Abstract
Investigation into the causes underlying the rapid, global amphibian decline provides critical insight into the effects of changing ecosystems. Hypothesized and confirmed links between amphibian declines, disease, and environmental changes are increasingly represented in published literature. However, there are few long-term amphibian studies that include data on population size, abnormality/injury rates, disease, and habitat variables to adequately assess changes through time. We cultured and identified microorganisms isolated from abnormal/injured and repressed tissue regeneration sites of the endangered Ozark Hellbender, Cryptobranchus alleganiensis bishopi, to discover potential causative agents responsible for their significant decline in health and population. This organism and our study site were chosen because the population and habitat of C. a. bishopi have been intensively studied from 1969–2009, and the abnormality/injury rate and apparent lack of regeneration were established. Although many bacterial and fungal isolates recovered were common environmental organisms, several opportunistic pathogens were identified in association with only the injured tissues of C.a. bishopi. Bacterial isolates included Aeromonas hydrophila, a known amphibian pathogen, Granulicetella adiacens, Gordonai terrae, Stenotrophomonas maltophilia, Aerococcus viridans, Streptococcus pneumoniae and a variety of Pseudomonads, including Pseudomonas aeruginosa, P. stutzeri, and P. alcaligenes. Fungal isolates included species in the genera Penicillium, Acremonium, Cladosporium, Curvularia, Fusarium, Streptomycetes, and the Class Hyphomycetes. Many of the opportunistic pathogens identified are known to form biofilms. Lack of isolation of the same organism from all wounds suggests that the etiological agent responsible for the damage to C. a. bishopi may not be a single organism. To our knowledge, this is the first study to profile the external microbial consortia cultured from a Cryptobranchid salamander. The incidence of abnormalities/injury and retarded regeneration in C. a. bishopi may have many contributing factors including disease and habitat degradation. Results from this study may provide insight into other amphibian population declines.
Introduction
The amphibian decline controversy has focused on many factors that affect amphibian populations to varying degrees [1], including habitat loss and degradation, climate change, pollution, increased ultraviolet B (UV-B) radiation, direct exploitation, introduced species and disease, including infectious disease [2], [3]. Although evidence of disease in amphibian populations is not new, early literature on amphibian health in natural populations is quite scattered, deals primarily with anurans and/or local problems, or was initiated because of concerns related to supply and demand and decline of commercial harvest [4]–[6]. In the late 1960's and early 1970's, various biological supply houses noted problems in the commercial supply of amphibians causing Maugh [7] to comment on “the apparent short supply and diseased state of amphibians collected in nature”.
The review of mycoses of amphibians by Reichenback-Klinke and Elkan [8] primarily focuses on Basidiobolus ranarum and Saprolegnia parasitica and indicates the dearth of knowledge relating to the distribution and importance of microfungi associated with amphibians. With few exceptions, this void continued for the next two decades [9]–[12]. The impetus for research on amphibian microbes became a major focal point of the 1st World Herpetology Congress in Canterbury, England in 1989, where herpetologists shared their observations on declining frog populations and developed initial strategies to investigate the potential problems. The discovery that the chytridiomycete Batrachochytrium dendrobatidis, a zoosporic fungus related to infectious oomycete water molds, Saprolegnia spp., was capable of causing lethal dermatitis in amphibians led to a proliferation of studies [13]. After reviewing studies of microbes implicated in amphibian population declines, including chytridiomycosis, Ranavirus disease, saprolegniosis and Ribieroia spp., Daszak et al. [14] concluded that “available data provide the clearest link for the fungal disease amphibian chytridiomycosis”. Herpetological Review dedicated an entire section to amphibian chytridiomycosis geographical distribution [15]–[17].
The advances in research on the secretions, structure, and functions of amphibian integument and their products reveal a remarkable complexity of bioactive secretions and diversity of amines, peptides, alkaloids, bufodienolides, and other compounds [18], [19]. The presence of antimicrobial agents in amphibian skin hypothesized by Csordas and Michl [20] and Croce et al. [21] has led to an increased interest in the relationship of the bacteria and fungi present in the skin of amphibians and the antimicrobial peptides and metabolites that they produce [22]–[30]. Some of these peptides and alkaloids can inhibit the growth of pathogenic fungi [31], [32] and common cutaneous bacteria from the terrestrial salamander Plethodon cinerus can inhibit pathogenic fungi [33]. Evidence also suggests that symbiotic bacteria may contribute to innate immune defense of some amphibians [30]. While evidence suggests that these antimicrobial compounds and symbiotic bacteria can provide some level of protection for amphibians against microbial invaders [30]–[33], the connection between disease and amphibian decline has been confirmed for some amphibian populations [34]. However, long-term amphibian studies, especially those including population and environmental data, are so rare that we have very few data to support many claims related to decline or changes in, or the health of, wild amphibian populations [35]. One long-term study subject, the Ozark Hellbender, Cryptobranchus alleganensis bishopi, and its habitat within a 4.6 km section of the North Fork of White River (NFWR), has been the subject of numerous investigations since the intensive 110 day surveys conducted during 1969–1971 [36], [37].
In 1969, the 4.6 km research section within the North Fork of White River (NFWR), Ozark County, Missouri, was a crystalline, substantially spring-fed stream located in the least densely human populated area of the second least densely populated county in the state (9 people/sq mi; [38]). Only one rarely used sportsman's cabin graced the banks of the research section in 1969. The springs and occasionally the river were used as drinking water by some locals and visitors. From 1969 to 1980, 169 days of skin-diving surveys, coupled with environmental sampling, were conducted in this section, including some in every year and within every calendar month, but not every month of every year [39]. Population and ecological studies of the aquatic Ozark Hellbender C. a. bishopi and its habitat were conducted in this section during 1969–1971 [36], [37]. Other ecological studies included year-round water quality, benthic habitat, macro-invertebrate structure, cottid fish diet studies, and numerous shorter-term studies [36], [40]. These early studies in the NFWR found an immense and healthy population of C. a. bishopi, as many as 428 individuals/km, and almost no abnormalities/injuries. Only 2.9% of 479 individuals observed in 1969 were abnormal/injured and they exhibited rapid regeneration capabilities [36], [39], [41]. Additional surveys between 1972 to 1980 continued to show immense and healthy populations of Ozark Hellbenders [27], [42]. All surveys during this time period ranged from between 9–12 individual Ozark hellbenders collected per hour per person (Nickerson et al, unpublished data). Reassessment of the ecological characteristics of the NFWR conducted in 2004–2007 revealed extensive habitat alteration and degradation, including increased land development, siltation, sedimentation, and water quality degradation [43]. Community changes included algal and nuisance aquatic vegetation blooms, otter establishment, and fish and macroinvertebrate community alterations [43]. Canoe use within the NFWR significantly increased [43]. Intensive surveys of the NFWR hellbender population conducted in 2005 yielded only 55 individuals, of which 26 (i.e., 47%) had visible abnormalities/injuries, including loss of limbs, limbs with exposed bones, and degeneration of other tissues which did not regenerate or had remarkably retarded regeneration (J. Briggler, unpublished data). The high prevalence of abnormalities/injuries and the lack of the historically characteristic (rapid) regeneration of injured/affected tissue in hellbenders in the NFWR was the impetus for our examination of the microbial community associated with the observed abnormalities/injuries.
