Abstract
Autophagy is a cellular response to starvation which generates autophagosomes to carry cellular organelles and long-lived proteins to lysosomes for degradation. Degradation through autophagy can provide an innate defense against virus infection, or conversely autophagosomes can promote infection by facilitating assembly of replicase proteins. We demonstrate that the avian coronavirus, infectious bronchitis virus (IBV), activates autophagy. A screen of individual IBV nonstructural proteins (nsps) showed that autophagy was activated by IBV nsp6. This property was shared with nsp6 of mammalian coronaviruses mouse hepatitis virus, and severe acute respiratory syndrome virus, and the equivalent nsp5–7 of the arterivirus porcine reproductive and respiratory syndrome virus. These multiple-spanning transmembrane proteins located to the endoplasmic reticulum (ER) where they generated Atg5 and LC3II -positive vesicles, and vesicle formation was dependent on Atg5 and class III PI3 kinase. The vesicles recruited double-FYVE-domain containing protein (DFCP) indicating localized concentration of phosphatidylinositol 3 phosphate, and therefore shared many features with omegasomes formed from the ER in response to starvation. Omegasomes induced by viral nsp6 matured into autophagosomes that delivered LC3 to lysosomes and therefore recruited and recycled the proteins needed for autophagosome nucleation, expansion, cellular trafficking and delivery of cargo to lysosomes. The coronavirus nsp6 proteins activated omegasome and autophagosome formation independently of starvation, but activation did not involve direct inhibition of mTOR signaling, activation of sirtuin 1 or induction of ER stress.
Key words: autophagy, omegasome, endoplasmic reticulum, coronavirus, nonstructural proteins
Introduction
Autophagy was discovered as a cellular response to starvation that degrades cytoplasmic organelles and long-lived proteins. Degradation is mediated by membrane-bound autophagosomes which engulf small portions of cytoplasm and fuse with lysosomes providing a short-term supply of amino acids to maintain cellular processes during nutrient depletion. Work over the past decade has shown that autophagy also provides an innate protection against pathogens since delivery to lysosomes can kill pathogens and increase presentation of microbial components to the immune system.1–4 Autophagosome formation starts with the production of phagophores which expand and close to form double-membrane autophagosomes. This process is initiated by sequential interactions between the nutrient-sensing mTOR kinase and ULK1 protein complex (UKC) which contains ULK1, Atg13 and FIP200. UKC acts downstream of mTOR and binds to a second protein complex containing Beclin 1/Atg6 and a class 3 PI3 kinase, called vps34. Activation of vps34 generates phosphatidylinositol-3-phosphate (Ptdlns(3)P) to initiate recruitment of autophagy proteins to sites of phagophore formation and expansion. The activated vps34/Beclin 1 complex also promotes ubiquitin-like reactions which generate an Atg12-Atg5:Atg16 conjugate, and add phosphatidylethanolamine (PE) to Atg8/LC3 (LC3). The LC3-PE conjugate, also known as LC3II, is anchored in the phagophore and remains with the autophagosome until fusion with the lysosomes.5–7
Viruses are obligate intracellular parasites and as such have to survive innate cellular defenses against infection. Activation of autophagy is emerging as a general response to viral infections but autophagy can have a positive or negative outcome on infection depending on the nature of the virus. Herpes simplex 1 (HSV1), vesicular stomatitis (VSV) and Sindbis viruses are susceptible to autophagy, and inhibition of autophagy leads to increased replication and virulence.4,8,9 Neurovirulence in HSV1 has been linked to the ICP34.5 protein which binds the Beclin 1/vps34 complex and inhibits autophagosome formation.10 In contrast, autophagy can promote infection by picornaviruses, such as poliovirus and coxsackieviruses, because autophagosomes provide sites for replication.11–13 Autophagosomes appear to be induced by picornavirus replicase proteins. Poliovirus 2BC expressed alone results in recruitment of LC3 to membranes but co-expression with nonstructural protein 3A is required to generate the double-membrane autophagosomes thought to house the replication complex.14
It is clear that autophagy can have an impact on virus infection in diverse ways. This has generated considerable interest in the identification of viral proteins able to modulate autophagy and in understanding how viral proteins influence autophagosome formation. The origins of the membranes providing lipids for autophagosomes remain controversial but recent studies suggest that lipids can originate from either the endoplasmic reticulum (ER),15,16 or from mitochondria.17 Thus it is possible that cells can access at least two sources of membrane to generate autophagosomes in response to virus infection. The ER has been implicated as a source of membrane for autophagosomes because ER targeting of Atg14, which regulates the vps34/Beclin 1 complex, is required for autophagy and Atg14 locates to both the ER and the autophagosome phagophore.18 Activation of autophagy recruits DFCP1 (double FYVE-domain containing protein 1) to the ER.19 DFCP1 binds PtdIns(3)P through its FYVE domains suggesting that PtdIns(3)P is generated from ER lipids. The ER domains containing DFCP1 have a characteristic morphology in electron micrographs and have been called omegasomes. The omegasomes recruit the ULK1, Atg14, Atg5 and LC3 proteins important for autophagy induction and it has been proposed that phagophores are generated from ER cisternae via this omegasome intermediate. Mitochondria have been implicated as an alternative source of lipid because Atg5, which is essential for both LC3II recruitment and autophagosome formation, locates to the outer membrane of mitochondria following starvation.17 A flow of lipids and membrane proteins from the mitochondrial outer membrane to autophagosomes in response to starvation was observed by live cell imaging.
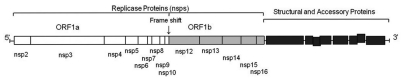
This paper has investigated the autophagy response to infectious bronchitis virus (IBV) nonstructural proteins. IBV is an avian gamma-coronavirus, subfamily Coronavirinae, family Coronaviridae, Order Nidovirales,20 which causes major economic loss to the poultry industry.21 In common with the mammalian β-coronaviruses such as severe acute respiratory syndrome coronavirus (SARS-CoV) and mouse hepatitis Vvirus (MHV), IBV is a large, positive-sense RNA virus in which two-thirds of the genome encodes 15–16 nonstructural proteins (nsps) required for virus replication.22 The nsps are auto-proteolytically processed from two overlapping polyprotein precursors, transcribed from the genomic RNA (Fig. 1), and assembled into replicase-transcriptase complexes associated with convoluted membranes or double-membrane vesicles produced as a result of coronavirus infection.23–27 In this study we have used IBV as a tool to identify viral proteins that activate autophagy and then determine the source of autophagosome membranes. A systematic screen of individual IBV replicase nonstructural proteins for an ability to generate LC3 puncta showed that the nsp6 protein activated autophagy. Nsp6 located to the ER and induced ER puncta containing DFCP1 and Atg5. Autophagosome production by nsp6 was also dependent on Atg5 and class 3 PI3 kinase activity, suggesting generation of PtdIns(3)P from ER lipids to initiate phagophore formation and expansion. The study was extended to the mammalian β-coronaviruses, SARS and MHV, and the porcine arterivirus porcine reproductive and respiratory syndrome virus (PRRSV), family Arteriviridae, Order Nidovirales. In each case the nsp6 orthologs located to the ER and generated autophagosomes. These results suggest that generation of autophagosomes from the ER via an omegosome intermediate is a general feature of the nsp6 proteins or orthologs encoded by viruses belonging to the Nidovirales.
