Abstract
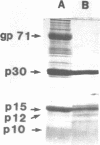
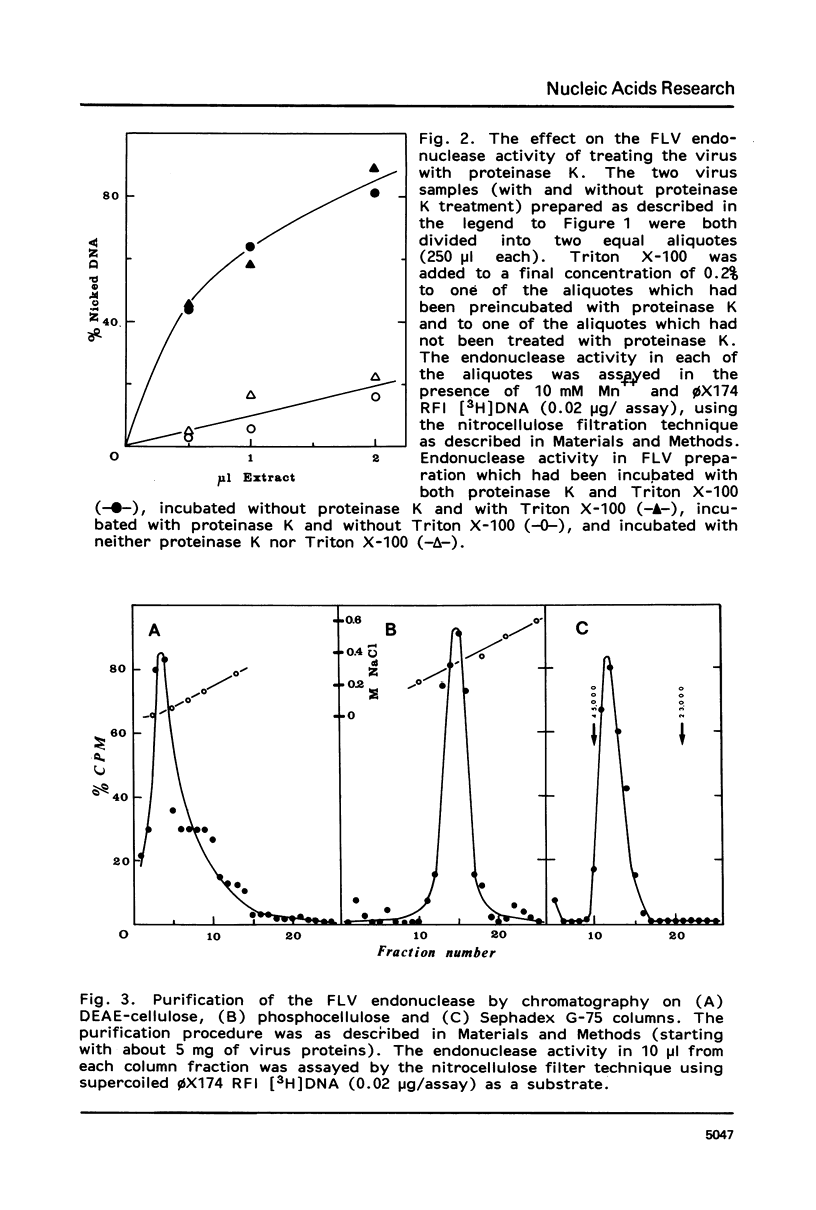
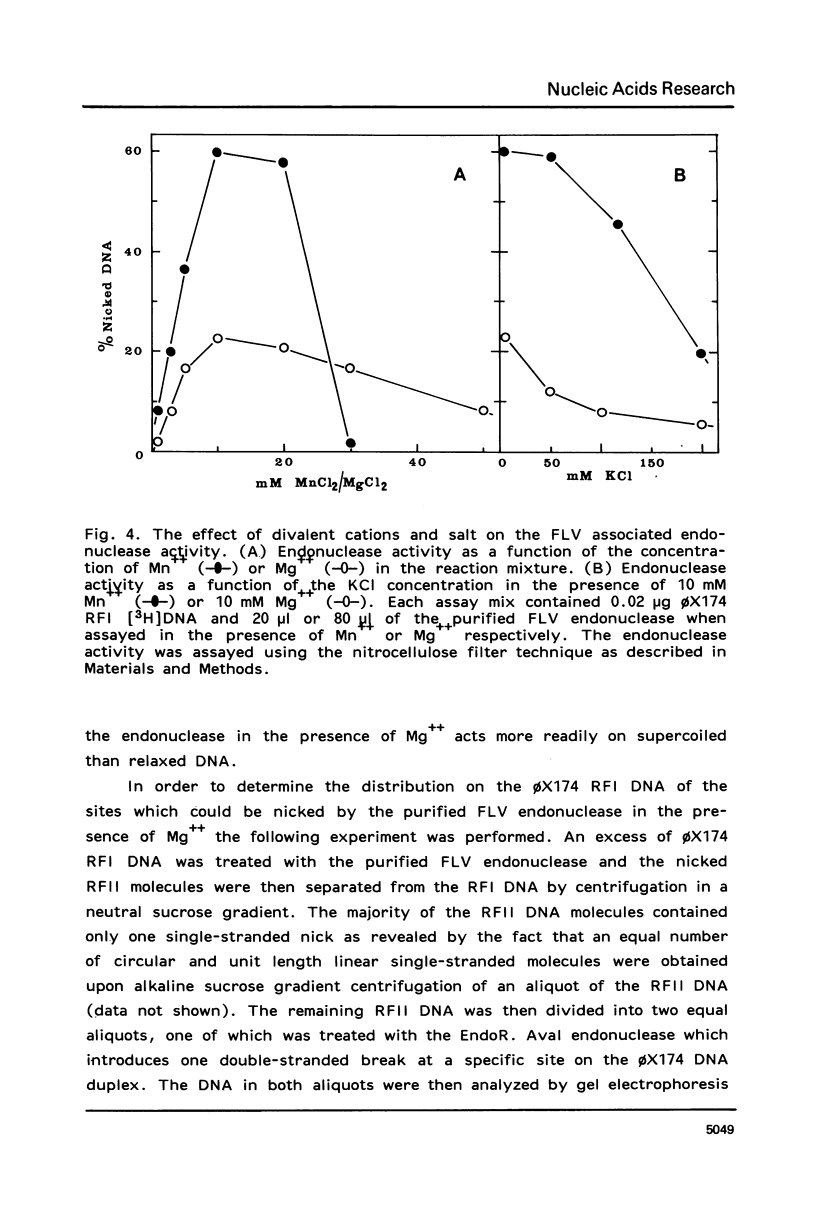
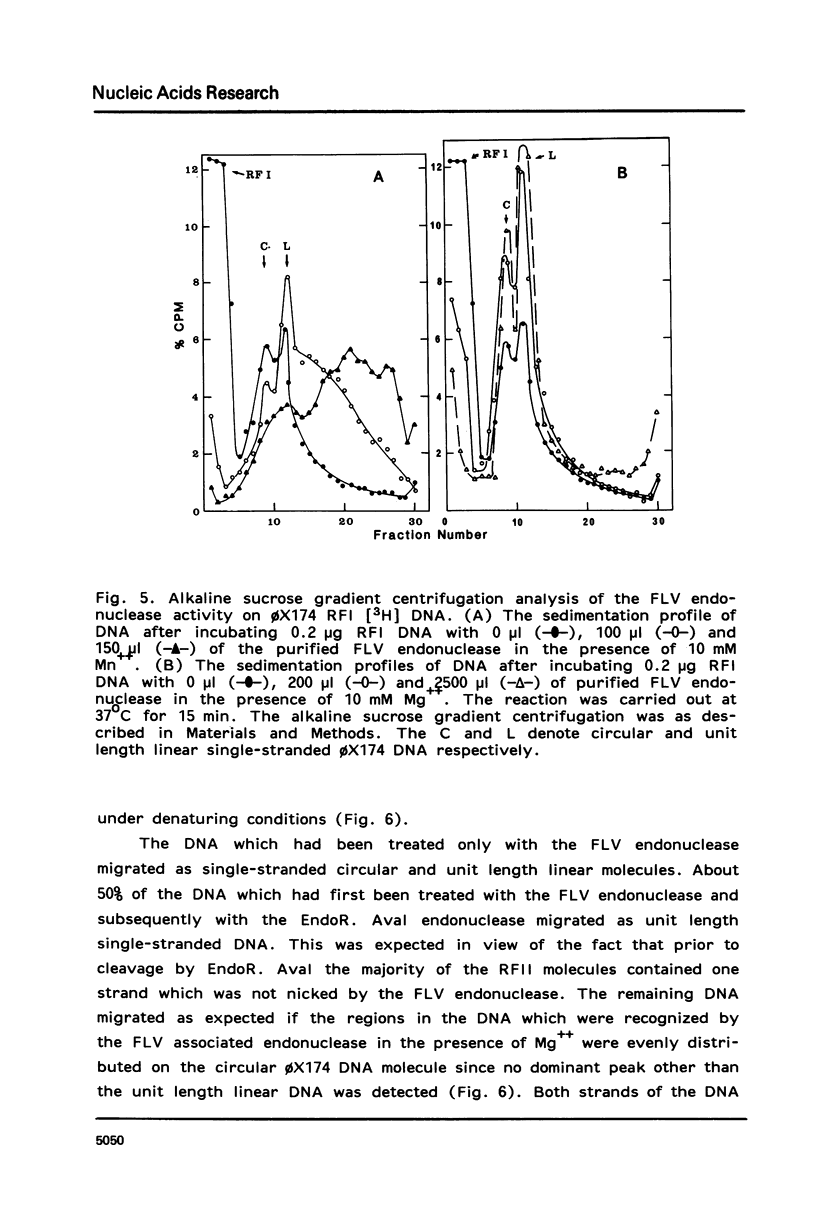
An endonuclease associated with the core of Friend leukemia virus (FLV) has been purified more than 10(3)-fold by ion exchange chromatography and gel filtration. Its molecular weight was determined by gel filtration to be about 40,000. Divalent cations were required for the endonuclease to function and KCl concentrations above 50 mM inhibited the enzyme activity. In the presence of Mg++ the purified enzyme nicked preferentially supercoiled circular DNA duplexes and in most of these molecules only one single-stranded nick was introduced per strand. The regions into which the nick could be introduced appeared to be randomly distributed on the circular molecule. When