Results
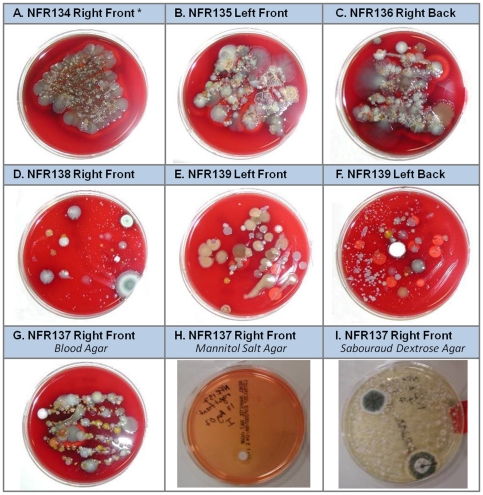
Our results reflect our strategy to optimize chances for successful culture of potentially pathogenic microorganisms by using three different media for each sample; blood agar (all purpose growth media), mannitol salts (differential and selective media), Sabouraud's (differential and selective media often used to isolate fungi) (Figure 1).
Figure 1. Representative microbial flora cultured from C. a. bishopi on three different media.
Swabs from injured (or uninjured control) tissues of six adult hellbenders were streaked onto three different microbiological culture media: Sheep's blood agar (A–G), Mannitol Salt Agar (H), and Sabouraud Dextrose Agar (I). * Indicates uninjured control sample.
An evaluation of the microbial flora sampled from C. a. bishopi indicated the presence of common environmental flora from both abnormal/injured and uninjured limbs. A wide variety of both Gram positive and Gram negative bacteria were isolated. While no consistent pattern of bacterial colonization was observed between uninjured and abnormal/injured body parts (Tables 1 and 2), several interesting microbial associations were observed. The genus Aeromonas was identified in 9 separate occasions, although the amphibian pathogenic species Aeromonas hydrophila, was identified only once from an abnormal/injured animal (NFWR 139 - lower lip). Likewise, certain organisms that can cause human infection were also identified, including the opportunistic pseudomonal pathogens Pseudomonas aeruginosa (NFWR 135 - right back limb), P. stutzeri (NFWR 134 - right back; NFWR- 135 right back), P. alcaligenes (NFWR – right front), and the pseudomonal-like pathogen, Stenotrophomonas maltophilia (NFWR 136 – left front limb). In addition, the poultry pathogen Riemerella anatipestifer [44] was also isolated only from abnormal/injured animals. On rare occasion, one bacterial species would represent the vast majority of the colonies from a given abnormal/injured sample as exemplified by the known human pathogen Granulicatella adiacens (formerly Streptococcus adiacens; [45]), which represented 91% of the 163 bacterial isolates found on the blood agar plate associated with the NFWR 139 lower lip sample. When using Sabouraud Dextrose Agar, only fungi were isolated, except for one plate (from the injured animal NFWR 135 - left front limb) where 4 colonies of Kocuria kristinae (previously Micrococcus) were found, which is a common inhabitant on human skin that has been increasingly associated with infectious disease in humans [46].
Table 1. Bacteria identified on Blood Agar.
| NFWR | Limb sampled | Colony Forming Units | Closest match |
| 134 | Right Front* | TNTC | Aeromonas sobria |
| Right Back | 15 | Aeromonas sobria; Pseudomonas stutzeri; Riemerella anatipestifer⧫; Unidentified Gram positive species | |
| 135 | Right Back | TNTC | Kocuria varians; Microbacterium luteolum⧫; Pseudomonas stutzeri; Pseudomonas aeruginosa |
| Right Front | TNTC | Roseomonas cervicalis⧫; Pseudomonas alcaligenes; Brevundimonas diminuta/vesicularis | |
| Left Back | TNTC | Aeromonas sobria (3 morphologically different colonies) | |
| Left Front | TNTC | Kocuria rosea; Kocuria kristinae; Aeromonas sobria; Pseudomonas stutzeri | |
| 136 | Left Front | TNTC | Pseudomonas stutzeri⧫; Acinetobacter baumannii; Kocuria varians; Stenotrophomonas maltophilia (2 morphologically different colonies) |
| Left Back | TNTC | Aerococcus viridans; Aeromonas veronii⧫; | |
| Delftia acidovorans; Streptococcus pneumoniae | |||
| Right Back | TNTC | Exiguobacterium acetylicum⧫; Acenitobacter baumannii; Aeromonas sobria; Kocuria varians (2 morphologically different colonies) | |
| 137 | Right Front | TNTC | Kocuria kristinae (2 morphologically different colonies); Kocurea rosea; Unidentified species - unable to isolate (3 morphologically different colonies); Unidentified Gram positive species |
| Right Back | TNTC | Roseomonas cervicalis⧫; Kocuria rosea; Kocuria varians; Myroides species | |
| Left Front | TNTC | Aeromonas sobria; Brevundimonas diminuta/vesicularis; Kocuria rosea | |
| 138 | Left Front | 10 | Sphingomonas aurantiaca; Unidentified species - unable to isolate; Unidentified Gram positive species |
| Left Back* | 12 | Brevundimonas diminuta/vesicularis; Kocuria varians; Sphingomonas paucimobilis | |
| 139 | Lower Lip | 163 | Granulicatella adiacens; Aeromonas hydrophila/caviae; Bacillus sphaericus/fusiformis; Rhizobium radiobacter |
| Left Front | 28 | Bacillus lentus; Geobacillus thermoglucosidasius/thermodenitrificans; Micrococcus luteus/lytae; Rhizobium radiobacter; Bacillus species | |
| Right Front | 11 | Sphingomonas paucimobilis; Unidentified Gram positive species; Kocuria kristinae; Brevundimonas diminuta/vesicularis; Dermacoccus/Kytococcus species | |
| Right Back | 19 | Aeromonas sobria; Micrococcus luteus⧫; Unidentified Gram negative species; Kocuria kristinae; Rhizobium radiobacter; Ewingella Americana; Brachybacterium alementarium⧫; Sphingomonas paucimobilis; Gordonia terrae⧫ |
Colony Forming Units represent the number of microbial colonies counted on each plate. Sample plates which had no growth are not listed. The appearance of different morphologies for singles species is noted. Asterisk (*) indicates control sample from uninjured limb. Diamond (⧫) indicates an isolate identified by 16S sequencing (percent similarity of greater than or equal to 98%). NFWR = North Fork of White River samples.