Figure 1.
Schematic of the IBV RNA genome. A schematic of the IBV RNA genome illustrating the locations of the structural and nonstructural virus proteins (nsps).
Results
Infectious bronchitis virus infection redistributes autophagosome marker LC3.
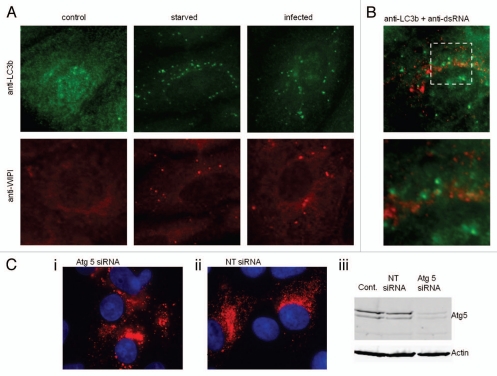
Cells infected with the Beaudette strain of IBV were fixed after 18 h and examined for evidence of autophagosome formation. Figure 2A shows immunofluoresence staining using antibodies specific for the autophagosome marker LC3, and the early autophagosome marker WIPI. In control cells LC3 and WIPI generated diffuse signals indicating even distribution within the cytoplasm. When cells were starved, or infected with IBV, the LC3 and WIPI signals redistributed to punctate vesicles indicative of autophagosomes. In Figure 2B, centers of virus replication were identified using antibodies against double-stranded RNA (dsRNA) and a merge of signals from dsRNA and LC3 in infected cells is shown. Although some co-localization was apparent, there are LC3-positive structures that are negative for dsRNA. Atg5 is a key component of the autophagy machinery. Figure 2Ci–iii show that IBV can infect cells following silencing of Atg5 demonstrating that autophagy is not essential for infection. This is consistent with infection time course studies which showed little change over control after Atg5 silencing, or in cells treated with wortmannin to inhibit the vps34 class3 PI3 kinase (data not shown). Taken together these results show that infection of cells with the avian coronavirus IBV induces autophagosomes, but the autophagosomes are unlikely to be required for virus replication or provide sites for replication.
Figure 2.
IBV infection results in redistribution of autophagy proteins. (A) Vero cells were infected with the Beaudette R isolate of IBV for 18 h, starved in HBSS media for 2 h or untreated. Cells were visualized by fluorescence microscopy. LC3-puncta were identified using antibodies against endogenous LC3b (green), WIPI-puncta were identified using antibodies against endogenous WIPI (red). (B) Vero cells infected with IBV for 18 h were fixed and visualized by fluorescence microscopy using antibodies recognizing dsRNA (red) and antibodies recognizing endogenous LC3 (green). The high power image shows the extent of colocalization of dsRNA and LC3. (C) (i) vero cells transfected with Atg5 targeted siRNA were infected with IBV for 18 h prior to antibody staining for dsRNA (red) and nuclear staining with DAPI, (ii) vero cells transfected with nontargeted siRNA were infected with IBV for 18 h prior to antibody staining for dsRNA (red) and nuclear staining with DAPI, (iii) protein gel blot analysis of control, nontargeted siRNA transfected and Atg5 siRNA transfected, blotting against Atg5 and actin as a loading control.
IBV nsp6 induces the formation of autophagosomes.
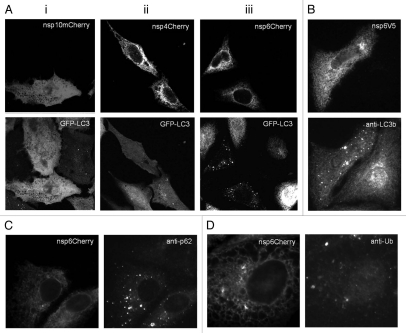
As our initial experiments indicated that IBV was able to induce the formation of autophagosomes, we decided to investigate whether a specific replicase associated nsp encoded by IBV could induce autophagy. The replicase nsps 4 through to 10, and 13 to 16 proteins from the Beau-R strain were fused to a C-terminal mCherry tag and screened by expression in CHO cells expressing GFP-LC3 as an assay system to determine if they could induce autophagosomes. Analysis of the mCherry signal showed most nsps were distributed diffusely throughout cells suggesting they were cytoplasmic proteins (data not shown). Interestingly, nsp4 and nsp6 showed a reticular distribution indicative of localization within the endoplasmic reticulum (ER). However, of the 10 mCherry tagged IBV nsps investigated, only nsp6 caused redistribution of GFP-LC3 to punctate structures indicative of autophagosomes. This is illustrated in Figure 3 which compares the two ER located IBV replicase proteins, nsp4 (Fig. 3Aii) and nsp 6 (Fig. 3Aiii), and nsp10 (Fig. 3Ai) that distributed to the cytosol. To ensure that the induction of autophagy was not due to the mCherry tag, we fused IBVnsp6 to a smaller V5 tag. As shown in Figure 3B this construct retained the ability to induce autophagosome formation. Sequestersome1/p62 is a linker protein that binds ubiquitinated proteins and LC3 and facilitates delivery of ubiquitinated proteins to autophagosomes. Figure 3C shows that IBV nsp6mCherry expression resulted in increased numbers of p62 positive puncta, again indicating induction of autophagosomes. Immunofluorescence signals for polyubiquitin were, however, separate from IBVnsp6mCherry suggesting that IBVnsp6 was not itself ubiquitinated (Fig. 3D). This is consistent with later experiments (Fig. 9) that show a homogeneous signal for nsp6 on protein gel blots indicating an absence of covalently attached chains of polyubiquitin.
Figure 3.
IBV nonstructural protein 6 induces vesicles containing LC3. (A) IBV nsps 10, 4 and 6 with C-terminal mCherry tags were expressed in CHO cells expressing GFP-LC3 and the distribution of the proteins was determined from their natural fluorescence. (i) IBVnsp10 locates to the cytosol and does not induce LC3 puncta, (ii) IBVnsp4 locates to reticular membranes and does not induce LC3 puncta, (iii) IBVnsp6 locates to reticular membranes and induces LC3 puncta. (B) IBVnsp6 with a C-terminal V5 tag was expressed in Vero cells. Cells were visualized by fluorescence with antibodies recognizing V5 and endogenous LC3. (C) Vero cells expressing IBVnsp6mCherry were stained with antibodies recognizing endogenous p62 and (D) polyubiquitin.
Figure 9.
