Performance of blind source separation algorithms for fmri analysis using a group ica method. Magnetic Resonance Imaging. 2007 June;25 (5):684–694. doi: 10.1016/j.mri.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DD, Savoy RL. Functional Magnetic Resonance Imaging (fMRI) “brain reading”: detecting and classifying distributed patterns of fMRI activity in human visual cortex. Neuroimage. 2003;19 (2):261–70. doi: 10.1016/s1053-8119(03)00049-1. [DOI] [PubMed] [Google Scholar]
- De Martino F, Valente G, Staeren N, Ashburner J, Goebel R, Formisano E. Combining multivariate voxel selection and support vector machines for mapping and classification of fmri spatial patterns. NeuroImage. 2008;43 (1):44–58. doi: 10.1016/j.neuroimage.2008.06.037. [DOI] [PubMed] [Google Scholar]
- Decharms R. Reading and controlling human brain activation using real-time functional magnetic resonance imaging. Trends in Cognitive Sciences. 2007 Nov;11 (11):473–481. doi: 10.1016/j.tics.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Demirci O, Clark VP, Calhoun VD. A projection pursuit algorithm to classify individuals using fmri data: Application to schizophrenia. NeuroImage. 2008;39 (4):1774–1782. doi: 10.1016/j.neuroimage.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci O, Stevens MC, Andreasen NC, Michael A, Liu J, White T, Pearlson GD, Clark VP, Calhoun VD. Investigation of relationships between fmri brain networks in the spectral domain using ica and granger causality reveals distinct differences between schizophrenia patients and healthy controls. NeuroImage. 2009;46 (2):419–31. doi: 10.1016/j.neuroimage.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Gibbon M, Williams JBW. Structured Clinical Interview for DSM-IV Axis I Disorders-Patient Edition (SCID-I/P, Version 2.0) Biometrics Research Department, New York State Psychiatric Institute; New York: 1995. [Google Scholar]
- Fjell AM, Walhovd KB. Structural brain changes in aging: courses, causes and cognitive consequences. Reviews in the neurosciences. 2010;21 (3):187–221. doi: 10.1515/revneuro.2010.21.3.187. [DOI] [PubMed] [Google Scholar]
- Ford J, Farid H, Makedon F, Flashman LA, McAllister TW, Mega-looikonomou V, Saykin AJ. Patient classification of fmri activation maps. Proc. of the 6th Annual International Conference on Medical Image Computing and Computer Assisted Intervention (MIC-CAI’03; 2003. pp. 58–65. [Google Scholar]
- Franco AR, Pritchard A, Calhoun VD, Mayer AR. Interrater and intermethod reliability of default mode network selection. Hum Brain Mapp. 2009;30 (7):2293–303. doi: 10.1002/hbm.20668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire L, Roche A, Mangin JF. What is the best similarity measure for motion correction in fmri time series? Medical Imaging, IEEE Transactions. 2002 May;21 (5):470–484. doi: 10.1109/TMI.2002.1009383. [DOI] [PubMed] [Google Scholar]
- Friston K, Ashburner J, Frith C, Poline J, Heather JD, Frackowiak R. Spatial registration and normalization of images. Human Brain Mapping. 1995;2:165–189. [Google Scholar]
- Friston K, Chu C, Mourao-Miranda J, Hulme O, Rees G, Penny W, Ashburner J. Bayesian decoding of brain images. Neuroimage. 2008 Jan;39 (1):181–205. doi: 10.1016/j.neuroimage.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Mechelli A, Turner R, Price CJ. Nonlinear responses in fmri: The balloon model, volterra kernels, and other hemodynamics. NeuroImage. 2000;12 (4):466–477. doi: 10.1006/nimg.2000.0630. [DOI] [PubMed] [Google Scholar]
- Garrity AG, Pearlson GD, McKiernan K, Lloyd D, Kiehl KA, Calhoun VD. Aberrant “Default Mode” Functional Connectivity in Schizophrenia. Am J Psychiatry. 2007;164 (3):450–457. doi: 10.1176/ajp.2007.164.3.450. [DOI] [PubMed] [Google Scholar]
- Guyon I, Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res. 2003;3:1157–1182. [Google Scholar]
- Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Mach Learn. 2002;46:1–3. [Google Scholar]
- Haynes JD, Rees G. Predicting the orientation of invisible stimuli from activity in human primary visual cortex. Nature Neuroscience. 2005;8 (5):686–691. doi: 10.1038/nn1445. [DOI] [PubMed] [Google Scholar]
- Kamitani Y, Tong F. Decoding the visual and subjective contents of the human brain. Nature Neuroscience. 2005;8 (5):679–685. doi: 10.1038/nn1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehl KA, Liddle PF. An event-related functional magnetic resonance imaging study of an auditory oddball task in schizophrenia. Schizophrenia Research. 2001;48 (2–3):159–171. doi: 10.1016/s0920-9964(00)00117-1. [DOI] [PubMed] [Google Scholar]
- Kim D, Burge J, Lane T, Pearlson G, Kiehl K, Calhoun V. Hybrid ica-bayesian network approach reveals distinct effective connectivity differences in schizophrenia. NeuroImage. 2008;42 (4):1560–1568. doi: 10.1016/j.neuroimage.2008.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Mathalon D, Ford JM, Mannell M, Turner J, Brown G, Belger A, Gollub RL, Lauriello J, Wible CG, O’Leary D, Lim K, Potkin S, Calhoun VD. Auditory Oddball Deficits in Schizophrenia: An Independent Component Analysis of the fMRI Multisite Function BIRN Study. Schizophr Bull. 2009;35:67–81. doi: 10.1093/schbul/sbn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloft M, Brefeld U, Sonnenburg S, Zien A. lp-norm multiple kernel learning. J Mach Learn Res. 2011 March;12:953–997. [Google Scholar]
