Summary
Background
Wee1 kinases delay entry into mitosis by phosphorylating and inactivating cyclin-dependent kinase 1 (Cdk1). Loss of this activity in many systems, including Drosophila, leads to premature mitotic entry.
Results
We report here that Drosophila Wee1 (dwee1) mutant embryos show mitotic-spindle defects that include ectopic foci of microtubule organization, formation of multipolar spindles from adjacent centrosome pairs, and promiscuous interactions between neighboring spindles. Furthermore, centrosomes are displaced from the embryo cortex in dwee1 mutants. These defects are not observed to the same extent in embryos in which nuclei also enter mitosis prematurely as a result of a lack of checkpoint control or in embryos with elevated Cdk1 activity. dWee1 physically interacts with members of the γ-tubulin ring complex (γTuRC), and γ-tubulin is phosphorylated in a dwee1-dependent manner in embryo extracts.
Conclusions
Some of the abnormalities in dwee1 mutant embryos cannot be explained by premature entry into mitosis or bulk elevation of Cdk1 activity. Instead, dWee1 is also required for phosphorylation of γ-tubulin, centrosome positioning, and mitotic-spindle integrity. We propose a model to account for these requirements.
Introduction
Cyclin-dependent kinases (Cdks) promote cell proliferation by driving the events of the cell-division cycle. Inhibitors of Cdks enforce checkpoints that halt cell cycle events until previous events have been properly completed. One such inhibitory activity is provided by the tyrosine kinase Wee1, which acts in diverse organisms to phosphorylate and inhibit the Cdk that promotes entry into mitosis (Cdk1) [1–3] [1, 4, 5]. This activity is important for coupling mitotic entry with the completion of interphase events such as DNA replication and cell growth [1, 6, 7].
Previous studies established that Drosophila wee1, dwee1, is essential for embryogenesis [8]. Embryogenesis in Drosophila begins with 13 nuclear divisions that are driven by maternally supplied products and consist only of S phase and mitosis. These divisions occur in a common cytoplasm called a syncytium [9]. The first nine syncytial nuclear divisions occur in the embryo interior. By cycle 10, nuclei have migrated to the embryo cortex, and the last four syncytial divisions occur in a monolayer beneath the cortex of the embryo (“cortical divisions”). The syncytial divisions are followed by cellularization of the embryo and the events of gastrulation.
Flies that are heterozygous for the strongest extant allele of dwee1, dwee1ES1, and for a small chromosomal deletion that removes dwee1 are viable, but embryos derived from such females (to be called “dwee1 mutant embryos” hereafter) do not progress beyond syncytial stages [8]. Thus, a maternal supply of dwee1 gene product is needed to complete syncytial divisions. In dwee1 mutant embryos, nuclei enter mitosis prematurely [7]. A similar phenotype is also caused by mutations in DNA checkpoint genes such as grapes and mei-41 (Drosophila Chk1 and ATR, respectively). It is thought that a gradual depletion of maternally supplied replication factors prolongs genome duplication and that grapes and mei-41 act to delay mitosis, i.e., lengthen interphase, to allow the completion of DNA replication [10–12]. As such, premature entry into mitosis in grp mutants is thought to occur with incompletely replicated DNA. This situation induces a checkpoint response that inactivates mitotic centrosomes [13, 14]. This checkpoint is also triggered by ionizing-radiation-induced DNA damage and requires the Chk2 kinase. Activation of the checkpoint results in dispersal of centrosomal proteins, such as the γTuRC components, loss of astral microtubules from mitotic spindles, and failure to fully segregate chromosomes [13, 14]. Nuclei exit mitosis nonetheless and enter the next interphase in a polyploid state. Another Chk2-dependent mechanism then causes detachment of nuclei from centrosomes and their removal from the cortical layer.
We reported previously that premature mitotic entry in dwee1 mutants also induces the Chk2-dependent centrosome-inactivation checkpoint. Here, we characterized mitotic-spindle abnormalities in dwee1 mutants in detail and found that not all can be explained by disrupted cell cycle timing or bulk elevation of Cdk1 activity. We also demonstrate that centrosomes and nuclei are displaced from the embryo cortex in dwee1 mutants, that dWee1 forms a complex with components of the γTuRC in vivo, and that γ-tubulin is phosphorylated in a dwee1-dependent manner in embryo extracts. On the basis of these data, we propose that dWee1 has a function in positioning centrosomes and nuclei at the embryo cortex and that loss of this activity leads to mitotic-spindle abnormalities.
Results and Discussion
dwee1-Specific Spindle Phenotypes
We previously reported that dwee1 mutant embryos enter mitosis prematurely and form abnormal mitotic spindles [7]. To determine whether premature mitotic entry and consequent centrosome inactivation account for the spindle abnormalities seen, we compared dwee1 mutants, grp mutants, and irradiated wild-type embryos. In all three cases, we saw evidence of centrosome inactivation: diminished astral microtubules and dispersal of both γ-tubulin and Dgrip84 from the centrosome in fixed embryos [7, 13]. Similar results were obtained from analyses of live dwee1 mutant embryos carrying the 17238-GFP transgene, in which GFP is inserted into a gene of unknown function and localizes to microtubules and centrosomes [15]. dwee1 and grp mutant embryos and irradiated wild-type embryos show the loss of GFP signal on astral microtubules and at spindle poles in M12 and M13 [7] (Figures 1A and 1B and data not shown). In addition, all three groups display monopolar spindles, the inability to form a central spindle in M12 and M13, and the failure to fully separate centrosomes during interphase (Table 1).
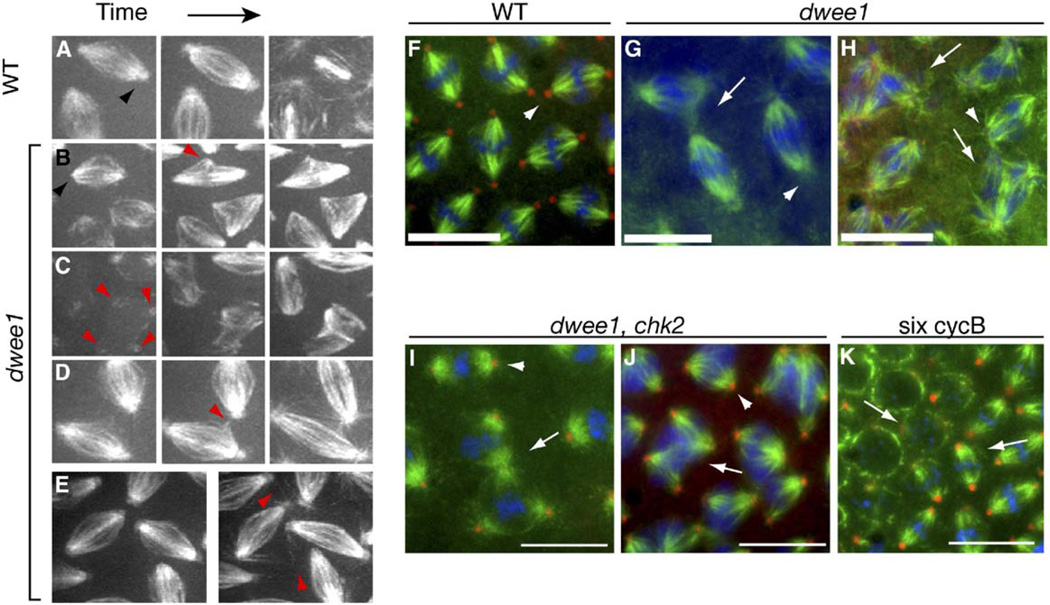
Figure 1. dwee1-Specific Spindle Defects.
(A–E) Live transgenic embryos expressing 17238-GFP, which labels microtubules and centrosomes (black arrowhead in [A]) were analyzed by time-lapse confocal microscopy. The centrosomal signal is lost in dwee1 mutants (black arrowhead in [B]). Other dwee1 phenotypes include the following: (B) 17238-GFP foci that appear to be ectopic microtubule-organizing centers (MTOC; red arrowhead) are seen moving around within an otherwise bipolar spindle in dwee1 mutants; (C) interactions between interphase centrosomes (arrowheads) that lead to formation of multi-polar spindles; and (D and E) adjacent spindles with promiscuous microtubule interactions that initiate in metaphase (D) or in anaphase ([E]; cycle 13). Spindle defects are quantified in Table 1.