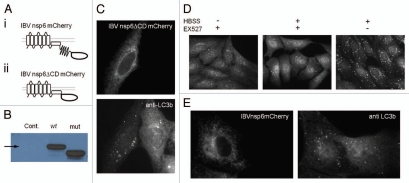
Deletion of hydrophilic cytoplasmic domain of IBVnsp6, and effect of EX527. (A) Schematic of protein structure of (i) mCherry tagged IBVnsp6 and (ii) mCherry tagged truncated IBVnsp6 termed ‘IBVnsp6ΔCDmCherry’. (B) SDS-PAGE protein gel blot analysis of wild-type and truncated protein using antibodies against mCherry. (C) IBVnsp6ΔCDmCherry was expressed in vero cells and stained with antibodies against endogenous LC3. (D) Vero cells were incubated with HBSS to induce starvation, EX527 to inhibit Sirtuin 1, or a combination. The cells were fixed and immunostained with antibodies against endogenous LC3. (E) Vero cells expressing IBVnsp6mCherry were incubated in the presence of 50 nM EX527 for 4 h prior to being fixed and immunostained with antibodies recognizing endogenous LC3.
Activation of autophagy by IBV nsp6 requires Atg5 and the recruitment of lipidated LC3II to autophagosomes.
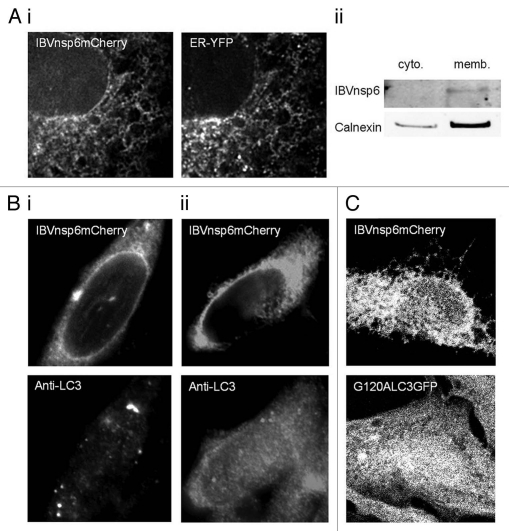
The IBV nsp6 contains seven predicted transmembrane domains, and gave a reticular staining pattern when expressed alone in cells. To further investigate the cellular location of the protein, IBV nsp6-mCherry was co-expressed with an ER-targeted YFP protein. Figure 4Ai shows co-localization of nsp6-mCherry with the ER-YFP marker, confirming that nsp6 was targeted to the ER. Cell homogenates prepared from cells expressing IBVnsp6mCherry were fractionated to separate cytoplasmic and membrane fractions which were probed by protein gel blot for IBVnsp6mCherry and integral ER membrane protein calnexin. Figure 4Aii shows that IBVnsp6mCherry was only visible within the membrane fraction enriched for calnexin. This evidence for ER location agrees with previous work studying the membrane orientation of the first 6 hydrophobic domains of MHVnsp6 which demonstrated that the protein is an integral transmembrane ER protein.28
Figure 4.
IBVnsp6 is localized to the ER and induces autophagosomes induced that require Atg5 and recruit LC3II. (A) (i) Vero cells expressing IBVnsp6 tagged with mCherry were fixed and processed for fluorescence microscopy. The distribution of IBVnsp6 with a co-expressed ER marker tagged with YFP in Vero cells. (ii) Cell lysates were separated into cytoplasmic and membrane fractions. Both fractions were analyzed by SDS-PAGE protein gel blot using antibodies against Calnexin (an endogenous ER resident protein) and RFP (to detect IBVnsp6mCherry). (B) (i) IBVnsp6 tagged with mCherry was expressed in MEF cells, antibodies recognizing endogenous LC3 were used to identify LC3 punctae, (ii) IBVnsp6 tagged with mCherry was expressed in ATG5-/- MEF cells, antibodies recognizing endogenous LC3 were used to identify LC3 puncta. (C) IBVnsp6 tagged with mCherry was expressed in CHO cells expressing GFP-LC3-G120A.
The mechanism of LC3 redistribution induced by IBVnsp6 was investigated further. Recent studies have shown the recruitment of LC3 to double-membrane vesicles (DMVs) formed from the ER during coronavirus infection. The work demonstrated that the DMVs induced by MHV recruited non-lipidated LC3 (LC3I) by a pathway that is independent of autophagy but is linked to the formation of vesicles, called EDEMosomes, which export ER chaperones from the ER to endosomes.29 These results raised the possibility that the structures induced by IBV nsp6 may be DMVs related to EDEMosomes, rather than autophagosomes. At first this assumption appeared unlikely since DMVs induced by coronaviruses do not recruit GFP-LC3, and our results indicated that the GFP-LC3 probe was recruited to vesicles induced by IBV nsp6. Atg5 is essential for autophagy but it is not required for DMV formation or mammalian coronavirus replication.30 To determine if Atg5 was required for the formation of autophagosomes induced by IBV nsp6, and to distinguish them from DMVs and edemosomes, the viral protein was expressed in Atg5-/- mouse embryo fibroblast (MEF) cells. As shown in Figure 4B, LC3-positive vesicles were induced by IBVnsp6 in the wild-type MEF cells (Fig. 4Bi) but they were not induced in MEF cells lacking Atg5 (Fig. 4Bii). Activation of autophagy results in the removal of C-terminal amino acids from LC3 and addition of PE generating LC3II which is then translocated to phagophores. This addition of PE is not required for recruitment of LC3I to DMVs in MHV infected cells.29 IBV nsp6 was expressed in CHO cells expressing GFP-LC3-G120A where the G120A substitution prohibits cleavage of LC3. Figure 4C shows that IBVnsp6 was unable to induce redistribution of LC3 carrying the G120A substitution. This demonstrated that conversion of LC3I to LC3II by cleavage and subsequent PE lipidation is required for recruitment of LC3 to vesicles induced by IBVnsp6. Taken together these results show that IBV nsp6 expressed in the ER induces the formation of autophagosomes rather than EDEMosomes.
IBV nsp6 induces PI3P-dependent omegasomes.