Table 2. Bacteria identified on Mannitol Salt Agar.
| NFWR | Limb sampled | Colony Forming Units | Closest match |
| 134 | Right Back | 4 | Exiguobacterium acetylicum⧫; Micrococcus luteus/lylae |
| Right Front* | 17 | Kocuria kristinae; Micrococcus luteus/lylae | |
| 135 | Right Back | 11 | Curtobacterium flaccumfaciens⧫; Bacillus megaterium; Unidentified Gram positive species (2 morphologically different colonies) |
| Right Front | 1 | Micrococcus luteus/lylae | |
| Left Back | 4 | Staphylococcus hominis | |
| Left Front | 18 | Brevibacterium casei⧫; Kocuria kristinae; Staphylococcus sciuri; Micrococcus luteus/lylae | |
| 136 | Left Front | TNTC | Bacillus licheniformis⧫; Micrococcus luteus/lylae; Staphylococcus warneri |
| Left Back | TNTC | Exiguobacterium acetylicum⧫; Bacillus megaterium; Staphylococcus hominus/novobisepticus; Unidentified Gram positive species | |
| 137 | Right Front | 1 | Micrococcus luteus/lylae |
| Right Back | 6 | Staphylococcus vitulinus; Micrococcus luteus/lylae (2 morphologically different colonies); Unidentified Gram positive species | |
| Left Front | TNTC | Dermacoccus species/Kytococcus species; Gemella morbillorum; Micrococcus luteus/lylae; | |
| Unidentified species - unable to isolate | |||
| 138 | Left Front | 2 | Dermacoccus/Kytococcus species |
| Left Back* | 1 | Bacillus megatarium | |
| 139 | Lower Lip | 1 | Micrococcus luteus/lytae |
| Left Back | TNTC | Brevibacterium casei⧫, Brevundimonas diminuta/vesicularis (2 morphologically different colonies); Brevundimonas diminuta/vesicularis; Micrococcus luteus/lylae | |
| Left Front | 7 | Bacillus lentus | |
| Right Front | 4 | Pantoea agglomerans | |
| Right Back | 2 | Pantoea species |
Colony Forming Units represent the number of colonies counted on each plate. Sample plates without growth are not listed. The appearance of different morphologies for singles species is noted. Asterisk (*) indicates control sample from uninjured limb. Diamond (⧫) indicates an isolate identified by 16S sequencing. NFWR = North Fork of White River samples.
Fungal isolates were consistent with common environmental flora from genera that included Penicillium, Streptomycetes, Cladosporium, Fusarium, Acremonium, Curvularia, and the Class Hyphomycetes (Tables 3, 4, and 5).
Table 3. Fungi identified on Blood Agar.
| NFWR | Limb sampled | Colony Forming Units | Closest match |
| 137 | Right Back | 60 | Streptomycetes species |
| Left Front | 180 | Fusarium species | |
| 138 | Left Front | TNTC | Penicillium species; Cladosporium species; Streptomycetes species |
| Left Back* | TNTC | Penicillium species (2 different species); Streptomycetes species | |
| 139 | Left Back | 510 | Hyphomycetes species; Penicillium species; |
| Cladosporium species (2 different species) | |||
| Right Front | 750 | Hyphomycetes species (2 different species); | |
| Cladosporium (2 different species); Penicillium species |
Colony Forming Units represent the number of colonies counted on each plate. Sample plates without growth were not listed. Genera with different morphological characteristics suggesting different species are noted. Asterisk (*) indicates control sample from uninjured limb. NFWR = North Fork of White River samples.
Table 4. Fungi identified on Mannitol Salt Agar.
| NFWR | Limb sampled | Colony Forming Units | Closest match |
| 134 | Right Back | 90 | Cladisporium species |
| Penicillium species | |||
| Hyphomycetes species | |||
| Right Front* | 30 | Cladosporium species | |
| 135 | Right Back | 30 | Penicillium species |
| Right Front | 30 | Hyphomycetes species | |
| Left Back | 690 | Cladosporium species (3 different species) | |
| Exophilia species | |||
| Hyphomycetes species | |||
| Left Front | 30 | Hyphomycetes species | |
| 136 | Left Front | 30 | Cladosporium species |
| Left Back | 30 | Penicillium species | |
| Right Back | 60 | Hyphomycetes species | |
| Cladosporium species | |||
| 137 | Right Front | 30 | Penicillium species |
| Right Back | 150 | Penicillium species | |
| Left Front | 180 | Cladosporium species (2 different species) | |
| Hyphomycetes species (2 different species) | |||
| Aureobasidium species | |||
| 138 | Left Back* | 60 | Acremonium species |
| 139 | Lower Lip | 30 | Cladosporium species |
| Left Back | 1100 | Streptomycetes species | |
| Cladosporium species (2 different species) | |||
| Left Front | 210 | Penicillium species | |
| Cladosporium species | |||
| Right Front | 960 | Penicillium species | |
| Cladosporium species | |||
| Streptomycetes species | |||
| Right Back | 30 | Penicillium species |
Colony Forming Units represent the number of colonies counted on each plate. Sample plates without growth are not listed. Genera with different morphological characteristics suggesting different species are noted. Asterisk (*) indicates control sample from uninjured limb. NFWR = North Fork of White River samples.
Table 5. Fungi identified on Sabouraud Dextrose Agar.
| NFWR | Limb sampled | Colony Forming Units | Closest match |
| 134 | Right Back | 1100 | Penicillium species (2 different species) |
| Cladosporium species | |||
| Right Front* | TNTC | Penicillium species | |
| Hyphomycetes species | |||
| 135 | Right Back | 120 | Penicillium species |
| Aspergillus species | |||
| Hyphomycetes species (2 different species) | |||
| Right Front | TNTC | Exophilia species | |
| Left Back | 600 | Wangiella species | |
| Hyphomycetes species | |||
| Penicillium species | |||
| Left Front | 150 | Cladosporium species (2 different species) | |
| Hyphomycetes species (2 different species) | |||
| 136 | Left Front | 60 | Cladosporium species |
| Streptomycetes species | |||
| Left Back | 30 | Acremonium species | |
| Right Back | 150 | Fusarium species | |
| Hyphomycetes species (2 different species) | |||
| Cladosporium species | |||
| Penicillium species | |||
| 137 | Right Front | TNTC | Penicillium species (2 different species) |
| Right Back | 60 | Curvularia species | |
| Cladosporium species | |||
| Left Front | TNTC | Penicillium species (3 different species) | |
| Cladosporium species | |||
| 138 | Left Front | 150 | Penicillium species |
| Cladosporium species | |||
| Hyphomycetes species | |||
| Left Back* | TNTC | Penicillium species | |
| Hyphomycetes species | |||
| 139 | Lower Lip | TNTC | Penicillium species |
| Cladosporium species | |||
| Left Back | 330 | Penicillium species (2 different species) | |
| Acremonium species | |||
| Sporothrix species | |||
| Left Front | TNTC | Cladosporium species (2 different species) | |
| Right Front | TNTC | Cladosporium species | |
| Right Back | 150 | Cladosporium species | |
| Penicillium species |
Colony Forming Units represent the number of colonies counted on each plate. Sample plates without growth are not listed. Genera with different morphological characteristics suggesting different species are noted. Asterisk (*) indicates control sample from uninjured limb. NFWR = North Fork of White River samples.
Discussion
The C. a. bishopi population decline in the NFWR is well documented and of significant concern [42], [47], [48]. Many reasons for the decline in population and health of the Ozark Hellbender have been suggested, including flooding [39], [49] amphibian harvesting [42], the use of the anesthetic MS-222 (Tricane) [50], [51], the reintroduction and introduction of species including otters and trout [47], [52], habitat alteration and degradation [53], disease including those having a genetic, chemical, or infectious etiology [36], [41], [48], [50], [51], [53]–[61], and the interaction of these factors [47]. Many of these hypothesized causal agents of decline have been investigated to various degrees, yet disease research has largely been limited to B. dendrobatidis [59]–[61], and prior to our study, the microbial community associated with the abnormalities typifying the affected hellbenders had not been assessed. As a variety of pathogenic microbes have been linked to amphibian declines, it is critical that the microbial community be examined for all potential disease agents and that research not be initially limited to a single potential infectious agent until causation has been properly evaluated and established.