(F–K) One- to two-hour embryos derived from mothers of the indicated genotypes were fixed and stained for DNA (blue), β-tubulin (green), and Dgrip 84 (red). (F) Wild-type (WT) metaphase 13 spindles are in close proximity to each other, but adjacent spindles do not interact. (G–H) dwee1 mutants show interactions between adjacent spindles (arrows in [G]) during metaphase 13. Multipolar spindles are also seen in metaphase 13 dwee1 mutants (arrows in [H]). (I–J) Mutation of chk2 rescues loss of Dgrip84 from centrosomes in dwee1 mutants (arrowheads in [F–J]) but not spindle interactions (arrow in [I]) or multipolar spindles (arrow in [J], multiple spindles displaying dwee1-specific phenotypes were seen in 11 of 12 chk2, dwee1 embryos analyzed). (K) Elevated cyclinB-Cdk1 activity does not produce dwee1-specific spindle phenotypes. six cycB embryos show expected mitotic problems such as loss of cell cycle synchrony (arrows indicate adjacent nuclei that are in different cell cycle stages) but do not contain interacting spindles or multipolar spindles like dwee1 mutants (0 spindles in 16 mitosis 12 embryos analyzed). Scale bars = 10 µm.
Table 1.
Quantification of Microtubule Defects in Live dwee1, grp, and Irradiated Embryos
| Wild-Type | dwee1 | grp | Wild-Type + Rad | |||||
|---|---|---|---|---|---|---|---|---|
| Defect | n = 97, cyc12 |
n = 167, cyc13 |
n = 65, cyc12 |
n = 109, cyc13 |
n = 149, cyc12 |
n = 249, cyc13 |
n = 56, cyc12 |
n = 44, cyc13 |
| Anastrala | 0 | 0 | 6.2 | 100 | 55.7 | 100 | 28.6 | 86.4 |
| Cent Spindleb | 0 | 0 | 1.5 | 67.9 | 21.5 | 76.7 | 53.6 | 34.1 |
| Cent sepc | 2.1 | 0 | 3.1 | 18.3 | 8.1 | 18.1 | 0 | 20.5 |
| Monopolard | 2.1 | 4.2 | 1.5 | 22.9 | 4.7 | 27.3 | 50 | 45.5 |
| Multi MTOCe | 0 | 0 | 0 | 5.5 | 0.7 | 1.6 | 0 | 0 |
| Multipolarf | 0 | 0 | 0 | 4.6 | 0 | 0 | 0 | 0 |
| MT intg | 0 | 0 | 27.7 | 40.4 | 0 | 0.8 | 0 | 0 |
n represents the number of nuclei assayed.
Lack 17238-GFP staining on astral microtubules and centrosomes.
Central spindles are disorganized or not formed.
Centrosomes do not separate completely during interphase.
Monopolar spindles.
Spindles appear to have extra MTOCs.
Multipolar spindles.
Adjacent spindles show MT interactions.
Live analysis revealed three additional, previously unreported phenotypes in cycles 12 and 13 of dwee1 mutants (Figures 1B–1E): (1) spindles with one to two ectopic microtubule-organizing centers (MTOC) within the single microtubule network (the ectopic sites change location dramatically within the confines of the spindle); (2) promiscuous interactions between adjacent spindles (in these situations, microtubules from one spindle appear to pull on a neighboring spindle); and (3) multipolar spindles that result from centrosome pairs of neighboring nuclei that interact. The last two phenotypes are also seen in fixed embryos (Figures 1F–1H). The first, however, was not; ectopic MTOC may be too dynamic to be preserved during fixation. Importantly, these three phenotypes were absent in live irradiated wild-type embryos and present only at low frequencies in live grp mutants (Table 1). We refer to these phenotypes as “dwee1-specific” spindle defects hereafter.
dwee1-Specific Spindle Defects Are Not Chk2 Dependent
Centrosome inactivation in irradiated or grp mutant embryos is dependent on Chk2 function [14]. Therefore, we analyzed dwee1, chk2 double mutants to further assess whether dwee1 spindle defects are caused by induction of the checkpoint. As reported previously, chk2 mutations rescue cellularization in dwee1 mutants [7]. As expected, mutation of chk2 also rescued the phenotypes, such as anastral spindles and the dispersal of γTURC in dwee1 mutants, that are characteristic of centrosome inactivation (Figures 1I and 1J). dwee1, chk2 double mutants, however, still displayed spindle interactions and formed multipolar spindles (11 of 12 embryos in M12 or M13; Figures 1I and 1J), which are the two dwee1-specific spindle defects discernable in fixed embryos. On the basis of these data and the finding that grp mutants or irradiated embryos do not display dwee1-specific spindle defects to the same extent, we conclude that spindle interactions and multipolar spindles in dwee1 mutants are not due to premature mitotic entry or to induction of chk2-dependent centrosome inactivation.
Increased Cdk1 Activity Does Not Cause dwee1-Specific Spindle Defects
Cdk1 activity is elevated during cortical syncytial cycles in dwee1 mutant embryos [7], and elevation of Cdk1 activity has been shown previously to affect spindle morphogenesis in precortical syncytial cycles [16, 17]. Therefore, we next addressed the possibility that elevated Cdk1 activity is the cause of dwee1-specific spindle defects. To this end, we analyzed fixed embryos from a fly stock with six copies of cyclin B, six cycB [17]. Previous studies have indicated that increasing cyclin B levels in embryos increases Cdk1 activity [16, 17]. Consistent with these studies, we found that six cycB embryos harbor higher CycB-Cdk1 activity than do wild-type embryos (Figures 2A–2D). More Cdk1 coprecipitated with cyclin B from six cycB embryos than from wild-type or dwee1 mutant embryos (Figure 2C, legend), suggesting that this increase in activity is due to the presence of more-active complexes, an idea that is consistent with previous observations that Cdk1 levels are not limiting in embryos [16, 18]. As previously described, six cycB embryos displayed defects such as asynchronous divisions [17], but dwee1-specific spindle defects were not detected (Figure 1K). We conclude that a bulk elevation of Cdk1 activity cannot account for dwee1-specific spindle defects.
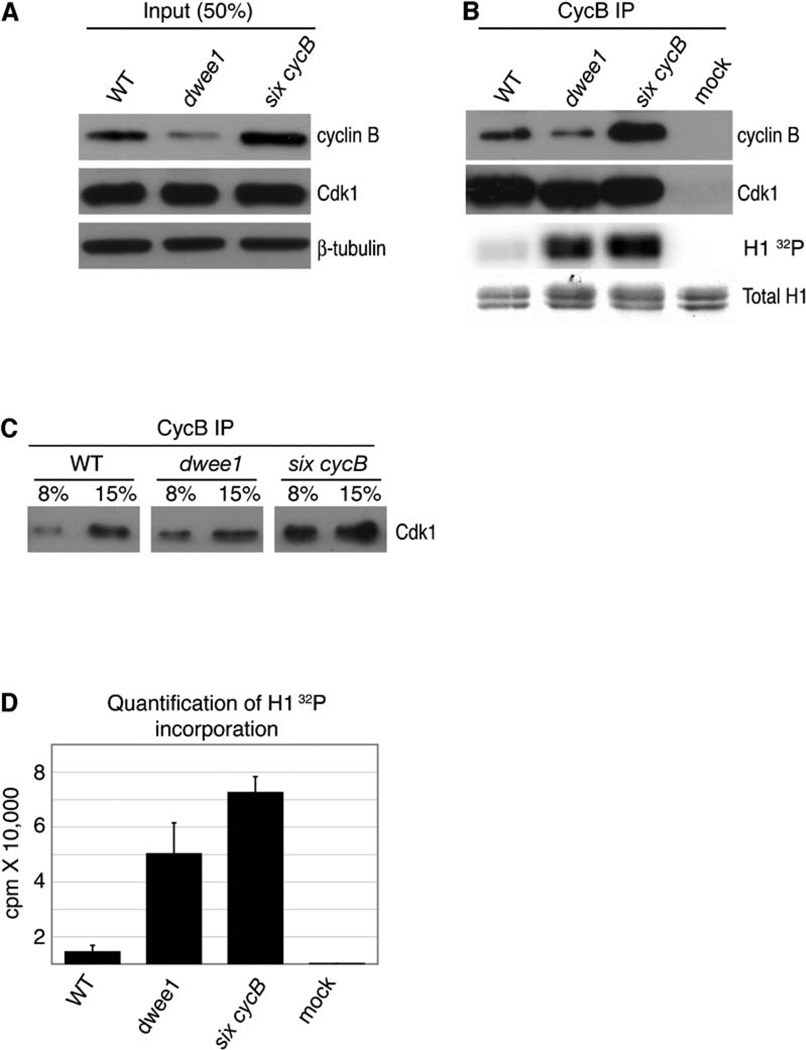
Figure 2. Elevated Cdk1 Activity Does Not Cause dwee1-Specific Spindle Defects.