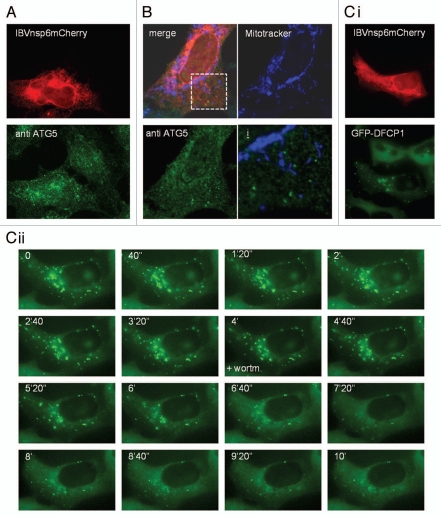
Recent work shows that autophagosome membranes can be derived from mitochondria17 or the ER.19 The source of autophagosome membranes can be inferred from the location of early autophagosome markers such as Atg5, or by searching for sites of Beclin 1/vps34 induced PtdIns(3)P. Staining of endogenous Atg5 in fixed cells expressing IBVnsp6mCherry showed Atg5 puncta aligned along reticular structures positive for nsp6 (Fig. 5A). The colocalization of nsp6 with ER markers (Fig. 4A) suggested that the Atg5 puncta originated from the ER. Immunofluorescence analysis within cells incubated with mitotracker showed that Atg5 puncta induced by nsp6 did not colocalize with mitochondria (Fig. 5B) arguing against mitochondria as a source of autophagosomes induced by nsp6. Activation of autophagy in response to amino acid starvation has also been shown to induce PtdIns(3)P in vesicles close to the ER called omegasomes.19 Omegasomes recruit Atg5 and LC3II and are thought to be sites for the generation of autophagosome phagophores. Formation of omegasomes can be followed using a DFCP1-GFP probe in which the double-FYVE domains binds PtdIns(3)P. Therefore we investigated the effect of IBV nsp6 on HEK cells expressing DFCP1-GFP (Fig. 5C). IBVnsp6 caused redistribution of DFCP1 to transient punctate structures aligned along ER membranes (Fig. 5Ci), suggesting that IBVnsp6 induces PtdIns phosphorylation at the ER consistent with the formation of autophagosomes via an ER-derived omegasome intermediate. Due to the fact that omegasome formation requires the PI3Kinase complex, we tested the requirement for PI3kinase activity using wortmannin. The time-lapse experiment in Figure 5Cii shows that addition of wortmannin 4 min into the experiment resulted in rapid reduction of omegasomes induced by IBVnsp6.
Figure 5.
Expression of IBV nsp6 leads to formation of omegasomes that are sensitive to wortmannin. (A) IBVnsp6 tagged with mCherry was expressed in HEK cells, which were immunostained for Atg5 to identify Atg5 puncta indicative of autophagosomes. (B) IBVnsp6 tagged with V5 was expressed in Vero cells were incubated with MitoTracker (to stain mitochondria) before being fixed and immunostained for V5 and Atg5. The merge of all three is shown, and the individual stains of the mitochondria (blue) and Atg5 (green). The bottom right image shows a high resolution merge of the mitochondrial stain and the Atg5 stain. (C) (i) IBVnsp6 tagged with mCherry was expressed in HEK cells constitutively expressing DFCP1-GFP, omegasomes were identified as punctate DFCP1-GFP signals. (ii) The cells expressing IBVnsp6 mCherry and constitutively expressing DFCP1-GFP were analyzed by live cell microscopy. DFCP1-GFP puncta were monitored using time lapse at the times indicated, wortmannin (+wortm) was added at 4 min (4′).
Autophagosomes induced by IBVnsp6 fuse with lysosomes.
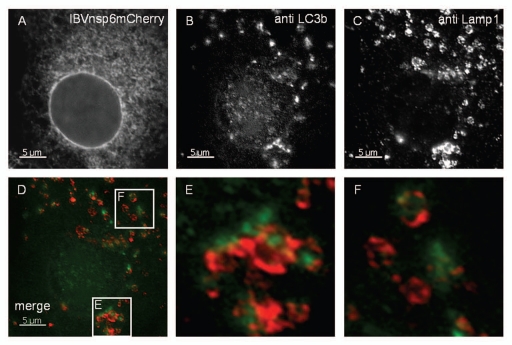
In the final stages of autophagy the autophagosomes generated in response to amino acid starvation engulf cytoplasmic contents and fuse with lysosomes. The fate of the LC3-positive vesicles induced by IBV nsp6 was investigated to determine whether they could mature into structures capable of completing the autophagy pathway to lysosome fusion. Cells expressing IBV nsp6 were probed with antibodies recognizing LC3 and lysosome marker Lamp1. Figure 6 shows LC3 puncta interspersed with Lamp1-positive vesicles. The cells were also incubated with bafilomycin to prevent degradation of LC3 in lysosomes. The regions of interest in Figure 6D are presented at high magnification in Figure 6E and F and show that the LC3 signal was often located within Lamp1-positive structures indicating fusion of autophagosomes with lysosomes. Given that IBVnsp6 is located to the ER and the autophagosomes originate from the ER, it was possible that IBVnsp6 may travel with the autophagosomes and be degraded in lysosomes. Close examination of LC3 positive or Lamp 1 positive vesicles, however, failed to show co-localization with IBVnsp6, even in the presence of bafilomycin. Taken together the results suggest that IBV nsp6 induces the formation of bona fide autophagosomes from the ER, but IBV nsp6 does not travel with the autophagosomes to the lysosomes.
Figure 6.
Autophagosomes induced by nsp6 fuse with lysosomes. Vero cells expressing IBVnsp6mCherry (A) were immunostained with antibodies recognizing endogenous LC3 (B) and LAMP1 (C). (E and F) show high magnification of merged images of the regions of interest indicated in (D). Endogenous LC3b is shown in green and LAMP1 immunostaining in red.
Nsp6 orthologs of mammalian coronaviruses and arteriviruses locate to the endoplasmic reticulum and activate autophagy.
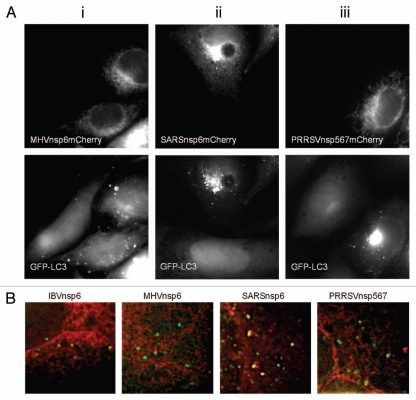
The nsp6 proteins of all coronaviruses share the 6–7 transmembrane topology predicted for their IBV counterpart suggesting that they may also locate to the ER. Arteriviruses also express a multiple transmembrane protein equivalent of the coronavirus nsp6 protein. This protein is termed nsp5–7 and is the predominant replicase-derived protein expressed during arterivirus infection, the nsp5 precursor, and has the membrane-spanning domains of nsp5 lying N-terminal to the nsp6 and 7 protein domains.31 PRRSV nsp5–7 precursor protein was chosen as a representative arterivirus nsp6 ortholog, and the nsp6 proteins of MHV and SARS-CoV were chosen as mammalian coronavirus representatives to investigate whether they located to the ER and activated LC3 positive autophagosomes in an analogous manner as identified for the IBV nsp6 protein. Each protein was fused to a C-terminal mCherry tag and expressed. As can be seen from Figure 7Ai–iii, all three viral proteins distributed to reticular structures spread throughout the cytoplasm. Separate experiments (Fig. S1) showed that the proteins co-localized with the ER marker PDI. The ability of these ER-localized viral proteins to induce autophagosomes was then investigated using the CHO GFP-LC3 reporter cell line as described for IBV nsp6. All three proteins induced translocation of LC3-GFP to punctate structures indicative of autophagosomes. Thus the ability of these multiple transmembrane domain proteins to locate to the ER and induce autophagy is retained throughout the Nidovirales order. Interestingly, merged images in Figure 7B showed that the IBV, MHV and PRRSV nsp6 signals were separate from LC3 suggesting that the viral proteins induce autophagosome formation from the ER, but remain in the ER and do not travel with autophagosomes released from the ER into the cell. In contrast the SARSnsp6mCherry partially colocalized with the LC3 positive structures suggesting it may enter autophagosomes.