While our microbiological evaluations in this study indicated common environmental organisms in both abnormal/injured and uninjured Ozark Hellbenders, several opportunistic pathogenic organisms were identified that were associated only with the abnormal tissue/injuries of C. a. bishopi, such as Aeromonas hydrophila, a known pathogen of amphibians [62], [63], Granulicatella adiacens [64], Stenotrophomonas malophilia [65], and a variety of opportunistic pathogen Pseudomonad species - P. aeruginosa [66], P. stutzeri [67] and P. alcaligenes [68]. These microbial pathogens are known to form biofilms in the environment and/or in vivo in the infected host. Several of the filamentous fungi isolated in this study, including Penicillium, Fusarium, and Cladosporidium are genera containing opportunistic pathogens that are known to be associated with environmental biofilms [69]–[72]. Multispecies biofilms may interact synergistically yielding an increased resistance to antibacterial agents [73]. While a possible cause and effect role for biofilms in disease progression observed in the Ozark Hellbenders is outside the scope of this study, this observation warrants investigation in future studies. The lack of isolation of the same organism from multiple wounds suggests that none of the organisms identified were the sole etiological agent responsible for the damage to C. a. bishopi. If the immune system of the injured C. a. bishopi were repressed, it is possible that a combination of the isolated opportunistic pathogens may have contributed to the observed tissue damage. The Gram positive opportunistic pathogen, Streptococcus pneumoniae, was isolated from one animal. While the presence of S. pneumoniae may be the result of contamination during collection or processing, the genus Streptococcus has been found by the EPA downstream of our NFWR research site.
Reintroduced or introduced species may not only be a source of pathogenic microbes in the NFWR, but in some cases may also increase injury, and subsequently infection rates by creating open sores. River otters (Lutra or Lontra canadensis) used in reintroduction programs are known to carry a suite of pathogenic microbes, including Gram positive Streptococcus spp. and Gram negative Pseudomonas spp. [74]. Otters reintroduced into Missouri were sourced from Louisiana and could carry many different microbes which are not found in the streams of the Ozark Highlands [75]. We speculate that reintroduced otters may introduce pathogenic microbes into the environment through fecal transmission or direct contact with hellbenders, crayfish (the primary prey of both otters and hellbenders), or other species. In addition, river otters are capable of killing or injuring C. a. bishopi. Non-lethal injuries such as bites or scratches yielding open sores may provide a pathway for pathogens. Non-native rainbow trout (Oncorhynchus mykiss) and brown trout (Salmo trutta) are stocked in the NFWR annually. These trout come from multiple hatcheries with multiple water sources and are transferred between hatcheries, which have known reoccurring water quality issues, including harboring pathogenic microorganisms [76]. While we know of no predation of C. a. bishopi by salmonids in the wild, hatchery-raised trout released into the NFWR and other water bodies may serve as a source for pathogens.
Increased recreational use of NFWR may also be a source of hellbender injury and pathogenic microbes. Canoeing and other water activities may disturb or dislodge habitat rocks, inadvertently injuring hellbenders located underneath [43]. Humans may also be a source of pathogens such as S. pneumoniae, one of the opportunistic bacteria found in this study.
The rapid regeneration that historically typified injured hellbenders was not apparent in recent studies of the NFWR population. Based on data collected in 1969, hellbender injuries (i.e., tail holes) induced by tagging healed completely with no sign of infection and no visible scars within two months (Nickerson unpublished data). The remarkable regenerative capacity of salamanders has been known since first reported by Spallanzani in 1769 [77]. Regenerative studies have included phylogenetic, seasonal, and environmental analysis of limb regeneration [77]–[79]. Environmental factors that have been considered to affect regeneration include temperature, diet, photoperiod, parasitism, infection, and quality of terrestrial and aquatic microhabitats [79]. Human-induced alterations to the NFWR and surrounding landscape have resulted in changes to the physical-chemical properties of the NFWR, including nutrient-loading, introduction of estrogenic chemical levels, algal blooms, and a microbial content deemed unsafe for full body contact by state and federal agencies [43], [80], [81]. Previous studies investigating the impact of human activities on NFWR water quality revealed relatively high concentrations of total phosphorus (6–52 µg L−1) and total nitrogen (0.35–3.06 mg L−1) in the 4.6 km research site originally investigated by Nickerson and Mays [80], [81]. The impact of this type of increased nutrient level on the hellbenders was investigated by Solis et al., which focused on a historically populated area, located 11.3 km downstream of the Nickerson and Mays site [80], [82]. Not unexpectedly, the site studied by Solis et al. indicated that nutrient concentrations (including total phosphorous and nitrogen) exceeded the EPA recommended criteria in two thirds of the samples [80]. However, a direct correlation between these elevated levels and abnormalities/disease of the hellbenders was not supported, as all individual concentrations of nutrients and organic chemicals were at much lower levels than any laboratory and field experiments shown to have deleterious effects on amphibians [80]. Likewise, serum samples from C. a. bishopi collected at the Solis et al. site were analyzed for possible endocrine disrupting chemicals, however, none were detected at levels above the EPA and Missouri Clean Water Commission criteria for aquatic organisms [82]. Thus, the direct impact of increased chemical levels on the hellbenders remains inconclusive.
The impact of eutrophication associated with human activity was further investigated in a periphyton survey of the NFWR in 2006 to determine if changes in the periphyton communities could be a factor in the Ozark Hellbender decline [81]. Some Periphyton, such as cyanobacteria (i.e., blue-green algae) may cause cutaneous damage, neural and hepatic effects, tumor induction, diarrhea, vomiting, respiratory dysfunction, convulsions and occasionally death [83]. The periphyton community within the NFWR 4.6 km Nickerson and Mays research section consisted of diatoms, chlorophytes, and cyanobacteria [81]. The green algae Cladophora spp. achieved relative abundances of >90% of the total periphyton community [81]. Blooms of the benthic, filamentous Cladophora spp. are a visible indicator of eutrophication and are linked to phosphorus concentration with 20 µg µL−1 being the threshold for Cladophora dominance [84], [85]. High nitrate concentrations are an issue with karst topography such as that in the NFWR drainage [81] and increases in human usage and poor sewage facilities. Large Cladophora spp. blooms have been a component of the NFWR since at least 1968, but have increased over the decades and large floating algal masses seen during recent summers were not a component of the NFWR 4.6 km section during the early surveys [36]. Increased algal levels are known to increase both biofilm formation and antimicrobial resistance, and Cladophora spp mats maintain higher E. coli densities than the surrounding aquatic habitat [86], [87]. Green algae (Cladophora sp.) mats may have almost ubiquitous populations of E. coli and enterococci, which may survive at least six months of drying [88]. A 2007 study of total coliform (TC) bacteria and Escherichia coli content was conducted at multiple sites and habitats in the NFWR between Mark Twain National Forest Campground Access and Norfork Reservoir, as well as springs which flow into the NFWR [89]. Total coliform levels exceeded the values deemed safe for full body contact by Missouri Department of Natural Resources (MDNR) in 70 of 94 individual water samples and 25 of the 94 samples also surpassed concentrations of E. coli deemed safe for full body contact [89].