Cyclin B immunoprecipitates from extracts of syncytial-blastoderm-stage wild-type (WT), dwee1 mutant, and six cycB embryos were analyzed for associated Cdk1 and kinase activity toward Histone H1 (H1).
(A) Western-blot analysis of input extracts from wild-type, dwee1 mutant, and transgenic six cycB embryos. Total Cdk1 levels are the same in all samples, whereas six cycB embryos show a ~2–3-fold increase in cyclin B.
(B) CycB immunoprecipitates were processed to detect CycB, Cdk1, phosphorylation of H1 (H1 32P), and amount of H1 added to each reaction (Total H1). “Mock” represents immunoprecipitations from wild-type extracts where anti-CycB antibody was omitted.
(C) Western-blot analysis of the indicated fractions of anti-cyclin B immunopreciptates indicates that the amount of Cdk1 recovered from six cycB embryos is ~2-fold higher than the amount recovered from wild-type or dwee1 mutant embryo extracts (the signal from 8% of the IP in six cycB sample is comparable to the signal from 15% of the IP in wild-type or dwee1 samples).
(D) Quantification of 32P incorporation into H1. Data from three independent experiments are shown. Cyclin B associated kinase activity toward H1 is 10-fold higher in precipitates from dwee1 extracts (p < .01) and 14-fold higher in precipitates from six cycB extracts (p < .001) compared to wild-type. Given that associated H1 kinase activity is only 1.5-fold higher in the six cycB sample than in the dwee1 sample, the specific activity of CycB-Cdk1 complexes must be higher in dwee1 mutants than in six cycB embryos. However, because CycB-Cdk1 complexes are about twice as abundant in six cycB extracts as in dwee1 extracts (C), total CycB-Cdk1 activity is likely higher in the former.
Each error bar indicates one standard deviation.
CycB levels are reproducibly lower in dwee1 embryos than in wild-type embryos. We do not know the reason for this finding, but we nonetheless ruled out the possibility that reduction of CycB levels is the cause of dwee1-specific phenotypes. We did so by analyzing embryos from mothers that are hemizygous for cycB and therefore have lower CycB levels [16, 17]. We found no evidence of spindle interactions or centrosome-positioning changes that resemble those of dwee1 mutant embryos (see Figure S1 in the Supplemental Data available with this article online).
Centrosomes and Nuclei Are Displaced in dwee1 Mutants
The interactions between neighboring centrosomes and spindles in dwee1 mutants are limited to cortical syncytial divisions. These are a subset of syncytial divisions that follow the migration of nuclei to the embryo cortex. During cortical divisions, nuclei and centrosomes closely abut the cortex, and their position is maintained by microtubule-filament- and actin-filament-dependent mechanisms [9]. Specifically, astral microtubules nucleated by the centrosomes are proposed to interact with cortical actin to mediate the attachment of centrosomes, along with their associated nuclei, to the cortex. Physical separation of mitotic spindles in a syncytium is thought to occur by reorganization of F-actin caps into pseudocleavage furrows that surround each dividing nucleus [19]. Pseudocleavage furrows form during prophase and metaphase, retract in anaphase, and are mostly absent in late anaphase and telophase [20].
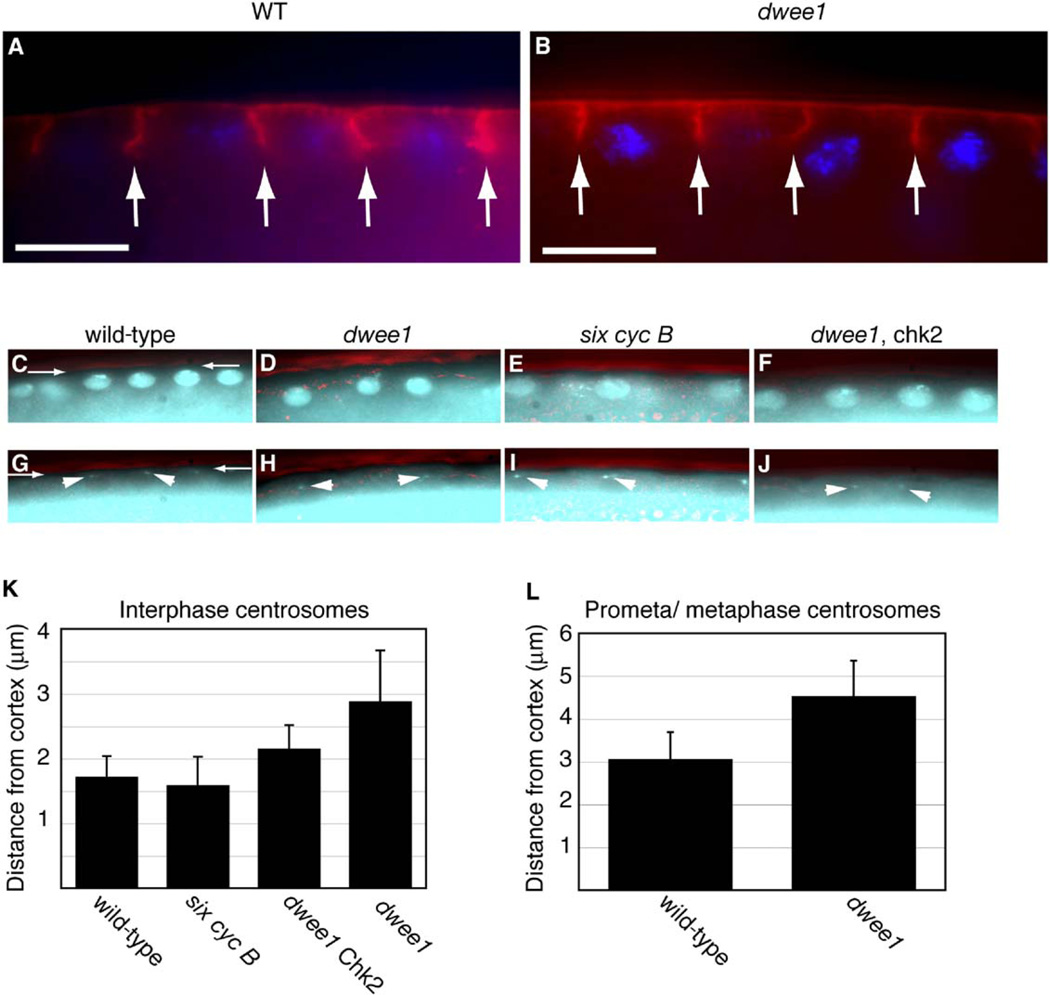
We find that pseudocleavage furrows in dwee1 mutants form with normal timing and reach similar depths as in wild-type embryos (Figure 3A and 3B). Nuclei in dwee1 mutants, however, are positioned beyond the deepest part of the furrows in metaphase (Figure 3B). Quantification of centrosome-cortex distance in wildtype embryos and dwee1 mutants in cycle 12 illustrates this phenotype (Figures 3C–3L). We chose this cycle because it occurs after completion of cortical nuclear migration but before the onset of the centrosome-inactivation checkpoint, as evidenced by anastral spindles (Table 1). Displacement of centrosomes from the cortex is observed in both interphase and mitosis of cycle 12 in dwee1 mutants and is only partially rescued by the chk2 mutation (Figures 3K and 3L). We suggest that centrosome and nuclear displacement in dwee1 mutants has two underlying components: One is a consequence of the Chk2-mediated checkpoint, and the other is a more direct result of loss of dwee1. That is, dwee1 is required to promote centrosome-cortex interaction, which is thought to be dependent on centrosomal microtubules and cortical actin. The displacement of centrosomes from the cortex in dwee1 mutants could distance mitotic spindles from the protection of pseudocleavage furrows in prophase and metaphase, thereby allowing interactions between adjacent spindles.