Figure 7.
Nsp6 orthologs from MHV, SARS-CoV and PRRSV induce vesicles containing LC3 and show differences in extent of incorporation into autophagosomes. (A) (i–iii) MHVnsp6mCherry, SARSnsp6mCherry and PRRSVnsp567mCherry, were expressed in CHO cells expressing GFP-LC3. The distribution of the proteins was determined from their natural fluorescence. GFP-LC3 signals are shown in the lower parts. (B) IBVnsp6mCherry, MHVnsp6mCherry, SARSnsp6mCherry and PRRSVnsp567mCherry, were expressed in CHO cells expressing GFP-LC3. The distribution of the proteins was determined from their natural fluorescence. High magnification images were merged to determine extent of colocalization between the nsp6 orthologs and GFP-LC3. Notably vesicular structures containing SARS-CoV colocalize with LC3-GFP.
Nsp6 orthologs do not suppress mTOR signaling or induce excessive ER stress.
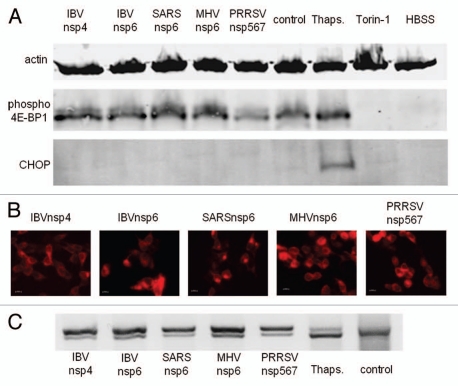
Autophagosomes were induced by nsp6 proteins in nutrient-rich conditions, which would be expected to suppress autophagy through mTOR kinase signaling. This raises the possibility that the nsp6 proteins may activate autophagosome formation by inhibiting the mTOR kinase directly. Therefore the activation of substrates downstream of mTOR was assessed in cells expressing nsp6 (Fig. 8A). Control experiments using torin-1 or HBSS showed that inhibition of mTOR resulted in loss of phosphorylated 4E-BP1. In contrast, expression of IBV, SARS-CoV, MHV or PRRSV nsp-6 orthologs did not affect levels of phosphorylated 4E-BP1 showing that they did not inhibit the mTOR kinase.
Figure 8.
Expression Nsp6 orthologs does not inhibit mTOR or induce CHOP. HEK cells expressing the nsp6 orthologs were lysed and analyzed by SDS PAGE followed by protein gel blot. (A) In parallel experiments HEK cells were transfected with (Lane 1) IBVnsp4mCherry, (Lane 2) IBVnsp6mCherry, (Lane 3) SARSnsp6mCherry, (Lane 4) MHVnsp6mCherry, (Lane 5) PRRSVnsp5–7mCherry, (Lane 6) left untreated, (Lane 7) incubated with thapsigargin to induce ER stress, (Lane 8) Torin-1 to inhibit mTOR, or (Lane 9) starved in HBSS to induce autophagy, as indicated. Protein gel blots were probed with antibodies to actin as a loading control, anti phospho4E-BP1 and CHOP. (B) Shows expression of transfected proteins at x40 magnification; (1) IBVnsp4mCherry, (2) IBVnsp6mCherry, (3) SARSnsp6mCherry, (4) MHVnsp6mCherry, (5) PRRSVnsp5–7mCherry. (C) Transfected cells, cells incubated with thapsigargin and control HEK293 cells were subjected to PCR analysis using primers specific for XBP1 mRNA.
Autophagy can be induced independently of starvation through ER stress. The observation that the nsp6 orthologs located to the ER raised the possibility that nsp6 may induce ER stress. The C/EBP homologous protein (CHOP) is known to be induced during ER stress32 and therefore the expression of CHOP was assessed in cells expressing the nsp6 proteins. Figure 8 shows that induction of ER stress by thapsigargin increased expression of CHOP but expression of CHOP was not induced by the nsp6 proteins. An upstream and earlier marker of ER stress is the splicing of XBP1 mRNA. This was also investigated through limited cycle PCR (Fig. 8C) where splicing is indicated by generation of a more rapidly migrating PCR product. Thapsigargin induced near complete splicing of XBP1 mRNA indicated by a major band migrating faster than the PCR product from control cells. The nsp6 orthologs were found to trigger some splicing of XBP1mRNA indicated by the doublet seen on the gels, however, the extent of splicing was less than that induced by thapsigargin. Additionally IBVnsp4, which locates to the ER but does not induce autophagosomes was found to induce similar levels of XBP1 mRNA splicing. Thus expression of nsp6 orthologs may trigger limited ER stress, but not to the extent required to induce autophagy.
Deletion of hydrophilic cytoplasmic domain does not abrogate the ability of IBVnsp6 to induce autophagy.
IBVnsp6 is predicted to contain 7 hydrophobic transmembrane domains, followed by a hydrophilic cytoplasmic domain. To determine whether or not the hydrophilic cytoplasmic domain was important for induction of autophagy by IBVnsp6, we constructed a deletion mutant that contained only the hydrophobic domains. Figure 9A shows a schematic of the deletion mutant and Figure 9B demonstrates the smaller molecular weight of the truncated protein as detected by protein gel blot. The truncated protein was expressed in Vero cells where it localized to the ER and induced autophagosomes indicated by increased numbers of LC3 puncta (Fig. 9C). The results show that the cytoplasmic domain of nsp6 is not required for induction of autophagy.
Sirtuin 1 is not required for IBV nsp6-induced autophagy.
Recent studies show that autophagy can be induced following sirtuin 1-mediated protein deacetylation.33 Sirtuin 1 deacetylates Atg5, Atg7 and LC3,34 and activates autophagy by a pathway that is independent of mTOR signaling.35 This raised the possibility that nsp6 proteins may activate autophagy through activation of sirtuin 1. The effects of pharmacological inhibition of sirtuin 1 by EX527 on autophagy induced by starvation or IBVnsp6 are compared in Figure 9D and E. EX527 inhibited autophagosome formation in response to starvation but did not prevent induction of autophagy by IBVnsp6. Therefore IBVnsp6 induces autophagy through a pathway independent of both sirtuin 1 deacetylation and mTOR signaling.