Our results do not preclude that an infectious agent caused or exacerbated the tissue damage observed in Ozark Hellbenders, as other microorganisms, which would not grow on the media used in this experiment, may have been present (i.e., the microbial diversity observed in this study is likely a subset of the total microbial diversity). Alternatively, if the immune system of the abnormal/injured C. a. bishopi was suppressed, many of the opportunistic pathogens that were isolated in this study, alone or in combination, may have caused infection which was responsible for or served to exacerbate the tissue damage. As such, the increase in incidence of abnormalities/injuries and retardation of tissue regeneration may have multiple contributing factors including changes in the Ozark Hellbenders' susceptibility to infection and exposure to microorganisms. The Ozark hellbender is a federally listed endangered species that has yet to have successfully reproduced in captivity. The unavailability of healthy Ozark hellbenders, small sample size, and conservation status precluded our ability to evaluate all of Koch's Postulates. However, this study provides the most complete analysis of potential microbial stressors on Ozark hellbenders to date and places these findings in the context of habitat alterations. Follow up studies are planned to identify causative mechanism(s) and environmental factors that are contributing to health and population declines in this endangered species.
Materials and Methods
The Ozark Hellbender, C. a. bishopi, is now listed as endangered and populations have been extirpated and face extinction in much of the former range. The NFWR currently supports only a very small population of C. a. bishopi, of which about 50% within the original NFWR research section of Nickerson and Mays [36], [37] have substantial abnormalities/injuries (J. Briggler unpublished data). On 17 August 2007, we methodically searched a portion of the NFWR by snorkeling and lifting rocks. We located and captured six adult hellbenders, all with abnormalities/injuries (Table 6). Each individual hellbender was placed into a clean bucket filled with river water and then measured, weighed, and individually photographed. All C. a. bishopi were visually inspected for the presence of leeches, injuries, or abnormalities. The feet/limbs showing signs of infection (e.g., lesions, sores or exposed bone) were swabbed with sterile, buffered swabs. In addition, the lower lip of one individual with a raw sore was swabbed (Figure 2). Swabs were then streaked onto three different microbiological culture media: sheep's blood agar (SBA), a general all purpose growth medium that supports the culture of a large number of microorganisms and also indicates hemolytic activity; Mannitol Salt Agar (MSA), primarily selective for halo-tolerant bacteria such as staphylococci; and Sabouraud Dextrose Agar (SDX), primarily selective for fungi (Figure 1). Given the very small population of C. a. bishopi currently existing in the NFWR and given that no animals without abnormality/injury were captured throughout the duration of this study, two feet showing no signs of infection from two C. a. bishopi were swabbed in the same manner and served as uninfected controls. The swabs were streaked onto the different agar plates, and sample plate lids were immediately added and secured with tape. Secured plates were immediately placed into styrofoam coolers with ice packs and transported by vehicle to St. Louis, MO, and flown to the Microbiology Laboratory at the NASA Johnson Space Center (Houston, TX) for microbial identification. Bacterial and fungal isolates were enumerated and then sub-cultured on the medium from the parent culture at room temperature. Bacterial isolates were identified using biochemical analysis with the Vitek 2 system (bioMérieux, Marcy l'Etoile, France). Bacterial isolates that could not be identified by the Vitek 2 system were identified by 16S ribosomal DNA sequencing using a MicroSeq 500 16S rDNA Bacterial Identification Kit (Applied Biosystems, Foster City, CA). Sequences were compared to those on the National Center for Biotechnology Information (NCBI) website for microorganisms. Speciation was reported for isolates having greater than 98% sequence similarity. Fungal isolates were identified by microscopic morphological characteristics [90].
Table 6. Ozark Hellbenders, Cryptobranchus alleganiensis bishopi, captured and swabbed for microbial flora from the North Fork of the White River, Ozark County, Missouri on 17 August 2007.
| Sample No. | Mass (g) | TL (cm) | SVL (cm) | Gender |
| NFWR 1341 | 559 | 45.5 | 31.0 | Male |
| NFWR 1352 | 610 | 46.0 | 30.5 | Male |
| NFWR 1363 | 569 | 45.5 | 32.5 | Female |
| NFWR 1374 | 690 | 47.5 | 47.5 | Male |
| NFWR 1385 | 971 | 53.0 | 53.0 | Female |
| NFWR 1396 | 545 | 48.5 | 48.5 | Male |
Two sample location (right back limb and right front limb) were swabbed.
Four sample locations (all limbs) were swabbed.
Three sample locations (right back limb, left back limb, and left front limb) were swabbed.
Three sample locations (right back limb, right front limb, and left front limb) were swabbed.
Two sample locations (left back limb and left front limb) were swabbed.
Five sample locations (all limbs and lower lip) were swabbed.
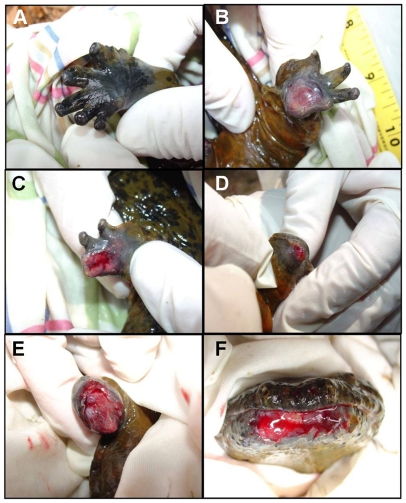
Figure 2. Representative samples of normal and abnormal lesions on Ozark Hellbenders, Cryptobranchus alleganiensis bishopi.
All individuals sampled were captured from the North Fork of the White River, Ozark County, Missouri on 17 August 2007. A shows a normal left back foot (NFWR 138), B shows lesion on palm of right back foot (NFWR 136), C shows lesion on toes of left front foot (NFWR 136), D shows lesion on right back limb with all toes missing (NFWR 135), E shows lesion on right back limb with all toes missing (NFWR 139), and F shows lesion on lower lip (NFWR 139).
Acknowledgments
Research was conducted in compliance with applicable animal care guidelines. We also thank the Missouri Department of Conservation for permits to conduct this research. Dr. Lawrence M. Page kindly read and commented on the manuscript. Special thanks to L. Davis for assistance with graphics.
Footnotes
Competing Interests: V. M. Garcia and T. C. Molina are employees of EASI, Wyle Laboratories. There are no patents, products in development or marketed products to declare. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials, as detailed online in the guide for authors.