Figure 3. Pseudocleavage Furrows Are Present, but Centrosomes and Nuclei Are Displaced, in dwee1 Mutants.
(A and B) Wild-type (WT; panel [A]) and dwee1 mutant (B) embryos were fixed and stained for DNA (blue) and phosphotyrosine to visualize pseudocleavage furrows (red).
(C–J) Embryos of the indicated genotypes were fixed and stained to visualize DNA (C–F) and with an antibody against Dgrip84 to visualize centrosomes (arrowheads in [G–J]). The embryo cortex is visualized under DIC optics (pseudocolored red) in all panels (for example, between arrows in [C] and [G]).
The shortest distance between the centrosome and the cortex is measured and shown for interphase 12 in (K) and prometaphase or metaphase 12 in (L). Student’s T test indicates that differences between wild-type and dwee1 and between wild-type and dwee1, chk2 in (K) and between wild-type and dwee1 in (L) are significant (p < 0.0001 in all cases). n = 24 for wild-type embryos, n = 63 for six cycB, n = 35 for dwee1, chk2, and n = 17 for dwee1 in at least 3 embryos each in (K). n = 24 in 3 embryos each for wild-type embryos and dwee1 in (L). Each error bar indicates one standard deviation.
six cycB embryos show normal localization of nuclei and centrosomes, suggesting that bulk elevation of Cdk1 activity in dwee1 mutants cannot account for the cortical detachment of centrosomes. Additionally, spindle interactions in dwee1 mutants also initiate when pseudocleavage furrows are normally absent (anaphase and telophase) (Figure 1E). Therefore, a furrow-independent mechanism may operate to keep spindles apart in anaphase and telophase in a dwee1-dependent fashion.
dWee1 Copurifies with γTuRC Components and Is Required for γ-Tubulin Phosphorylation
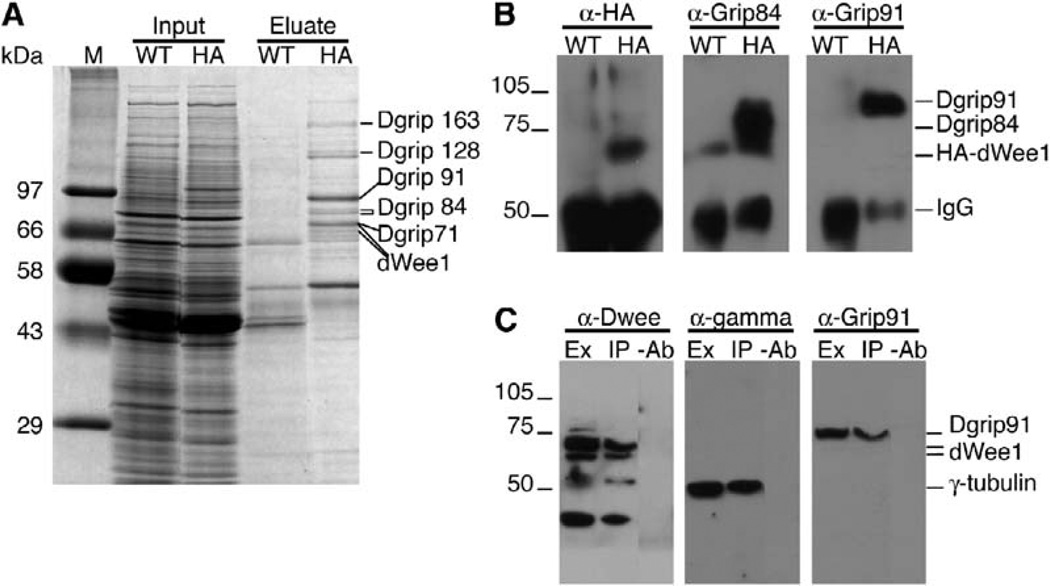
To further address the requirement for dWee1 in centrosome and spindle function, we purified dWee1-containing protein complexes and identified dWee1-interacting proteins by mass spectrometry. The heat-inducible HA-dWee1 transgene used as a source of dWee1 was shown previously to partially rescue dwee1 mutant embryos when expressed in the mothers, indicating that the product is functional [8]. HA-dWee1 was induced in embryos, purified on an anti-HA antibody column, and eluted with a HA-dipeptide. Analysis of eluates by SDS-PAGE and mass spectrometry identified peptides that matched dWee1 and Drosophila γ-tubulin ring proteins (Dgrips) 163, 128, 91, 84, and 71 (Figure 4A). The identities of Dgrip84, Dgrip91, and HA-dWee1 were confirmed by Western blotting (Figure 4B). Note that we did not detect γ-tubulin in the HA-dWee1 eluates (Figure 4A) because a strong background band in the 50 kDa range (the MW of γ-tubulin), likely the IgG heavy chain, prevented us from analyzing that region of the gel.
Figure 4. dWee1 Forms a Complex with γTURC Components.
(A) Coomassie-stained gel showing that Dgrips 71, 84, 91, 128, and 163 copurify with HA-tagged dWee1 (HA-dWee1). Soluble extracts (Input) from 0–8-hr-old wild-type embryos (WT) or 1–10-hr-old transgenic embryos expressing HA-dWee1 (HA) were passed over anti-HA affinity columns. Bound proteins were competitively eluted with HA-dipeptide (Eluate) and separated by SDSPAGE. Proteins that specifically copurified with HA-dWee1 were visualized by Coomassie stain and identified by MALDI mass spectrometry.
(B) Western blots with antibodies specific for HA (α-HA), Dgrip84 (α-Grip84), and Dgrip91 (α-Grip91) confirmed that these proteins were present in the HA-dWee1 eluate (HA) but not the wild-type control eluate (WT).
(C) Endogenous dWee1 coimmunoprecipitates with endogenous γ-tubulin and Dgrip91. Extracts were prepared from wild-type 0–8-hr-old embryos. Anti-dWee1 peptide antibodies (α-dWee1), anti-γ-tubulin (α–γ) antibodies, and anti-Dgrip91 (α-Grip91) antibodies detect these proteins in anti-dWee1 immunoprecipitates (IP) but not mock immunoprecipitates (−Ab).
Because the above experiments were performed with overexpressed, tagged dWee1, we needed to ensure that endogenous dWee1 interacts with γTuRC. We were able to detect Dgrip91 and γ-tubulin readily in immunoprecipitates by using an antibody against dWee1 (Figure 4C). We were, however, unable to detect dWee1 in immunoprecipitates by using an antibody against γ-tubulin or Dgrip91. This may be because we can at best precipitate approximately 5% of total protein present with each antibody (data not shown). The presence of γ-tubulin, a structural component of the cytoskeleton, at a higher concentration than dWee1, a regulatory kinase, is a likely scenario, and it could explain why we do not see dWee1 in immunoprecipitates of γ-tubulin.