Discussion
This study has investigated the ability of individual replicase nonstructural proteins encoded by coronaviruses to activate autophagy. The first experiment showed that IBV infection induced redistribution of LC3 to vesicular structures indicative of autophagosomes. A series of 10 IBV-derived replicase nonstructural proteins derived from the IBV replicase polyprotein were tagged with m-Cherry and expressed in a GFP-LC3 reporter cell line to determine whether they could induce autophagosome formation. Eight proteins showed diffuse fluorescence and were unable to change the distribution of GFP-LC3. However, two of the proteins that contained membrane-spanning motifs, nsp4 and nsp6, localized to the ER, and nsp6 caused redistribution of LC3 to punctate structures indicative of autophagosomes. Recent studies showed that DMVs formed from the ER during coronavirus MHV infection can recruit LC3.29 These DMVs recruited the non-lipidated LC3 (LC3I) by a pathway that is independent of autophagy and linked to the export of ER chaperones from the ER to endosomes.36 The MHV-derived DMVs therefore differ from autophagosomes where recruitment of LC3II to membranes is dependent on autophagy, and requires the Beclin 1/vps34 class III PI3 kinase complex, Atg12-Atg5 and Atg16 and the addition of PE to the C-terminus of LC3. Our results showed that these three stages of autophagy were required for the redistribution of LC3 by IBV nsp6. The LC3 puncta were positive for Atg5, and were not formed in cells lacking Atg5, or when class III PI3 kinase activity was inhibited by wortmannin. In addition, LC3 puncta induced by IBV nsp6 were unable to incorporate a modified G120A LC3 that lacked the glycine required for addition of PE. These observations show that LC3 puncta generated by IBV nsp6 are autophagosomes rather than DMVs related to EDEMosomes. Further evidence for formation of bona fide autophagosomes by IBV nsp6 comes from the observation that the LC3 puncta induced by nsp6 complete the autophagosome-lysosome pathway and deliver LC3 to the interior of lysosomes for degradation. This demonstrates that the autophagosomes generated by IBV nsp6 therefore recruit and recycle the proteins needed for autophagosome nucleation, expansion, cellular trafficking and delivery of cargo to the lysosomes.
The origins of the membranes that generate autophagosomes in response to starvation remain the subject of intense research, and organelles as diverse as the Golgi, ER and mitochondria have been implicated.17,19 Our observation that IBV nsp6 located to the ER raised the possibility that the IBV nsp6 could drive autophagosome formation from the ER. Live cell imaging of autophagosomes formed from the ER in response to starvation has shown that a ring of membrane enriched for PtdIns(3) P, called an omegasome, expands from the ER and recruits LC3 to its interior.19 The LC3 signal subsequently moves from the omegasome and the Ptdlns(3)P signal decays rapidly. The formation of autophagosomes by IBV nsp6 also involved an omegasome intermediate. Nsp6 generated wortmannin-sensitive vesicular structures associated with the ER that recruited Atg5 and LC3. Importantly, the structures induced by nsp6 recruited the DFCP1-GFP probe specific for Ptdlns(3)P showing they contained high levels of the Ptdlns(3)P necessary to nucleate autophagosome formation. The half-life of DFCP-1-GFP recruitment was short and decayed at a similar rate to omegasomes in the presence of wortmannin. The results suggest that IBV nsp6 can increase Ptdlns(3)P concentration at the ER, independently of starvation, and induce omegasomes which then seed formation of autophagosomes. It is important to note that while we showed that exogenously expressed nsp6 activates autophagy, we do not have direct evidence that nsp6 activates autophagy during virus infection. It is possible that during infection autophagy is activated by other viral components, for example dsRNA, or complexes of viral proteins that we have not reconstituted in our experiments. It is also possible that nsp6 is part of a protein complex during viral infection and this may modulate the ability of nsp6 to activate autophagy.
The seven transmembrane domain topology of the IBV nsp6 protein is shared with the nsp6 protein of the mammalian coronaviruses SARS-CoV and MHV, and the nsp5–7 protein of the arterivirus, PRRSV. The ability to generate autophagosomes directly from the ER was retained by these nsp6 orthologs throughout the Nidovirales order. All four proteins located to the ER where they induced the formation of autophagosomes. Notably the coronavirus nsp6 protein described in this study is the first identified ER resident protein that induces omegasome/autophagosome progression. This provides additional support for the idea that at least some autophagosomes are formed from ER-connected omegasomes and that this pathway can be activated by viruses. Electron micrographs of cells expressing a defective form of the Atg4 protease (Atg4C74A) required for addition of PE to LC3 reveal crescent-shaped phagophores (IM) sandwiched between ER cisternae.37 These ER-IM complexes contain high concentrations of DFCP1 and are interconnected by narrow membrane bridges. It has been suggested that these specialized domains of the ER facilitate the formation of phagophores.38 It will be interesting to see if similar ER-IM domains are generated by IBV nsp6 to seed autophagosome formation.
The omegasomes and autophagosomes induced by IBV nsp6 formed independently of starvation. A potential explanation was that nsp6 protein could mimic starvation by inhibiting mTOR kinase signaling directly or, as seen for the inhibition of Akt by VSV G protein,39 inhibit nutrient-sensing pathways upstream of mTOR. mTOR signaling, however, appeared to be unaffected since phosphorylation of 4E-BP1 did not change in cells expressing nsp6. Given that all the coronavirus nsp6 ortholog proteins analyzed located to the ER, it was possible that they activated autophagy by inducing ER stress. This appeared unlikely since it was not possible to detect increased levels of CHOP in cells expressing nsp6. Splicing of XBP1 mRNA is an early upstream ER stress response. The nsp6 proteins activated splicing of XBP1 mRNA, however the degree of splicing was less than seen in response to thapsigargin and the same as splicing induced by nsp4, which was unable to induce autophagy. Autophagy can be induced independently of mTOR signaling by activation of sirtuin 1 but inhibition of the sirtuin 1 deacetylase failed to inhibit autophagy induced by nasp6 and the precise mechanism of activation remains unknown. IBV nsp6 did however recruit DFCP-GFP to the ER indicating that induction of autophagosome formation by nsp6 may involve increased production of Ptdlns(3)P. Nsp6 may mimic Atg14 and recruit the Vps34/p150/Beclin 1 complex directly to the ER. Alternatively, nsp6 may inhibit the phosphatase Jumpy that can inhibit autophagosome nucleation by removing phosphate from Ptdlns(3) P.40 Deletion analysis showed that the cytoplasmic domain of nsp6 is not required for activation of autophagy and does not therefore play a role in recruiting proteins to the cytoplasmic face ER.