Funding: NASA student grant NNX07AM16G, NASA grant NCC2-1362, The St. Louis Zoological Park, and The Reptile and Amphibian Conservation Corps. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lannoo M. Amphibian declines: The conservation status of United States species. Berkley and Los Angeles: University of California Press; 2005. 1094 [Google Scholar]
- 2.Halliday T. Diverse phenomena influencing amphibian population declines. In: Lannoo M, editor. Amphibian declines: The conservation status of United States species. Berkeley and Los Angeles: University of California Press; 2005. pp. 3–6. [Google Scholar]
- 3.Beebee TJC, Griffiths RA. The amphibian decline crisis: a watershed for conservation biology? Biological Conservation. 2005;125:271–285. [Google Scholar]
- 4.Anonymous Where have all the frogs gone? Modern Medicine. 1973;41:20–24. [Google Scholar]
- 5.Gibbs E, Nace G, Emmons M. The live frog is almost dead. Bioscience. 1971;21:1027–1034. [Google Scholar]
- 6.National Academy of Sciences. Amphibians: guidelines for the breeding, care and management of lab animals. Washington D.C.: 1974. 162. [PubMed] [Google Scholar]
- 7.Maugh TH. Frog shortage possible this winter. Science. 1972;178:387. doi: 10.1126/science.178.4059.387. [DOI] [PubMed] [Google Scholar]
- 8.Reichenbach-Klinke H, Elkan E. Amphibia. In: Reichenbach-Klinke H, Elkan E, editors. The principle diseases of lower vertebrates. New York: Academic Press; 1965. pp. 209–384. [Google Scholar]
- 9.Hutchison JA, Nickerson MA. Comments on the distribution of Basidiobolus ranarum. Mycologia. 1970;62:585–587. [PubMed] [Google Scholar]
- 10.Nickerson MA, Hutchison JA. A study of the distribution of the fungus Basidiobolus ranarum Eidam in fish, amphibians and reptiles. American Midland Naturalist. 1971;86:500–502. [Google Scholar]
- 11.Nickerson MA, King D, Hutchison JA. Mexican isolates of Basidobolus ranarum Eidam. Southwestern Naturalist. 1973;18:93–94. [Google Scholar]
- 12.Tills D, Nickerson MA, Hutchison JA. The distribution of the fungus, Basidobolus ranarum Eidam in fish, amphibians, and reptiles of the southern Appalachian Region of the United States. Transactions Kansas Academy of Science. 1977;80:75–78. [Google Scholar]
- 13.Taylor SK. Chapter 14: Mycoses. In: Wright K, Whitaker BR, editors. Amphibian medicine and captive husbandry. Malabar, Florida: Kreiger Publishing Company; 2001. pp. 181–191. [Google Scholar]
- 14.Daszak P, Cunningham AA, Hyatt AD. Infectious disease and amphibian population declines. Diversity and Distribution. 2003;9:141–150. [Google Scholar]
- 15.Greenbaum E, Kusamba C, Aristote MM, Reed KD. Amphibian chytrid fungus infections in Hyperolius (Anura: Hyperoliidae) from eastern Democratic Republic of Congo. Herpetological Review. 2008;39:70–73. [Google Scholar]
- 16.Reeves MK. Batrachochytrium dendrobatidis in wood frogs (Rana sylvatica) from three national wildlife refuges in Alaska, USA. Herpetological Review. 2008;39:68–70. [Google Scholar]
- 17.Woodhams DC, Hyatt AD, Boyle DG, Rollins-Smith LA. The northern leopard frog Rana pipiens is a widespread reservoir species harboring Batrachochytrium dendrobatidis in North America. Herpetological Review. 2008;39:66–68. [Google Scholar]
- 18.Erspamer V. Bioactive secretions of the amphibian integument. In: Heatwole H, Barthalmus GT, editors. Amphibian biology volume 1: The integument. Chipping Norton, NSW: Surry Beatty and Sons; 1994. pp. 178–350. [Google Scholar]
- 19.Heatwole H, Barthalmus GT, editors. Amphibian biology volume 1: The integument. Chipping Norton, NSW: Surry Beatty and Sons; 1994. 418 [Google Scholar]
- 20.Csordas A, Michl H. Primary structure of two oligopeptides of the toxin of Bombina variegata. Toxicon. 1969;7:103–108. doi: 10.1016/0041-0101(69)90072-5. [DOI] [PubMed] [Google Scholar]
- 21.Croce G, Giglioli N, Bolognani L. Antimicrobial activity in the skin of Bombina variegata pachypus. Toxicon. 1973;11:99–100. doi: 10.1016/0041-0101(73)90159-1. [DOI] [PubMed] [Google Scholar]
- 22.Becker MH, Brucker RM, Schwantes CR, Harris RN, Minbiole KPC. The bacterially-produced metabolite violacein is associated with survival in amphibians infected with a lethal disease. Appl Environ Microbiol. 2009;75:6635–6638. doi: 10.1128/AEM.01294-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brucker R, Baylor C, Walters R, Lauer A, Harris R, et al. The identification of 2,4-diacetylphloroglucinol as an antifungal metabolite produced by cutaneous bacteria of the salamander Plethodon cinereus. J Chem Ecol. 2008;34:39–43. doi: 10.1007/s10886-007-9352-8. [DOI] [PubMed] [Google Scholar]
- 24.Brucker RM, Harris RN, Schwantes CR, Gallaher TN, Flaherty DC, et al. Amphibian chemical defense: antifungal metabolites of the microsymbiont Janthinobacterium lividum on the salamander Plethodon cinereus. J Chem Ecol. 2008;34:1422–1429. doi: 10.1007/s10886-008-9555-7. [DOI] [PubMed] [Google Scholar]
- 25.Fedewa L, Lindell A. Inhibition of growth for select Gram-negative bacteria by tricaine methane sulfonate (MS-222). Journal of Herpetological Medicine and Surgery. 2005;15:13–17. [Google Scholar]
- 26.Lauer A, Simon MA, Banning JL, Lam BA, Harris RN. Diversity of cutaneous bacteria with antifungal activity isolated from female four-toed salamanders. The ISME Journal. 2008;2:145–157. doi: 10.1038/ismej.2007.110. [DOI] [PubMed] [Google Scholar]
- 27.Peterson CL. Age and growth of the Ozark hellbender. Springfield, Missouri: Southwest Missouri State University; 1979. 52 [Google Scholar]
- 28.Rollins-Smith LA, Conlon JM. Antimicrobial peptide defenses against chytridiomycosis, an emerging infectious disease of amphibian populations. Dev Comp Immunol. 2005;29:589–598. doi: 10.1016/j.dci.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 29.Sheafor B, Davidson EW, Parr L, Rollins-Smith L. Antimicrobial peptide defenses in the salamander, Ambystoma tigrinum, against emerging amphibian pathogens. J Wildl Dis. 2008;44:226–236. doi: 10.7589/0090-3558-44.2.226. [DOI] [PubMed] [Google Scholar]
- 30.Woodhams DC, Vredenburg VT, Simon M-A, Billheimer D, Shakhtour B, et al. Symbiotic bacteria contribute to innate immune defenses of the threatened mountain yellow-legged frog, Rana muscosa. Biological Conservation. 2007;138:390–398. [Google Scholar]
- 31.Rollins-Smith LA, Doersam JK, Longcore JE, Taylor SK, Shamblin JC, et al. Antimicrobial peptide defenses against pathogens associated with global amphibian declines. Developmental & Comparative Immunology. 2002;26:63–72. doi: 10.1016/s0145-305x(01)00041-6. [DOI] [PubMed] [Google Scholar]
- 32.Simmaco M, Mignogna G, Barra D. Antimicrobial peptides from amphibian skin: What do they tell us? Peptide Science. 1998;47:435–450. doi: 10.1002/(SICI)1097-0282(1998)47:6<435::AID-BIP3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 33.Lauer A, Simon MA, Banning E, Duncan AK, Harris RN. Common cutaneous bacteria from the eastern red-backed salamander can inhibit pathological fungi. Copeia. 2007;3:630–640. [Google Scholar]
- 34.Collins JP, Crump ML. Extinction in our times: Global amphibian declines. Oxford: Oxford University Press; 2009. 273 [Google Scholar]
- 35.Blaustein AR, Wake DB, Sousa WP. Amphibian declines: judging stability, persistence, and susceptibility of populations to local and global extinctions. Conservation Biology. 1994;8:60–71. [Google Scholar]
- 36.Nickerson MA, Mays CE. The hellbenders: North American “giant salamanders”. Milwaukee Public Museum Publications in Biology and Geology. 1973;1:1–106. [Google Scholar]
- 37.Nickerson MA, Mays CE. A study of the Ozark hellbender, Cryptobranchus alleganiensis bishopi. Ecology. 1973;54:1164–1165. [Google Scholar]
- 38.OSEDA. 2011. Available: http://www.oseda.missouri.edu/historicdata/popsqmi/29153.htm. Accessed 2011 July 20.