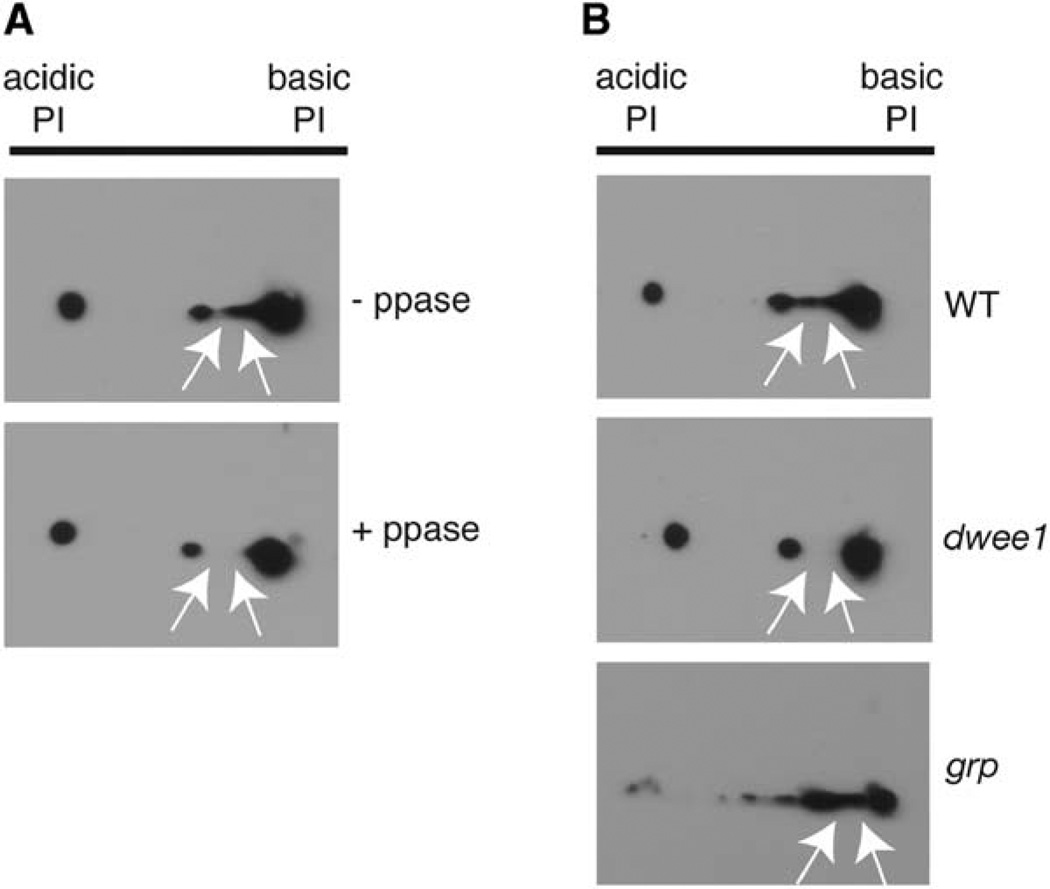
To determine whether dWee1, a kinase, influences the phosphorylation status of proteins it binds to, we examined γ-tubulin, which is known to be phosphorylated in budding yeast [21]. Two-dimensional (2D) gel electrophoresis followed by Western blotting revealed that Drosophila γ-tubulin separated as a series of five spots in the first dimension (Figure 5A). Two of the more acidic isoforms are phosphatase sensitive, suggesting that Drosophila γ-tubulin is a phosphoprotein (Figure 5A). The phosphatase-sensitive acidic forms are absent or severely diminished in extracts from dwee1 mutant embryos (Figure 5B).
Figure 5. dwee1 Is Required for γ-Tubulin Phosphorylation In Vivo.
(A) γ-tubulin is a phosphoprotein in vivo. Extracts from 0–2-hr-old wild-type embryos were incubated with (+ppase) or without (−ppase) Lambda phosphatase, separated in 2D gels, and Western blotted with mouse (shown) or rabbit (not shown) anti-γ-tubulin antibodies. Five isoforms of γ-tubulin are reproducibly detected (See WT in [E] also). Two isoforms are not detected after treatment with Lambda phosphatase (arrows).
(B) γ-tubulin phosphorylation requires dwee1 but not grp. Extracts from 0–2-hr-old wild-type, dwee1 mutant, or grp mutant embryos were separated in 2D gels and Western blotted for γ-tubulin. Phosphatase-sensitive isoforms of γ-tubulin are not detected in dwee1 mutants but are seen in grp mutants (arrows).
Given that interphase is truncated in dwee1 mutants, we addressed the possibility that loss of γ-tubulin phosphorylation is a consequence of changes in cell cycle profile. However, extracts from grp mutant embryos that exhibit truncated interphases retained the phosphatase-sensitive γ-tubulin isoforms (Figure 5B). We conclude that interphase shortening does not lead to loss of γ-tubulin phosphorylation and that dwee1 is required for γ-tubulin phosphorylation in vivo. Despite testing a range of conditions, we have not been able to phosphorylate γ-tubulin with recombinant dWee1 in vitro, although GST-dWee1 readily autophosphorylates and phosphorylates Cdk1 in our assays [7]. Either dWee1 regulates γ-tubulin phosphorylation indirectly or γ-tubulin phosphorylation by dWee1 requires a cofactor.
The known role of Wee1 homologs in cell cycle regulation is accomplished through a single substrate, Cdk1. Elevation of bulk Cdk1 activity in an otherwise wild-type background, however, did not produce dwee1-specific phenotypes. Therefore, if dWee1 influences spindle organization or positioning via Cdk1, it would have to regulate Cdk1 locally, at the embryo cortex for example. The attachment of centrosomes to the cortex is mediated by microtubules. In the absence of dWee1, Cdk1 activity would be higher locally, i.e., between the centrosome and the cortex, and could inhibit microtubule growth in this region, leading to the displacement of centrosomes. This idea is consistent with observations that increased Cdk1 activity destabilizes microtubules during nuclear migration in Drosophila embryos [16]. In six cycB embryos, dWee1 could still inhibit Cdk1 locally to allow normal nuclear and spindle positioning. This possibility can be addressed with a Cdk1 mutant that cannot be phosphorylated by dwee1 and should mimic the loss of dwee1. We have attempted to introduce into syncytial embryos (which are prezygotic transcription) such a mutant in which Y14 and T15 have been altered, Cdk1AF, by expressing it in females. Unfortunately, females fail to lay eggs after induction of Cdk1AF, suggesting disruption of oogenesis and precluding further analysis (J.S., D.R.K., K.K., and T.T.S., unpublished data).
An alternate approach to introduction of Cdk1AF is to induce dWee1-antagonizing phosphatases, Cdc25string and Cdc25twine, in embryos. Indeed, such experiments have been described before [22]. Increasing the maternal Cdc25 gene dose by up to 4-fold in various combinations of the two Drosophila Cdc25 homologs produced increased mRNA and protein in embryos and led to an extra syncytial nuclear division before cellularization. This division and preceding syncytial divisions, however, were normal in examination of both fixed and live embryos. No mitotic abnormalities, which are readily apparent throughout dwee1 mutant embryos, were seen in embryos with elevated Cdc25. This is consistent with our observation that bulk elevation of Cdk1 activity does not produce dwee1-specific phenotypes. Instead, localized regulation of Cdk1 by dWee1, which would still be present in embryos harboring extra Cdc25, could explain the apparently normal divisions in these embryos.
Another explanation for spindle phenotypes in dwee1 mutants is suggested by our finding that dWee1 shows physical interaction with components of the γTuRC and that dwee1 influences the phosphorylation status of γ-tubulin in vivo. In this model, dWee1 promotes the phosphorylation of γ-tubulin, either directly or indirectly. In dwee1 mutants, loss of γ-tubulin phosphorylation could compromise microtubule-dependent attachments between centrosomes and the cortex. A test of this model will require identification and mutation of dwee1-dependent phosphoacceptor residues in γ-tubulin. Interestingly, the budding-yeast γ-tubulin homolog, Tub4p, is phosphorylated on a tyrosine residue during G1, but the responsible kinase is yet to be identified [21]. A phosphomimetic mutation of Tub4p affects the number and organization of microtubules and causes transient nuclear-positioning abnormalities [21]. Thus, it is possible that in both yeast and in fly, the phosphorylation status of γ-tubulin plays a role in centrosome and nuclear positioning via interactions with the cortex.
At present, we cannot distinguish between the two above explanations for dwee1 phenotypes; the explanations are not mutually exclusive. Neither, however, is predicted by previous models that describe how Wee1 homologs act to regulate entry into mitosis. Human Wee1, for example, resides in the nucleus during interphase and is proposed to prevent nuclear accumulation of Cdk1 activity and nuclear-envelope breakdown, an initiating event in mitosis. Such models can explain Wee1’s role in regulating when mitosis occurs, but our results indicate that Wee1 can also regulate where (relative to the cortex) mitosis occurs.
The first 11 nuclear divisions proceed normally in dwee1 mutants. Therefore, dwee1-dependent regulation of spindle organization or positioning is not essential for mitosis per se. In cycle 12, nuclei are at the cortex and at twice the density (i.e., closer together) compared to those in cycle 11. We reason that manifestation of dwee1-specific spindle interactions in later cortical cycles is a consequence of increasing nuclear density with each cycle that brings neighboring spindles closer together. In such a situation, protection offered by actin furrows may be essential to keep spindles apart. Detachment of centrosomes from the cortex would distance the spindles from furrows, allowing neighbors to interact (Figure 6B).
