Abstract
Docosahexaenoic acid (DHA) has been reported to induce tumor cell death by apoptosis. However, little is known about the effects of DHA on autophagy, another complex well-programmed process characterized by the sequestration of cytoplasmic material within autophagosomes. Here we show that DHA increased both the level of microtubule-associated protein 1 light chain 3 and the number of autophagic vacuoles without impairing autophagic vesicle turnover, indicating that DHA induces not only apoptosis but also autophagy. We also observed that DHA-induced autophagy was accompanied by p53 loss. Inhibition of p53 increased DHA-induced autophagy and prevention of p53 degradation significantly led to the attenuation of DHA-induced autophagy, suggesting that DHA-induced autophagy is mediated by p53. Further experiments showed that the mechanism of DHA-induced autophagy associated with p53 attenuation involved an increase in the active form of AMP-activated protein kinase and a decrease in the activity of mammalian target of rapamycin. In addition, compelling evidence for the interplay between autophagy and apoptosis induced by DHA is supported by the findings that autophagy inhibition suppressed apoptosis and further autophagy induction enhanced apoptosis in response to DHA treatment. Overall, our results demonstrate that autophagy contributes to the cytotoxicity of DHA in cancer cells harboring wild-type p53.
Key words: DHA, autophagy, apoptosis, p53, cancer, mTOR, AMPK, p27
Introduction
It has been observed that docosahexaenoic acid (DHA), a member of the omega-3 family polyunsaturated fatty acids, causes cancer cell death and that apoptosis is the mechanism of DHA-induced cytotoxicity in tumor cells.1–3
Autophagy or autophagic process is characterized by the sequestration of bulk cytoplasm and organelles in double- or multi-membrane autophagic vesicles and their subsequent degradation by lysosomes for macromolecular synthesis and ATP generation.4 Autophagy is a dynamic process highly controlled by autophagy-related (ATG) genes which encode proteins necessary for autophagosome formation, cargo degradation and reuse of degraded materials contained in autophagosomes.5,6 Upstream of the ATGs, mammalian target of rapamycin (mTOR) kinase has the most potent impact on autophagy. It integrates and coordinates different sensory inputs from upstream pathways to regulate the autophagic process.6,7 Once mTOR is activated, it inhibits autophagy via phosphorylation of the Atg proteins.8 Recently, it has been shown that AMP-activated protein kinase (AMPK) activation leads to autophagy through negative regulation of mTOR and that many other factors involved in the autophagic process govern autophagy through AMPK/mTOR signaling.6,9–11 For example, the tumor suppressor protein p53 was recently found to trigger autophagy by phosphorylation of AMPK at Thr172 and further inactivation of mTOR.12–15
Although autophagy is induced as a survival response to either growth factor or nutrient deprivation, it is also an important mechanism of tumor cell death.5 One of the first lines of evidence suggesting the role of autophagy in cancer cell death comes from the study of Atg5, an ATG product required for autophagosome formation, which also provokes apoptotic cell death in cancer cells.16 Moreover, an increasing number of studies demonstrate that apoptosis and autophagy share some common signaling pathways and are mutually regulated.17 Although it is strongly believed that DHA kills tumor cells by apoptosis, the role of DHA in the induction of the autophagic pathway in cancer cells has not yet been examined. It is not known whether autophagy is induced in DHA-caused cancer cell death and, if so, how autophagy contributes to cell death. In the present study, we investigated the modes and molecular mechanisms of cell death that are involved in the cytotoxic effect of DHA on cancer cells. To the best of our knowledge, this study provides the first evidence that autophagy is induced in tumor cells treated with DHA. We showed that DHA treatment leads to autophagy via p53-mediated AMPK/mTOR signaling and that DHA-induced autophagy sensitizes tumor cells to apoptosis. Overall, our results develop a better understanding of a unique mechanism of the anticancer action of DHA.
Results
DHA induces caspase-3-mediated apoptosis as well as autophagic activation in SiHa cells.