- Kriegeskorte N, Goebel R, Bandettini P. Information-based functional brain mapping. Proceedings of the National Academy of Sciences of the United States of America. 2006;103 (10):3863–3868. doi: 10.1073/pnas.0600244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaConte S, Strother S, Cherkassky V, Anderson J, Hu X. Support vector machines for temporal classification of block design fmri data. Neuroimage. 2005 March;26:317–329. doi: 10.1016/j.neuroimage.2005.01.048. [DOI] [PubMed] [Google Scholar]
- LaConte S, Strother S, Cherkassky V, Hu X. Predicting motor tasks in fmri data with support vector machines. ISMRM Eleventh Scientific Meeting and Exhibition; Toronto, Ontario, Canada. Jul, 2003. [Google Scholar]
- Lancaster J, Summerln J, Rainey L, Freitas C, Fox P. The talairach daemon, a database server for talairach atlas labels. NeuroImage. 1997;5:S633. [Google Scholar]
- Lancaster J, Woldorff M, Parsons L, Liotti M, Freitas C, Rainey L, Kochunov P, Nickerson D, SAM, Fox P. Automated talairach atlas labels for functional brain mapping. Hum Brain Mapp. 2000;10:120–131. doi: 10.1002/1097-0193(200007)10:3<120::AID-HBM30>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y-OO, Adali T, Calhoun VDD. Estimating the number of independent components for functional magnetic resonance imaging data. Hum Brain Mapp. 2007 February; doi: 10.1002/hbm.20359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldjian J, Laurienti P, Kraft R, Burdette J. An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fmri data sets. NeuroImage. 2003;19:1233–1239. doi: 10.1016/s1053-8119(03)00169-1. [DOI] [PubMed] [Google Scholar]
- Martínez-Ramón M, Koltchinskii V, Heileman GL, Posse S. fmri pattern classification using neuroanatomically constrained boosting. Neuroimage. 2006a Jul;31 (3):1129–1141. doi: 10.1016/j.neuroimage.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Martínez-Ramón M, Rojo-Álvarez JL, Camps-Valls G, Muñoz-Marí J, Navia-Vázquez A, Soria-Olivas E, Figueiras-Vidal A. Support vector machines for nonlinear kernel ARMA system identification. IEEE Transactions on Neural Networks. 2006b Nov;17 (6):1617–1622. doi: 10.1109/TNN.2006.879767. [DOI] [PubMed] [Google Scholar]
- Mourão-Miranda J, Bokde AL, Born C, Hampel H, Stetter M. Classifying brain states and determining the discriminating activation patterns: Support vector machine on functional mri data. NeuroImage. 2005;28 (4):980–995. doi: 10.1016/j.neuroimage.2005.06.070. [DOI] [PubMed] [Google Scholar]
- Mourão-Miranda J, Reynaud E, McGlone F, Calvert G, Brammer M. The impact of temporal compression and space selection on svm analysis of single-subject and multi-subject fmri data. NeuroImage. 2006;33 (4):1055–1065. doi: 10.1016/j.neuroimage.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Reed MC, Simon B. Functional Analysis. Vol. I of Methods of Modern Mathematical Physics. Academic Press; 1980. [Google Scholar]
- Ryali S, Supekar K, Abrams DA, Menon V. Sparse logistic regression for whole-brain classification of fmri data. Neuroimage. 2010 doi: 10.1016/j.neuroimage.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkareva SV, Ombao HC, Sutton BP, Mohanty A, Miller GA. Classification of functional brain images with a spatio-temporal dissimilarity map. NeuroImage. 2006 October;33 (1):63–71. doi: 10.1016/j.neuroimage.2006.06.032. [DOI] [PubMed] [Google Scholar]
- Sonnenburg S, Rätsch G, Schölkopf B, Rätsch G. Large scale multiple kernel learning. J Mach Learn Res. 2006 December;7:1531–1565. [Google Scholar]
- Spitzer RL, Williams JBW, Gibbon M. Structured Clinical interview for DSM-IV: Non-patient edition (SCID-NP) Biometrics Research Department, New York State Psychiatric Institute; New York: 1996. [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in spm using a macroscopic anatomical parcellation of the mni mri single-subject brain. NeuroImage. 2002 January;15 (1):273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Ungar L, Nestor PG, Niznikiewicz MA, Wible CG, Kubicki M. Color stroop and negative priming in schizophrenia: An fmri study. Psychiatry Research: Neuroimaging. 2010;181 (1):24–29. doi: 10.1016/j.pscychresns.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnik V. Statistical Learning Theory, Adaptive and Learning Systems for Signal Processing, Communications, and Control. John Wiley & Sons; 1998. [Google Scholar]
- Wang X, Hutchinson R, Mitchell TM. Training fmri classifiers to discriminate cognitive states across multiple subjects. In: Thrun S, Saul L, Schölkopf B, editors. Advances in Neural Information Processing Systems. Vol. 16. MIT Press; Cambridge, MA: 2004. [Google Scholar]