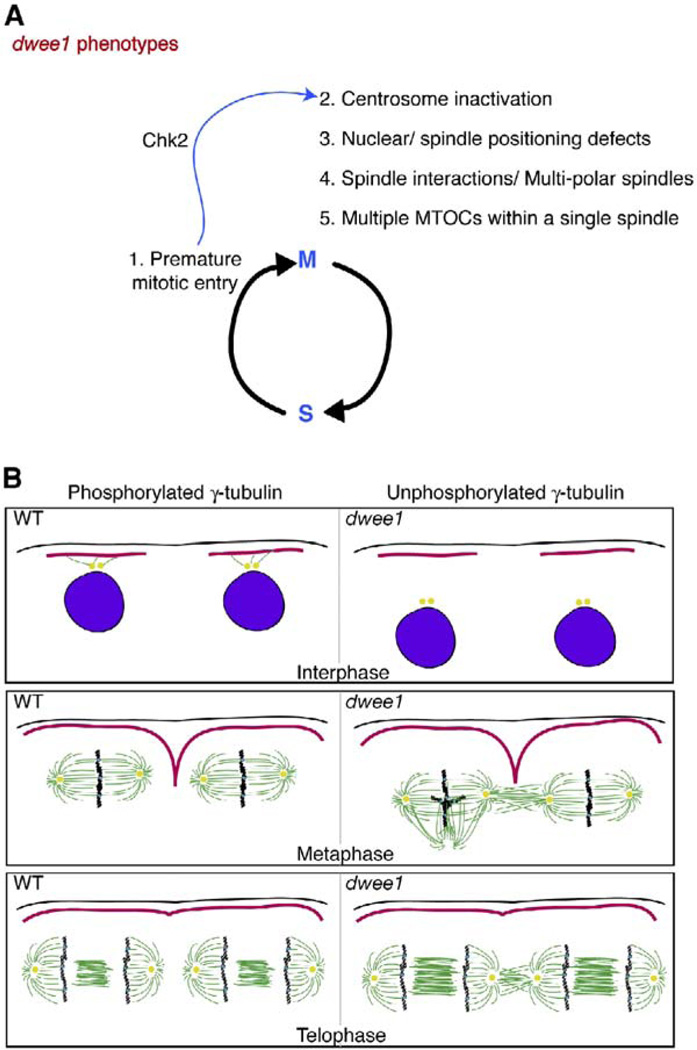
Figure 6. Model for dwee1 Functions during Cortical Syncytial Cycles.
(A) A summary of phenotypes in dwee1 mutant embryos. dWee1 inhibits Cdk1. In dwee1 mutants, Cdk1 is not inhibited and nuclei enter mitosis prematurely (phenotype 1). Consequently, a Chk2-dependent checkpoint is invoked, inactivates the centrosome (2), and prevents cellularization of the embryo (not shown); mutations in chk2 rescue these phenotypes [1]. Additional phenotypes include defects in nuclear and spindle positioning (3), spindle interactions (4), and multiple MTOCs that arise during mitosis within a single spindle (5). Phenotype 3 is partially rescued by chk2 mutations, and phenotypes 4 and 5 are not.
(B) A model to explain how nuclear and centrosome displacement from the cortex could lead to spindle collisions. In dwee1 mutants, centrosomes (yellow) and mitotic spindles are displaced from the cortex (black) and the protection of pseudocleavage furrows (red), allowing spindles to interact. γ-tubulin phosphorylation is dwee1 dependent (this study) and could play a role in nucleation of microtubules that attach centrosomes to the cortex. Pseudocleavage furrows have retracted by late anaphase or telophase. Therefore, another dwee1-dependent mechanism may exist to prevent spindle interactions during later stages in mitosis.
We do not know whether dwee1 also plays a role in spindle morphogenesis and centrosome positioning in cell cycles beyond cortical syncytial cycles. We do know, however, that dwee1 is needed to ensure fidelity of cell division in larvae; larval neuroblasts in dwee1 mutants show elevated mitotic index and ploidy [7]. It would be interesting to determine the basis for this requirement and whether dwee1 has a role in the positioning of the spindle in cell divisions where a specific cortical attachment of the spindle is required, such as in the asymmetric cell divisions of neuroblast lineages.
Conclusions
We present several lines of data that collectively suggest a requirement for dWee1 in centrosome function and spindle morphogenesis. Importantly, these roles translate into a requirement for dWee1 in not only temporal but also spatial regulation of mitosis. We offer two mechanistic models, which are not mutually exclusive, to account for these results: localized regulation of Cdk1 by dWee1 and phosphoregulation of γTuRC. Further analysis will be needed to test these models, but it is clear that the requirement for dWee1 cannot be explained by simple regulation of bulk Cdk1 activity. In this regard, Wee1 homologs may be likened to other kinases, such as Plk and Aurora B, that have multiple roles in mitosis through multiple substrates. Localized activity of master regulatory kinases such as these is likely to coordinate many distinct cell-division events—such as spindle movements, chromosome segregation, and cytokinesis—to allow faithful segregation of genetic information into daughter cells.
Experimental Procedures
Stocks
Wild-types are of Sevelin strain. The mutant alleles used here have been described before: dwee1ES1 and Df(2L)dwee1W05 [8]; grp1 [23]; dd42 [24]; and mnk6006 (DmChk2) [14]. grp, chk2, dwee1, and dwee1, chk2 double mutants were balanced over a CyO balancer carrying the actin-gfp transgene; homozygous mutants were identified by the lack of a GFP signal or curly wings. 17238-GFP, six cycB, and HA-dWee1 transgenic stocks have been described before [8, 15, 17, 25]. 17238-GFP transgenes were expressed constitutively, whereas HA-dWee1 is expressed from a heat-inducible hsp70 promoter. Fly stocks carrying dwee1 or grp mutations and 17238-GFP transgenes were constructed with standard Drosophila genetics.
Immunofluorescence
Fixation and antibody staining of syncytial blastoderm embryos to visualize microtubules and centrosomes was carried out as previously described [7]. To visualize F-actin, we fixed embryos as described by Foe [20] and stained them with phalloidin (1:10) or actin-furrow-marking antibodies against phosphorylated tyrosine [26]. Embryos were stained with the following primary antibodies: mouse monoclonal anti-phosphotyrosine 1:100 (Upstate Biotechnology), mouse monoclonal anti-β-tubulin 1:100 (Iowa developmental hybridoma bank), rabbit polyclonal anti- γs-tubulin 1:100 (Sigma), rabbit polyclonal anti-Cnn 1:1000 [27], and rabbit polyclonal anti-Dgrip84 (1:100) [28]. Secondary antibodies conjugated to fluorescein isothiocyanate (FITC) or rhodamine (Jackson) were used at 1:250. DNA was visualized by staining with 10 µg/ml bisbenzimide (Hoechst 33258 from Molecular Probes) for 10 min. Embryos were analyzed on a Leica DMR microscope with a Sensicam CCD camera and Slidebook software (Intelligent Imaging Innovations). The shortest distance between the center of the Dgrip84 dot and the embryo cortex was measured with the scale tool in Slidebook.
Time-Lapse Confocal Microscopy
Embryos derived from strains expressing 17238-GFP were analyzed by time-lapse confocal microscopy with a Leica DMRXA microscope equipped with a spinning-disk confocal laser system (Perkin Elmer) and CCD camera (Hamamatsu) as previously described [7]. To induce DNA damage, we identified embryos in interphase of cycle 11 under the microscope, removed them from the microscope, and irradiated them while they were still on coverslips at 2.2 rads/s in a TOR-REX120D X-ray generator (Astrophysics Research) set at 5 mA and 115 kV. For 17238-GFP detection, stacks of 12 to 15 images at 1.0 micron distance in the z direction were captured every 30 or 60 s at 250 ms exposures. We find that relying on collapsed images can give false indication of microtubules being shared by neighboring spindles; analysis in 3D by inspection of individual z sections was essential.
H1 Kinase Assays
Kinase assays were performed on extracts from 1–2-hr-old embryos as previously described [7]. Blots were stained with Ponceau-S (Sigma) to determine total histone H1 levels. 32P incorporation was detected by a Phosphorimager (Molecular Dynamics). 32P signal was quantified by liquid scintillation counting of excised, Ponceau-S-stained H1 bands.