- 39.Nickerson MA, Pitt AL, Prysby MD. The effects of flooding on hellbender salamander, Cryptobranchus alleganiensis Daudin, 1803, populations. Salamandra. 2007;43:111–117. [Google Scholar]
- 40.Cooper HR. Food and feeding selectivity of two cottid species in an Ozark stream. Masters Thesis. Jonesboro, Arkansas: Arkansas State University; 1975. 45 [Google Scholar]
- 41.Hiler WR, Wheeler BJ, Trauth SE. Abnormalities in the Ozark hellbender (Cryptobranchus alleganiensis bishopi) in Arkansas: A comparison between two rivers with a historical perspective. Journal of the Arkansas Academy of Science. 2005;59:88–94. [Google Scholar]
- 42.Nickerson MA, Briggler JT. Harvesting as a factor in population decline of a long-lived salamander, the Ozark hellbender, Cryptobranchus alleganiensis bishopi Grobman. Applied Herpetology. 2007;4:207–216. [Google Scholar]
- 43.Nickerson MA, Pitt AL, Tavano JJ. Decline of the Ozark hellbender (Cryptobranchus alleganiensis bishopi) in the North Fork of White River, Ozark County, Missouri: A historical perspective. 2009. 53 Final report to the St. Louis Zoo and the Reptile and Amphibian Conservation Corps.
- 44.Yu CY, Liu YW, Chou SJ, Chao MR, Weng BC, et al. Genomic diversity and molecular differentiation of Riemerella anatipestifer associated with eight outbreaks in five farms. Avian Pathol. 2008;37:273–279. doi: 10.1080/03079450802056546. [DOI] [PubMed] [Google Scholar]
- 45.Siqueira J, Rôças IN. Catonella morbi and Granulicatella adiacens: new species in endodontic infections. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology, and Endodontics. 2006;102:259–264. doi: 10.1016/j.tripleo.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 46.Ma E, Wong C, Lai K, Chan E, Yam W, et al. Kocuria kristinae infection associated with acute cholecystitis. BMC Infectious Diseases. 2005;5:60. doi: 10.1186/1471-2334-5-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Briggler J, Utrup J, Davidson C, Humphries J, Groves J, et al. Hellbender population and habitat viability assessment: final report, IUCN/SSC Conservation Breeding Specialist Group. 2007. 46 Apple Valley, Minnesota.
- 48.Wheeler BA, McCallum ML, Trauth S. Abnormalities in the Ozark hellbender. Journal of the Arkansas Academy of Science. 2002;56:250–252. [Google Scholar]
- 49.Nickerson MA, Krysko KL, Owen RD. Habitat differences affecting age class distributions of the hellbender salamander, Cryptobranchus alleganiensis. Southeastern Naturalist. 2003;2:619–629. [Google Scholar]
- 50.Byram J. Effects of nitrogen ammonia and MS-222 on Xenopus laevis development and foraging behavior. Masters Thesis. Gainesville: University of Florida; 2008. 49 [Google Scholar]
- 51.Byram JK, Nickerson MA. The use of Tricaine (MS-222) in amphibian conservation. 2009. pp. 1–15. Reptile and Amphibian Conservation Corps Occasional Papers in Reptile and Amphibian Conservation.
- 52.Gall BG, Mathis A. Innate predator recognition and the problem of introduced trout. Ethology. 2010;116(1):47–58. [Google Scholar]
- 53.Trauth SE, Robison HW, Plummer MV. The amphibians and reptiles of Arkansas. Fayetteville: University of Arkansas Press; 2004. 421 [Google Scholar]
- 54.Harshbarger JC, Trauth SE. Squamous cell carcinoma upgrade of the epidermal papilloma reported in an Ozark hellbender (Cryptobranchus alleganiensis bishop). In: McKinnell RB, Carlson DL, editors. Proceedings of the sixth international symposium on the pathology of reptiles and amphibians. Minneapolis: University of Minnesota Printing Services; 2002. pp. 43–48. [Google Scholar]
- 55.Miller BT, Miller JL. Prevalence of physical abnormalities in eastern Hellbender (Cryptobranchus alleganiensis alleganiensis) populations of middle Tennessee. Southeastern Naturalist. 2005;4:513–520. [Google Scholar]
- 56.Pfingsten RA. The status and distribution of the hellbender, Cryptobranchus alleganiensis in Ohio. Herpetological Review. 1990;21:48–51. [Google Scholar]
- 57.Smith BG. A case of defensive self-mutilation in Cryptobranchus. Bulletin of the Wisconsin Natural Society. 1911;9:64–65. [Google Scholar]
- 58.Trauth SE, Harshbarger JC, Daniel P. Epidermal papilloma in an Ozark hellbender (Cryptobranchus alleganiensis bishopi) from the Spring River of northern Arkansas. Journal of the Arkansas Academy of Science. 2002;56:190–197. [Google Scholar]
- 59.Briggler J, Ettling M, Wanner C, Schuette M, Duncan M, et al. Cryptobranchus alleganiensis (hellbender). Chytrid fungus. Herpetological Review. 2007;38:174. [Google Scholar]
- 60.Briggler J, Larson K, Irwin K. Presence of the amphibian chytrid fungus (Batrachochytrium dendrobatidis) on hellbenders (Cryptobranchus alleganiensis) in the Ozark Highlands. Herpetological Review. 2008;39:443–444. [Google Scholar]
- 61.Bodinof CM, Briggler JT, Duncan MC, Beringer J, Millspaugh JJ. Historic occurrence of the amphibian chytrid fungus Batrachochytrium dendrobatidis in hellbender Cryptobranchus alleganiensis populations from Missouri. Diseases of Aquatic Organisms. 2011;96:1–7. doi: 10.3354/dao02380. [DOI] [PubMed] [Google Scholar]
- 62.Julia Manresa M, Vicente Villa A, Gene Giralt A, Gonzalez-Ensenat MA. Aeromonas hydrophila folliculitis associated with an inflatable swimming pool: mimicking Pseudomonas aeruginosa infection. Pediatr Dermatol. 2009;26:601–603. doi: 10.1111/j.1525-1470.2009.00993.x. [DOI] [PubMed] [Google Scholar]
- 63.Barribeau SM, Villinger J, Waldman B. Major histocompatibility complex based resistance to a common bacterial pathogen of amphibians. PLoS One. 2008;3:e2692. doi: 10.1371/journal.pone.0002692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Del Pozo JL, Garcia-Quetglas E, Hernaez S, Serrera A, Alonso M, et al. Granulicatella adiacens breast implant-associated infection. Diagn Microbiol Infect Dis. 2008;61:58–60. doi: 10.1016/j.diagmicrobio.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 65.Denton M, Kerr KG. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin Microbiol Rev. 1998;11:57–80. doi: 10.1128/cmr.11.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rumbaugh KP, Griswold JA, Hamood AN. Pseudomonas aeruginosa strains obtained from patients with tracheal, urinary tract and wound infection: variations in virulence factors and virulence genes. Journal of Hospital Infection. 1999;43:211–218. doi: 10.1053/jhin.1999.0252. [DOI] [PubMed] [Google Scholar]