Mn++ was substituted for Mg++ the number of nicks introduced into DNA by the purified enzyme was greatly increased, and both relaxed circular and linear DNA duplexes were nicked as well as supercoiled circular DNA duplexes. Prior to its purification, however, in the presence of Mn++ the endonuclease activity in the virus extract was able to differentiate between circular and linear DNA duplexes, since both supercoiled and relaxed circular duplexes were nicked much more readily than linear duplexes. Single-stranded DNA functioned poorly as a substrate for the purified enzyme.
Full text
PDF




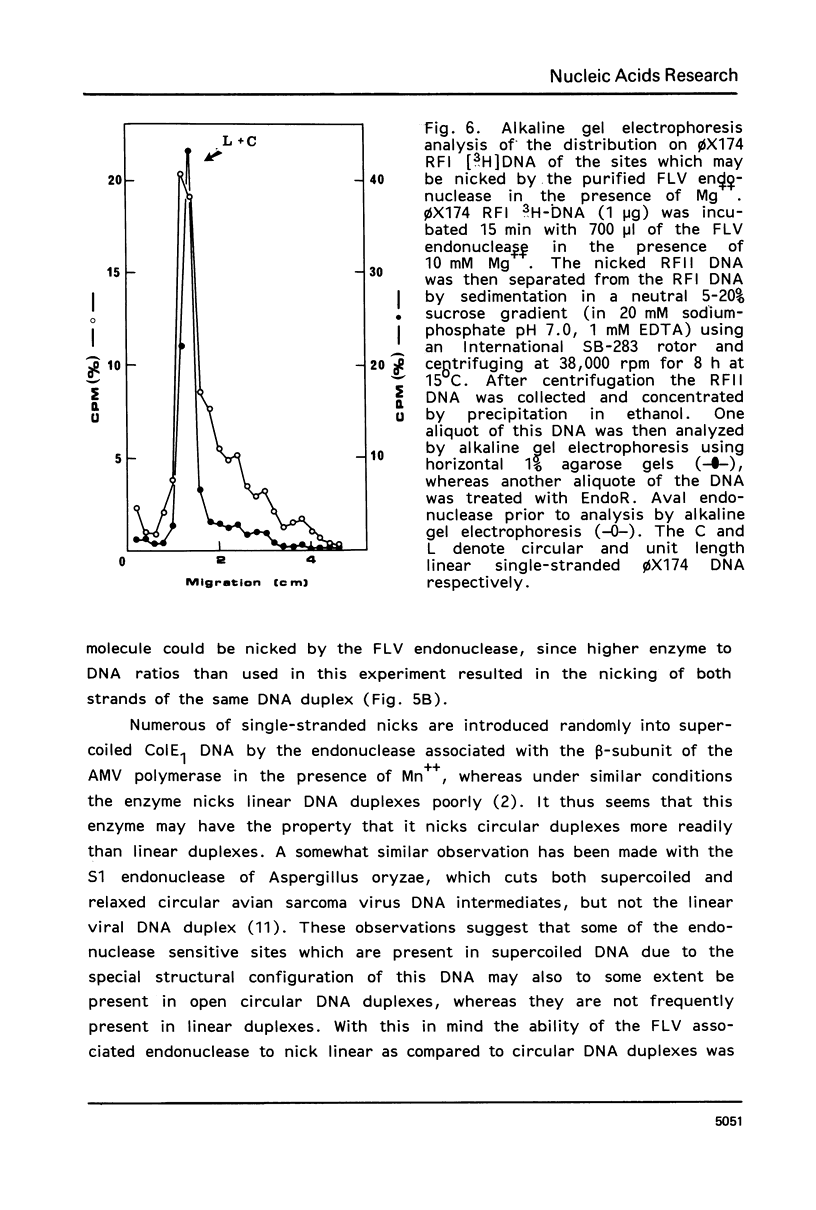
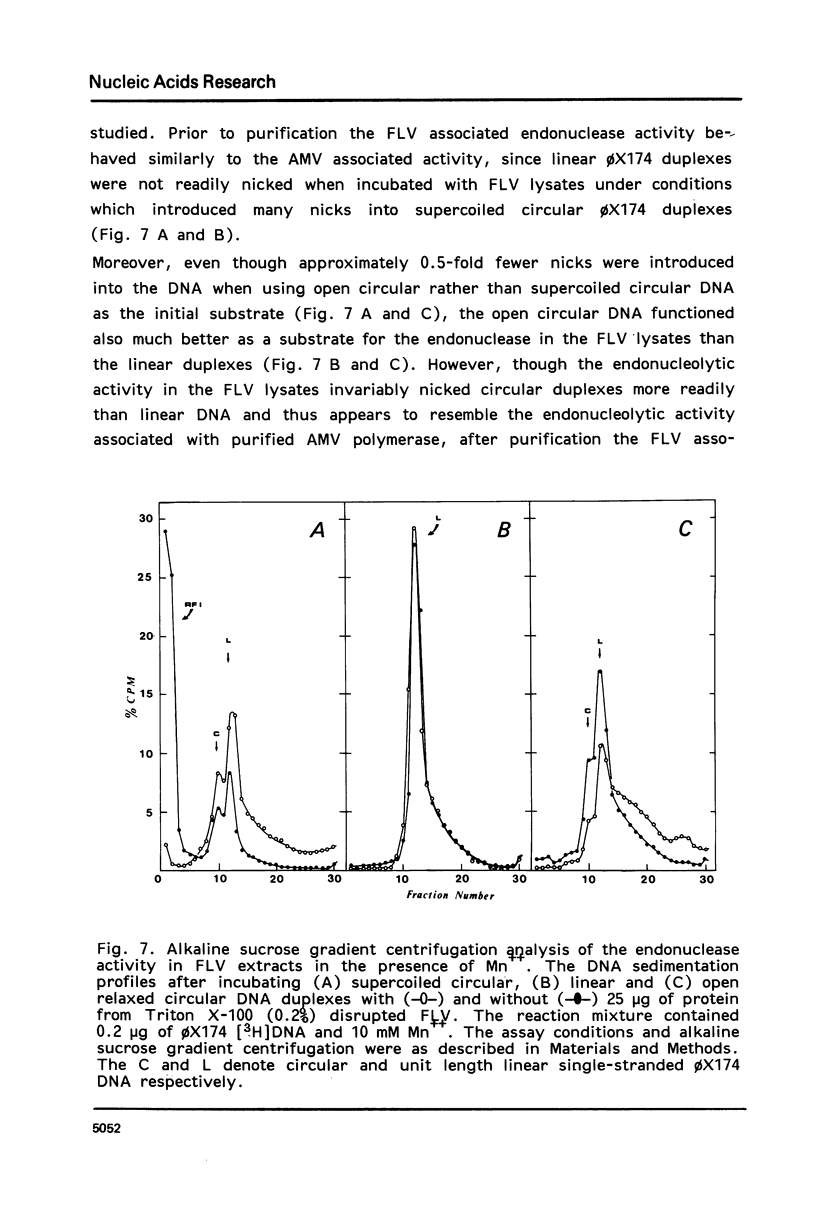



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clayman C. H., Mosharrafa E. T., Anderson D. L., Feras A. J. Circular forms of DNA synthesized by Rous sarcoma virus in vitro. Science. 