Heat-Shock Expression of HA-dWee1
Flies homozygous for the hs HA-dWee1 transgene were subjected to heat shock at 37°C for 1 hr. Embryos were collected for 10 hr after heat shock. Embryos were then heat-shocked for an additional hour at 37°C. Embryos were dechorionated in 50% bleach, frozen in liquid nitrogen, and stored at −80°C. For negative controls, wild-type embryos were colleted for 8 hr, dechorionated, and frozen similarly.
Affinity Purification of HA-dWee1
Fifteen grams each of frozen 1–10 hr embryos from hs HA-dWee1 flies and 0–8 hr embryos from wild-type flies were ground to a powder under liquid nitrogen in a Krupp’s coffee grinder as described [29]. Embryo powder was placed in a glass beaker that was previously frozen in liquid nitrogen and allowed to warm at room temperature until the powder started to thaw. Twenty-five milliliters of room-temperature extract buffer (50 mM Hepes-KOH [pH 7.6], 100 mM β-glycerolphosphate, 50 mM NaF, 1 mM MgCl2, 1 mM EGTA, 5% glycerol, 0.25% Tween-20, and 2 mM PMSF) was then added, and the powder was immediately stirred with a spatula to resuspend it. Embryo powder was further dissolved by stirring with a stir bar for 10 min at 4°C. Next, extracts were spun at 10,000 × g for 10 min and 100,000 × g for 1 hr at 4°C. The following steps were carried out in a 4°C cold room unless otherwise noted. Extract was added to 0.45 ml of protein A beads (BioRad) bound to 0.45 mg of rabbit polyclonal anti-HA antibody [29] in 15 ml tubes and incubated for 3 hr. Beads were washed twice in 15 ml wash buffer (25 mM NaF, 0.15% Tween 20, 200 µg/ml BSA) and transferred to Bio- Spin chromatography columns (BioRad). Columns were washed with 5 ml of wash buffer without BSA. Four elutions were performed at room temperature by adding 250 µl elution buffer (50 mM Hepes- KOH [pH 7.6], 100 mM β-glycerolphosphate, 1 mM MgCl2, 1 mM EGTA, 5% glycerol, 25 mM NaF, 0.01% Tween-20 and 0.46 mg/ml HA dipeptide). The first elution was discarded and the next three elutions were pooled. Proteins were precipitated by the addition of trichloroacetic acid to 10% final volume. Precipitate was resuspended in 1× SDS-sample buffer (65 mM Tris-HCl [pH 6.8], 3% SDS, 10% glycerol, bromophenol blue, and 5% β-mercaptoethanol). Samples were boiled for 5 min and run on 12% SDS-PAGE gels. Gels were stained with Coomassie blue (BioRad) according to the manufacturer’s instructions and destained overnight in 25% MeOH and 7% acetic acid. Coomassie-stained bands specific to the HA-dWee1 eluate along with gel slices of corresponding molecular weight from the wild-type control lane were excised with a razor blade and stored in eppendorf tubes at −80°C until trypsin digestion was performed.
In-Gel Trypsin Digestion
All steps were performed at room temperature unless otherwise noted. Proteins that were from excised bands and specific to HA-dWee1-expressing embryos and the corresponding MW bands from wild-type controls were rocked for 30 min at 60°C in 150 µl reduction solution (45 mM DTT, 100 mM ammonium bicarbonate) in .65 µl eppendorf tubes and were then cooled to room temperature. Ten microliters of alkylation solution (0.5 M Iodoacetamide, 100 mM ammonium bicarbonate) was then added to the reduction solution, and gel slices were rocked for 30 min in the dark. Gel slices were equilibrated by vortexing in 500 µl 100 mM ammonium bicarbonate for 30 min, followed by aspiration and then addition of 500 µl 50% Acetonitrile/50 mM ammonium bicarbonate for 30 min. This solution was removed, 50 µl 100% acetonitrile was added, and tubes were left open in a hood for 10 min. Gel slices were transferred to clean .65 µl eppendorf tubes and dried in a speed vac for 20 min. A 15 µl digestion solution (.02 g/ml modified trypsin [Promega] in 25 mM ammonium bicarbonate) was added to each dried gel slice in 5 µl increments, with 10 min between each addition. Gel slices were then covered with 25 mM ammonium bicarbonate over 20 min. Tubes were closed and incubated at 37°C for 24 hr with gentle motion. After digestion, 1 µl 88% formic acid was added to each tube, and samples were sonicated for 20 min. Recovered peptides were desalted and concentrated with C18Zip-Tips (Millipore), and peptides were eluted in 50% (v/v) acetonitrile: water. Samples were then cocrystallized with matrix (α-cyano-4-hydroxy-trans-cinamic acid, Hewlett Packard) on gold-coated plates for mass-spectrometric analysis.
Mass Spectrometry and Protein Identification
Matrix-assisted laser desorption ionization time of flight (MALDITOF) mass spectrometry (Voyager-DE STR, Perkin Elmer) was used to obtain peptide mass information for each unknown sample. Peptide masses obtained from control gel slices were subtracted from those of HA-dWee1 gel slices before peptide-mass fingerprinting (PMF) analysis. Identification of unknown proteins from HA-dWee1 eluates was then achieved by peptide-mass fingerprinting with ProFound (http://prowl.rockefeller.edu/profound_bin/WebProFound.exe) and ProteinProspector (http://prospector.ucsf.edu/), searching against Drosophila entries in the National Center for Biotechnology (NCBI) nonredundant protein database.
Immunoprecipitation
Embryo extracts were prepared by crushing 500 µl 0–3 hr or 0–8 hr embryos in Extract buffer (see affinity purification) with a glass homogenizer at 4°C. Extracts were cleared by spinning two times at 15,000 × g at 4°C in a microcentrifuge. Extracts were precleared with 15 µl protein A beads (Amersham), which were recovered by centrifugation at 5,000 × g for 1 min and discarded. Five micrograms of rabbit polyclonal anti-dWee1 antibody [8] was added to the precleared extract and incubated for 1 hr at 4°C. Fifteen microliters of protein A beads were then added and incubated for another 1 hr at 4°C. Beads were recovered by centrifugation as described above, washed three times in extract buffer at room temperature, and then boiled in 1× SDS-sample buffer (see Affinity Purification of HA-dWee1).
Electrophoresis and Western Blots
Protein samples were separated on 10% SDS-PAGE gels and transferred to PVDF membrane (Millipore) on a Multiphor II semidry transfer apparatus (Amersham). Membranes were blocked in PBT + 5% milk for 1 hr. The following primary antibodies were added in block for 1 hr: mouse monoclonal anti-γ-tubulin 1:1,000 (Sigma), rabbit polyclonal anti-γ-tubulin 1:1,000 (Sigma), rabbit polyclonal anti-Grip84 1:500 [28], rabbit polyclonal anti-Grip91 1:1000 [28], rabbit polyclonal anti-dWee1 1:1,000 [8], mouse anti-PSTAIR (Cdk1) 1:1,000 (Sigma), and mouse anti-β-tubulin 1:500. HRP-conjugated anti-mouse and anti-rabbit secondary antibodies (Amersham) were added at 1:5,000 and 1:10,000, respectively, for 1 hr in block. Pierce Supersignal and Kodak X-Omat film were used for detection of bound antibodies.
Preparation of Extracts for 2D Analysis
Embryos from Sevelin, ES1/Df, or grp1 flies were homogenized in extract buffer and cleared as described for immunoprecipitation. Protein was precipitated by addition of nine volumes ice-cold acetone and incubation for 2 hr at −20°C. Protein was pelleted by centrifugation at 8000 × g for 10 min, and pellets were resuspended in urea lysis buffer (6 M urea, 2 M thiourea, 2% Chaps, 10 mM Na-Hepes [pH 8.0], 20 mM DTT). For phosphatase treatment, 0–3 hr Sevelin embryos were homogenized in Lambda phosphatase buffer + MnCl2 (NEB) ± phosphatase inhibitors (1 mM o-Vanadate, 5 mM NaF, 20 mM β-glycerolphosphate) and were cleared as described for immunoprecipitation. Fifty milliliters of extract was incubated at 30°C with 100 units lambda phosphatase (NEB) for 30 min. The reactions were then subjected to acetone precipitation, and protein pellets were resuspended in urea lysis buffer.