Large numbers of DMVs are generated in the cytoplasm of cells infected with mammalian coronaviruses and arteriviruses. Electron tomography shows that DMVs generated by SARS-CoV are connected to the ER, with the interior of the DMVs labeling for dsRNA.24 The observation that MHV is unable to generate DMVs in mouse embryonic stem cells lacking Atg5, and that replication is reduced 1,000-fold in the absence of Atg5, suggested a functional link between DMVs, autophagy and virus replication.41 Other studies suggest that MHV does not require Atg5,30 or Atg7,29 for replication in primary fibroblasts or macrophages and question a role for autophagosomes in virus replication. Our study of the avian coronavirus IBV also failed to provide a link between autophagosomes and virus replication. Double-stranded RNA did not colocalize with LC3 and virus replication was unaffected by inhibitors of autophagy or silencing of Atg5. The DMVs induced by mammalian coronavirus and arteriviruses are approximately one-third the diameter of cellular autophagosomes, suggesting that they are modified autophagosomes, or independent structures induced by assembly of viral proteins. Recent work suggests that the DMVs are related to EDEMosomes, rather than autophagosomes, and recruit LC3I by a pathway independent of autophagy. Autophagy may not, therefore, be required to generate DMVs for replication but, in common with other virus infections, the activation of autophagy seen during coronavirus and arterivirus replication may represent an innate defense against infection to clear virus particles, or a modification of the adaptive immune response by increasing MHC class II antigen presentation. In addition, nsp6 may alter adaptive immune responses by directing immunomodulatory proteins synthesized by the ER into autophagosomes for degradation.
Methods
Cells and virus.
The CHO cell line stably expressing GFPLC3 (CHO-GFPLC3) and the CHO cell line expressing GFP-LC3G120A (CHO GFP-LC3G120A) were described previously in reference 42. African green monkey kidney epithelial cells or Vero cells (ECACC 84113001), were obtained from European Collection of Cell Cultures (ECACC) (Porton Down, UK). MEF wild-type (WT) and MEF Atg 5-/- cell lines were described previously in reference 43. HEK 293 cells stably expressing the double FYVE domain-containing protein 1 (DFCP1) were described previously in reference 19. All cells were grown at 37°C and 5% CO2. IBV, the Beaudette Beau-R isolate,44 was provided by the Institute for Animal Health (IAH), Compton Laboratory, and PRRSV RNA, was provided by Wilhelm Gerner, University of Veterinary Medicine Vienna.
Plasmid construction and transfection.
Virus RNA was extracted using TRIzol (Invitrogen, 15596-026), and reverse transcribed with Superscript II (Invitrogen, 18064022). PCR primers were designed to amplify a specific IBV nsp (Table S1), and products were amplified using the Expand Hi-fidelity system (Roche, 11 732 641 001). The individual nsps of IBV Beau-R were cloned into pmCherryN1 (Clontech, 632523) using Xho1 and BamH1 sites within the multiple cloning site. The IBVnsp6 PCR product was also cloned into the pcDNA3.1 vector (Invitrogen, V790-20) that contains a V5 tag. The SARS-CoV and MHV nsp6 sequences were synthesized and cloned into pmCherryN1 by GENEART. The truncated IBVnsp6 construct (IBVnsp6ΔCDmCherry) was amplified by PCR using primers described in Table S1, and then cloned into pmCherryN1. For immunofluorescence, plasmids were transfected into cells using calcium phosphate (Sigma CAPHOS-1KT), and the transfection mix was washed away after 8 h. For protein analysis, plasmids were transfected using jetPRIME (Polyplus transfection, 114-01) according to the manufacturer's protocol. The pEYFP-ER construct contains the calreticulin ER target sequence and a KDEL sequence (Biosciences, 6906-1BD). Atg5 siRNA (L-004374-00-0005, Thermo Scientific Dharmacon), was transfected using jetPRIME at a final concentration of 50 nM.
Antibodies and fluorescent stains.
Antibodies used for immunofluorescence were as follows; rabbit anti-LC3b and rabbit anti-ATG5 (Sigma, A0731), mouse anti-dsRNA J2 (English and Scientific Consulting, 10010200), rabbit anti-PDI was raised against the c-terminal peptide of PDI, chicken anti-IBV (Charles River, 10100604), mouse anti-WIPI was a gift from S. Tooze, anti-V5 (Invitrogen R960-25), Guinea pig anti-p62 (PROGEN Biotechnik, GP62-C). MitoTracker® RedCMXRos (Molecular Probes, Invitrogen, M7512) was used at a final concentration of 400 nM.
Mouse anti β-actin (Sigma, A5441), mouse anti-CHOP (Cell Signaling Technology, 2895), mouse anti-RFP (AbCam ab65856), rabbit anti-phospho (Ser65)4E-BP1 (Cell Signaling Technology, 9451), rabbit anti-ATG5 (Cell Signaling Technology, 2630) were all used as primary antibodies for protein gel blot analysis.
Drug treatments.
Wortmannin (Sigma, W1628) was used at a final concentration of 67–100 nM in complete media. Thapsigargin (Sigma, T9033) was used at a final concentration of 1 nM in complete media. TORIN-1 (Whitehead Institute for Biomedical Research and Dana-Farber Cancer Institute) was used at a final concentration of 0.5 µM in complete media. Cells were starved in Hank's Balanced Salt Solution (HBSS, Invitrogen, 14025-076). EX527 (Tocris Bioscience, 2780) was used at a final concentration of 50 µM.
Fractionation.
HEK 293 cells were homogenized in 8% sucrose (Sigma, 84097) solution by passing through a number 25 needle 20 times. The lysate was then centrifuged at 6,000 rpm for 2 min to remove the nuclear fraction. The post-nuclear fraction was then centrifuged at 15,000 rpm for 20 min to separate the membranes from the cytoplasmic fraction. The membrane pellet was then resuspended in SDS buffer.
XBP1 PCR.
RNA was extracted using TRIzol (Invitrogen, 11596-026) and 2 µg RNA was reverse transcribed with Superscript II. 3 µl of cDNA was amplified using GoTaq® Hot Start Polymerase (Promega, M5005), in a limited PCR reaction of 25 cycles annealing at 55°C, elongating for 30 sec using primers described previously in reference 45. PCR products were run on a 2% agarose gel at 100 V for 1 h.
Protein gel blot analysis.
Cell lysates were denatured using hot SDS lysis buffer (1% SDS, 10 mM Tris pH 6.8), and sonicated before running 50 µg of total protein lysate on 12% acrylamide gels using the BioRad system. Proteins were transferred on to PVDF membranes and probed using specific primary antibodies at 1:1,000 dilution, and IRDye®-labeled secondary antibodies (Li-Cor Biosciences, 926-322, 926-680) at 1:5,000 dilution or HRP conjugated antibodies (Jacksons Laboratories, 111-035-003). Proteins detected by the labeled secondary antibodies were visualized on the Odyssey infrared system, or enhanced chemiluminescence detection of horseradish peroxidase activity. Membranes were stripped and reblotted using Re-blot plus strong solution (10x) (Millipore, 2504).