1979 Nov 2;206(4418):582–584. doi: 10.1126/science.91199. [DOI] [PubMed] [Google Scholar]
- Golomb M., Grandgenett D. P. Endonuclease activity of purified RNA-directed DNA polymerase from avian myeloblastosis virus. J Biol Chem. 1979 Mar 10;254(5):1606–1613. [PubMed] [Google Scholar]
- Grandgenett D. P., Vora A. C., Schiff R. D. A 32,000-dalton nucleic acid-binding protein from avian retravirus cores possesses DNA endonuclease activity. Virology. 1978 Aug;89(1):119–132. doi: 10.1016/0042-6822(78)90046-6. [DOI] [PubMed] [Google Scholar]
- Hsu T. W., Guntaka R. V., Taylor J. M. Specific site of action for single-strand specific nuclease on the double-stranded circular DNA intermediates of an avian RNA tumor virus. J Virol. 1978 Dec;28(3):1015–1017. doi: 10.1128/jvi.28.3.1015-1017.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopchick J. J., Karshin W. L., Arlinghaus R. B. Tryptic peptide analysis of gag and gag-pol gene products of Rauscher murine leukemia virus. J Virol. 1979 May;30(2):610–623. doi: 10.1128/jvi.30.2.610-623.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K., Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol. 1973 Nov 15;80(4):575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Moelling K. Characterization of reverse transcriptase and RNase H from friend-murine leukemia virus. Virology. 1974 Nov;62(1):46–59. doi: 10.1016/0042-6822(74)90302-x. [DOI] [PubMed] [Google Scholar]
- Nes I. F., Nissen-Meyer J. Endonuclease activities from a permanently established mouse cell line that act upon DNA damaged by ultraviolet light, acid and osmium tetroxide. Biochim Biophys Acta. 1978 Aug 23;520(1):111–121. doi: 10.1016/0005-2787(78)90012-6. [DOI] [PubMed] [Google Scholar]
- Nissen-Meyer J., Nes I. F. Characterization of an endonuclease activity associated with Friend-murine leukemia virus. Biochim Biophys Acta. 1980 Aug 26;609(1):148–157. doi: 10.1016/0005-2787(80)90208-7. [DOI] [PubMed] [Google Scholar]
- Samuel K. P., Papas T. S., Chirikjian J. G. DNA endonucleases associated with the avian myeloblastosis virus DNA polymerase. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2659–2663. doi: 10.1073/pnas.76.6.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert E., Claviez M., Frank H., Hunsmann G., Schwarz H., Schäfer W. XII. Produktion grösserer Mengen von Friend-Virus durch eine permanente Zell-Suspensions-Kultur (Eveline-Suspensions-Zellen) Z Naturforsch C. 1975 Sep-Oct;30(5):698–700. [PubMed] [Google Scholar]