- 67.Lalucat J, Bennasar A, Bosch R, Garcia-Valdes E, Palleroni NJ. Biology of Pseudomonas stutzeri. Microbiol Mol Biol Rev. 2006;70:510–547. doi: 10.1128/MMBR.00047-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Valenstein P, Bardy GH, Cox CC, Zwadyk P. Pseudomonas alcaligenes endocarditis. American Journal of Clinical Pathology. 1983;79:245–247. doi: 10.1093/ajcp/79.2.245. [DOI] [PubMed] [Google Scholar]
- 69.De Lucca A. Harmful fungi in both agriculture and medicine. Revista Iberoamericana Micrologia. 2007;24:3–13. [PubMed] [Google Scholar]
- 70.Gonçalves A, Santos IM, Paterson RR, Lima N. FISH and Calcofluor staining techniques to detect in situ filamentous fungal biofilms in water. Revista Iberoamericana Micrologia. 2006;23:194–198. doi: 10.1016/s1130-1406(06)70044-4. [DOI] [PubMed] [Google Scholar]
- 71.Suihko M-L, Alakomi H-L, Gorbushina A, Fortune I, Marquardt J, et al. Characterization of aerobic bacterial and fungal microbiota on surfaces of historic Scottish monuments. Systematic and Applied Microbiology. 2007;30:494–508. doi: 10.1016/j.syapm.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 72.Sun Y, Chandra J, Mukherjee P, Szczotka-Flynn L, Ghannoum MA, et al. A murine model of contact lens-associated Fusarium keratitis. Invest Ophthalmol Vis Sci. 2009;51:1511–1516. doi: 10.1167/iovs.09-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Burmølle M, Webb JS, Rao D, Hansen LH, Sorensen SJ, et al. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl Environ Microbiol. 2006;72:3916–3923. doi: 10.1128/AEM.03022-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kimber KR, Kollias GV. Infectious and parasitic diseases and contaminant-related problems of North American river otter (Lontra canadensis): a review. Journal of Zoo and Wildlife Medicine. 2000;31:452–472. doi: 10.1638/1042-7260(2000)031[0452:IAPDAC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 75.Low J. Problems provide measure of otter program's success. 1996 Missouri Department of Conservation, Available: http://www.mdc.state.mo.us/news/out/1996/out04196.html#problems%20provide. Accessed 2004 October 20. [Google Scholar]
- 76.Missouri Department of Conservation Trout Plan Committee. A plan for Missouri trout fishing. Jefferson City: Missouri Department of Conservation; 2003. 34 [Google Scholar]
- 77.Scadding SR. Phylogenic distribution of limb regeneration capacity in adult amphibia. Journal of Experimental Zoology. 1977;202:57–67. [Google Scholar]
- 78.Schauble MK. Seasonal variation of newt forelimb regeneration under controlled environmental conditions. Journal of Experimental Zoology. 1972;181:281–286. doi: 10.1002/jez.1401810215. [DOI] [PubMed] [Google Scholar]
- 79.Young HE, Bailey CF, Dalley BK. Environmental conditions prerequisite for complete limb regeneration in the postmetamorphic adult land-phase salamander, Ambystoma. Anat Rec. 1983;206:289–294. doi: 10.1002/ar.1092060307. [DOI] [PubMed] [Google Scholar]
- 80.Solis ME, Liu CC, Nam P, Niyogi DK, Bandeff JM, et al. Occurrence of organic chemicals in two rivers inhabited by Ozark hellbenders (Cryptobranchus alleganiensis bishopi). Arch Environ Contam Toxicol. 2007;53:426–434. doi: 10.1007/s00244-006-0208-y. [DOI] [PubMed] [Google Scholar]
- 81.Quinlan EL, Phlips EJ. Preliminary investigation of periphyton in the North Fork branch of the White River, Missouri. 2007. 7 St. Louis Zoological Park and the Reptile and Amphibian Conservation Corps Report.
- 82.Solis ME, Bandeff JM, Huang Y-W. Hematology and serum chemistry of Ozark and eastern hellbenders (Cryptobranchus alleganiensis). Herpetologica. 2007;63:285–292. [Google Scholar]
- 83.Communicable Disease Center: Centers for Disease Control and Prevention. Harmful algal blooms. 2011. Available: http://www.cdc.gov/hab/cyanobacteria/facts.htm#cynolabs. Accessed 2001 May 13.
- 84.Cattaneo A. Periphyton in lakes of different trophy. Canadian Journal of Fisheries and Aquatic Sciences. 1987;44:296–303. [Google Scholar]
- 85.Chetelat J, Pick FR, Morin A, Hamilton PB. Periphyton biomass and community composition in rivers of different nutrient status. Canadian Journal of Fisheries and Aquatic Sciences. 1999;56:560–569. [Google Scholar]
- 86.Caramujo M-J, Mendes C, Cartaxana P, Brotas V, Boavida M-J. Influence of drought on algal biofilms and meiofaunal assemblages of temperate reservoirs and rivers. Hydrobiologia. 2008;598:77–94. [Google Scholar]
- 87.Englebert ET, McDermott C, Kleinheinz GT. Effects of the nuisance algae, Cladophora, on Escherichia coli at recreational beaches in Wisconsin. Science of The Total Environment. 2008;404:10–17. doi: 10.1016/j.scitotenv.2008.05.025. [DOI] [PubMed] [Google Scholar]
- 88.Whitman RL, Shively DA, Pawlik H, Nevers MB, Byappanahalli MN. Occurrence of Escherichia coli and enterococci in Cladophora (Chlorophyta) in nearshore water and beach sand of Lake Michigan. Appl Environ Microbiol. 2003;69:4714–4719. doi: 10.1128/AEM.69.8.4714-4719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pitt AL, Nickerson MA, Tavano JJ. Coliform bacterial content of the North Fork of White River, Ozark County, Missouri. 2008. 12 St. Louis Zoological Gardens and Reptile and Amphibian Conservation Corps Report.
- 90.Castro VA, Trasher AN, Healy M, Ott CM, Pierson DL. Microbial characterization during the early habitation of the International Space Station. Microbial Ecology. 2004;47:119–126. doi: 10.1007/s00248-003-1030-y. [DOI] [PubMed] [Google Scholar]