Imaging.
Cells were fixed using 100% ice-cold methanol for anti-LC3b staining, or in 4% paraformaldehyde (Sigma, P6148) with Triton-X100 (Sigma, T8787) mediated permeabilization for other antibodies. Cells were stained using antibodies at a 1:1,000 dilution in 2% bovine serum albumin (Sigma, A9418)/phosphate buffered saline. The fixed cell images were obtained at x63 magnification on a Zeiss Axioplan 2, unless otherwise stated. Images were analyzed using the Axioplan software version 4.7.1.
Acknowledgements
The work reported here was funded by BBSRC grant BB/E018521/1. We would like to thank Dr. Zvulun Elazar of the Weizmann Institute of Science for the CHO GFP-LC3 cells, and Noboru Mizushima of Tokyo Medical and Dental University for the MEF wild-type (WT) and MEF Atg 5-/- cell lines. We are also grateful to Dr. Sharon Tooze of Cancer Research UK London Research Institute for the WIPI antibody and Dr. David Sabatini, Whitehead Institute, MIT for Torin-1.
Abbreviation
- IBV
infectious bronchitis virus
- SARS CoV
severe acute respiratory syndrome coronavirus
- MHV
mouse hepatitis virus
- PRRSV
porcine respiratory and reproductive syndrome virus
- DFCP1
double FYVE-domain containing protein 1
- Ptdlns(3)P
phosphatidylinositol 3-phosphate
- ER
endoplasmic reticulum
- DMV
double-membrane vesicle
- nsp
nonstructural protein
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 2.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 3.Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 4.English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol. 2009;10:480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burman C, Ktistakis NT. Regulation of autophagy by phosphatidylinositol-3-phosphate. FEBS Lett. 2010;584:1302–1312. doi: 10.1016/j.febslet.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki K, Ohsumi Y. Current knowledge of the pre-autophagosomal structure (PAS) FEBS Lett. 2010;584:1280–1286. doi: 10.1016/j.febslet.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Cherry S. VSV infection is sensed by Drosophila, attenuates nutrient signaling, and thereby activates antiviral autophagy. Autophagy. 2009;5:1062–1063. doi: 10.4161/auto.5.7.9730. [DOI] [PubMed] [Google Scholar]
- 9.Orvedahl A, MacPherson S, Sumpter R, Jr, Talloczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leib DA, Alexander DE, Cox D, Yin J, Ferguson TA. Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T-cell responses. J Virol. 2009;83:12164–12171. doi: 10.1128/JVI.01676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cottam E, Pierini R, Roberts R, Wileman T. Origins of membrane vesicles generated during replication of positive-strand RNA viruses. Future Virology. 2009;4:473–485. doi: 10.2217/fvl.09.26. [DOI] [Google Scholar]
- 12.Kirkegaard K. Subversion of the cellular autophagy pathway by viruses. Curr Top Microbiol Immunol. 2009;335:323–333. doi: 10.1007/978-3-642-00302-8_16. [DOI] [PubMed] [Google Scholar]
- 13.Wileman T. Aggresomes and autophagy generate sites for virus replication. Science. 2006;312:875–878. doi: 10.1126/science.1126766. [DOI] [PubMed] [Google Scholar]
- 14.Suhy DA, Giddings TH, Jr, Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J Virol. 2000;74:8953–8965. doi: 10.1128/JVI.74.19.8953-65.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for auto-phagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 16.Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 17.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–521. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol-3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Groot RJ, Ziebuhr J, Poon LL, Woo PC, Talbot PJ, Rottier PJM, et al. Taxonomic proposal of the ICTV Executive Committee: Revision of the family 25 Coronaviridae. 2008 [Google Scholar]
- 21.Cavanagh D, Gelb J. Infectious bronchitis. In: Saif YM, Fadly AM, Glisson JR, McDougald LR, Nolan LK, Swayne DE, editors. Diseases of Poultry Ames. Blackwell Publishing Professional; 2008. pp. 117–135. [Google Scholar]
- 22.Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- 23.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziebuhr J. The coronavirus replicase. Curr Top Microbiol Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ulasli M, Verheije MH, de Haan CA, Reggiori F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell Microbiol. 2010;12:844–861. doi: 10.1111/j.1462-5822.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Britton P, Cavanagh D. Nidovirus genome organization and expression mechanisms. In: Perlman S, Gallagher T, Snijder EJ, editors. Nidoviruses. Washington, DC: ASM Press; 2008. pp. 29–46. [Google Scholar]
- 28.Baliji S, Cammer SA, Sobral B, Baker SC. Detection of nonstructural protein 6 in murine coronavirus-infected cells and analysis of the transmembrane topology by using bioinformatics and molecular approaches. J Virol. 2009;83:6957–6962. doi: 10.1128/JVI.00254-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reggiori F, Monastyrska I, Verheije MH, Cali T, Ulasli M, Bianchi S, et al. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Z, Thackray LB, Miller BC, Lynn TM, Becker MM, Ward E, et al. Virgin HWt. Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007;3:581–585. doi: 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]
- 31.Ziebuhr J, Snijder EJ, Gorbalenya AE. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 32.Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11:381–389. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 33.Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, et al. The life span-prolonging effect of sirtuin-1 is mediated by autophagy. Autophagy. 2010;6:186–188. doi: 10.4161/auto.6.1.10817. [DOI] [PubMed] [Google Scholar]
- 34.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010;1:10. doi: 10.1038/cddis.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Haan CA, Molinari M, Reggiori F. Autophagyindependent LC3 function in vesicular traffic. Autophagy. 2010;6:994–996. doi: 10.4161/auto.6.7.13309. [DOI] [PubMed] [Google Scholar]
- 37.Fujita N, Noda T, Yoshimori T. Atg4B(C74A) hampers autophagosome closure: a useful protein for inhibiting autophagy. Autophagy. 2009;5:88–89. doi: 10.4161/auto.5.1.7183. [DOI] [PubMed] [Google Scholar]
- 38.Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 39.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30:588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vergne I, Deretic V. The role of PI3P phosphatases in the regulation of autophagy. FEBS Lett. 2010;584:1313–1318. doi: 10.1016/j.febslet.2010.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prentice E, Jerome WG, Yoshimori T, Mizushima N, Denison MR. Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fass E, Shvets E, Degani I, Hirschberg K, Elazar Z. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem. 2006;281:36303–36316. doi: 10.1074/jbc. M607031200. [DOI] [PubMed] [Google Scholar]
- 43.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casais R, Thiel V, Siddell SG, Cavanagh D, Britton P. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J Virol. 2001;75:12359–12369. doi: 10.1128/JVI.75.24.12359-69.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samali A, Fitzgerald U, Deegan S, Gupta S. Methods for monitoring endoplasmic reticulum stress and the unfolded protein response. Int J Cell Biol. 2010;2010:830307. doi: 10.1155/2010/830307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.