Two-Dimensional Electrophoresis
One hundred micrograms of embryo extracts was separated in the 1st dimension on 18 cm Immobilone dry strips (pH 4–7; Amersham) run on a Multiphor II (Amersham) for 80 kVh. Focused strips were either frozen at −80°C or processed for the 2nd dimension immediately. Strips were reduced in Equilibration buffer (50 mM Tris [pH 6.8], 6 M Urea, 30% glycerol, 2% SDS) + 10 mM DTT for 15 min and then alkylated in Equilibration buffer + 200 mM iodoacetamide and trace bromophenol blue for 15 min. Strips were then loaded onto 10% SDS-PAGE gels for separation in the 2nd dimension. Electrophoresis and Western blots were carried out as described above.
Supplementary Material
Acknowledgments
We thank Yixiang Zheng for antibodies against γTURC proteins; Shelagh Campbell for dWee1 antibodies; Gerald Schubiger for six cycB flies; Mark Winey, Christine Weise, Chad Pearson, and Jackie Vogel for critical reading of the manuscript; and Tod Duncan, Yi-xiang Zheng, Bill Sullivan, and Bill Theurkauf for helpful comments. This work was supported by grants from the American Cancer Society and the National Institutes of Health (NIH) to T.T.S., a grant from the NIH to D.R.K, and an NIH grant for training in signal transduction and cell cycle regulation to J.S.
Footnotes
Supplemental Data
Supplemental Data include one figure and are available with this article online at http://www.current-biology.com/cgi/content/full/15/17/1525/DC1/.
References
- 1.Russell P, Nurse P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 1987;49:559–567. doi: 10.1016/0092-8674(87)90458-2. [DOI] [PubMed] [Google Scholar]
- 2.Gould KL, Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- 3.Nurse P. Genetic control of cell size at cell division in yeast. Nature. 1975;256:547–551. doi: 10.1038/256547a0. [DOI] [PubMed] [Google Scholar]
- 4.Campbell SD, Sprenger F, Edgar BA, O’Farrell PH. Drosophila Wee1 kinase rescues fission yeast from mitotic catastrophe and phosphorylates Drosophila Cdc2 in vitro. Mol. Biol. Cell. 1995;6:1333–1347. doi: 10.1091/mbc.6.10.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGowan CH, Russell P. Human Wee1 kinase inhibits cell division by phosphorylating p34cdc2 exclusively on Tyr15. EMBO. J. 1993;12:75–85. doi: 10.1002/j.1460-2075.1993.tb05633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harvey SL, Kellogg DR. Conservation of mechanisms controlling entry into mitosis: Budding yeast wee1 delays entry into mitosis and is required for cell size control. Curr. Biol. 2003;13:264–275. doi: 10.1016/s0960-9822(03)00049-6. [DOI] [PubMed] [Google Scholar]
- 7.Stumpff J, Duncan T, Homola E, Campbell SD, Su TT. Drosophila Wee1 kinase regulates Cdk1 and mitotic entry during embryogenesis. Curr. Biol. 2004;14:2143–2148. doi: 10.1016/j.cub.2004.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price D, Rabinovitch S, O’Farrell PH, Campbell SD. Drosophila wee1 has an essential role in the nuclear divisions of early embryogenesis. Genetics. 2000;155:159–166. doi: 10.1093/genetics/155.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foe VE, Odell GM, Edgar BA. Mitosis and morphogenesis in the Drosophila embryo. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor: CSHL Press; 1993. pp. 149–300. [Google Scholar]
- 10.Yu KR, Saint RB, Sullivan W. The Grapes checkpoint coordinates nuclear envelope breakdown and chromosome condensation. Nat. Cell Biol. 2000;2:609–615. doi: 10.1038/35023555. [DOI] [PubMed] [Google Scholar]
- 11.Sibon OC, Stevenson VA, Theurkauf WE. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature. 1997;388:93–97. doi: 10.1038/40439. [DOI] [PubMed] [Google Scholar]
- 12.Sibon OC, Laurencon A, Hawley R, Theurkauf WE. The Drosophila ATM homologue Mei-41 has an essential checkpoint function at the midblastula transition. Curr. Biol. 1999;9:302–312. doi: 10.1016/s0960-9822(99)80138-9. [DOI] [PubMed] [Google Scholar]
- 13.Sibon OC, Kelkar A, Lemstra W, Theurkauf WE. DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nat. Cell Biol. 2000;2:90–95. doi: 10.1038/35000041. [DOI] [PubMed] [Google Scholar]
- 14.Takada S, Kelkar A, Theurkauf WE. Drosophila checkpoint kinase 2 couples centrosome function and spindle assembly to genomic integrity. Cell. 2003;113:87–99. doi: 10.1016/s0092-8674(03)00202-2. [DOI] [PubMed] [Google Scholar]
- 15.Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stiffler LA, Ji JY, Trautmann S, Trusty C, Schubiger G. Cyclin A and B functions in the early Drosophila embryo. Development. 1999;126:5505–5513. doi: 10.1242/dev.126.23.5505. [DOI] [PubMed] [Google Scholar]
- 17.Ji JY, Haghnia M, Trusty C, Goldstein LS, Schubiger G. A genetic screen for suppressors and enhancers of the Drosophila cdk1-cyclin B identifies maternal factors that regulate microtubule and microfilament stability. Genetics. 2002;162:1179–1195. doi: 10.1093/genetics/162.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar BA, Lehman DA, O’Farrell PH. Transcriptional regulation of string (cdc25): A link between developmental programming and the cell cycle. Development. 1994;120:3131–3143. doi: 10.1242/dev.120.11.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan W, Theurkauf WE. The cytoskeleton and morphogenesis of the early Drosophila embryo. Curr. Opin. Cell Biol. 1995;7:18–22. doi: 10.1016/0955-0674(95)80040-9. [DOI] [PubMed] [Google Scholar]
- 20.Foe VE, Field CM, Odell GM. Microtubules and mitotic cycle phase modulate spatiotemporal distributions of F-actin and myosin II in Drosophila syncytial blastoderm embryos. Development. 2000;127:1767–1787. doi: 10.1242/dev.127.9.1767. [DOI] [PubMed] [Google Scholar]
- 21.Vogel J, Drapkin B, Oomen J, Beach D, Bloom K, Snyder M. Phosphorylation of gamma-tubulin regulates microtubule organization in budding yeast. Dev. Cell. 2001;1:621–631. doi: 10.1016/s1534-5807(01)00073-9. [DOI] [PubMed] [Google Scholar]
- 22.Edgar BA, Datar SA. Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila’s early cell cycle program. Genes Dev. 1996;10:1966–1977. doi: 10.1101/gad.10.15.1966. [DOI] [PubMed] [Google Scholar]
- 23.Fogarty P, Campbell SD, Abu-Shumays R, Phalle BS, Yu KR, Uy GL, Goldberg ML, Sullivan W. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr. Biol. 1997;7:418–426. doi: 10.1016/s0960-9822(06)00189-8. [DOI] [PubMed] [Google Scholar]
- 24.Barbosa V, Yamamoto RR, Henderson DS, Glover DM. Mutation of a Drosophila gamma tubulin ring complex subunit encoded by discs degenerate-4 differentially disrupts centrosomal protein localization. Genes Dev. 2000;14:3126–3139. doi: 10.1101/gad.182800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobs HW, Knoblich JA, Lehner CF. Drosophila Cyclin B3 is required for female fertility and is dispensable for mitosis like Cyclin B. Genes Dev. 1998;12:3741–3751. doi: 10.1101/gad.12.23.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grevengoed EE, Fox DT, Gates J, Peifer M. Balancing different types of actin polymerization at distinct sites: Roles for Abelson kinase and Enabled. J. Cell Biol. 2003;163:1267–1279. doi: 10.1083/jcb.200307026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Megraw TL, Kao LR, Kaufman TC. Zygotic development without functional mitotic centrosomes. Curr. Biol. 2001;11:116–120. doi: 10.1016/s0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- 28.Gunawardane RN, Martin OC, Cao K, Zhang L, Dej K, Iwamatsu A, Zheng Y. Characterization and reconstitution of Drosophila gamma-tubulin ring complex subunits. J. Cell Biol. 2000;151:1513–1524. doi: 10.1083/jcb.151.7.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mortensen EM, McDonald H, Yates J, 3rd, Kellogg DR. Cell cycle-dependent assembly of a Gin4-septin complex. Mol. Biol. Cell. 2002;13:2091–2105. doi: 10.1091/mbc.01-10-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.