Studies have shown that DHA induces apoptosis in cancer cells by activating both intrinsic and extrinsic apoptotic pathways.18 We confirmed the ability of DHA to induce apoptosis using SiHa cervical cancer cells. TUNEL (terminal deoxyribonucleotidyl transferase-mediated dUTP nick-end labeling) staining was performed to detect apoptotic nuclear DNA breaks in cells due to DHA treatment. As shown in Figure S1A, DHA treatment significantly increased the number of TUNEL-positive cells. Since caspase activity is necessary to induce apoptosis,19 using a fluorometric assay we assessed the activity of caspase-3, an executor caspase that is activated through both intrinsic and extrinsic pathways.20 DHA treatment induced dose- and time-dependent activation of caspase-3 in SiHa cells (Fig. S1B). Once the proapoptotic effect of DHA was established, we quantified apoptosis by flow cytometry. There was at least a 10-fold increase in the number of apoptotic cells at 24 h after supplementation with 75 µM DHA in relation to control cells (Fig. S1C).
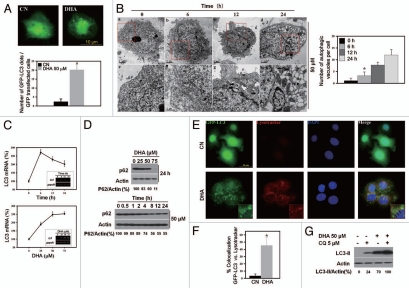
Recently, interesting functional links have been revealed between cell death and autophagy.5,17 To determine whether autophagy is involved in DHA-induced cell death, SiHa cells were transfected with green fluorescent protein-microtubuleassociated protein 1 light-chain 3 (GFP-LC3), a specific marker of autophagic vesicles and autophagic activity,4 and then treated with 50 µM DHA. After DHA treatment, the number of GFP-LC3 fluorescent dots dramatically increased (Fig. 1A), suggesting that autophagic vacuolization occurs in response to DHA treatment. To confirm this, transmission electron microscopy was used to visualize autophagic vacuoles. We observed a time-dependent accumulation of numerous lamellar structures with cytosolic autophagic vacuoles in SiHa cells starting at 6 h after treatment with 50 µM DHA (Fig. 1B).
Figure 1.
Autophagy is induced in SiHa cells after treatment with DHA. (A) GFP-LC3 puncta induced by DHA. Top part, representative images of GFP-LC3 staining in SiHa cells with or without 50 µM DHA treatment (scale bar, 10 µm); bottom part, the number of GFP-LC3 dots per transfected cells was quantified. Each bar represents the mean of 10 determinations repeated in three separate experiments. *p < 0.05. (B) Left, representative electron micrographs of cells incubated with 50 µM DHA for 0, 6, 12 or 24 h. Bottom part (e–h) is taken from the top part insets (a–d), respectively. Autophagosome (ap) and late autophagic compartment (ac) including partially degraded material are visible in DHA-treated cells. N indicates nucleus; arrow indicates autophagosome including the lamellar structure. Also, note the presence of nuclear envelope blebbing (b–d) and the formation of nuclear fragments (h), which are characteristic features of cells undergoing apoptosis. Scale bar, 2 µm (a–d); 1 µm (e–h); right, morphometric analysis of autophagic vacuoles per cell was performed. More than 20 cells were analyzed for each condition. (C) Expression of LC3 mRNA following DHA treatment. Time course (top part; 50 µM DHA) and dose course (bottom part; 6 h) of LC3 mRNA expression in response to DHA were examined by quantitative real-time PCR and RT-PCR analysis. Each bar represents the mean of four determinations repeated in two separate experiments. *p < 0.05. The insets shown are representative of three similar ones analyzed by agarose gel electrophoresis after RT-PCR. (D) DHA decreases p62 expression in a dose- (top part; 24 h) and time-dependent (bottom part; 50 µM DHA) manner as examined by protein gel blotting. (E) Representative images of Lysotracker Red staining in GFP-LC3-transfected SiHa cells with or without DHA treatment. SiHa cells were transfected with GFP-LC3 plasmids for 18 h and then incubated for an additional 24 h with 50 µM DHA followed by Lysotracker Red (50 nM) staining before counterstaining with DAPI. Insets show the higher magnification view of the boxed area. Arrows point to ring-shaped autophagic vesicles that retain GFP staining and Lysotracker Red fluorescence (scale bar, 10 µm). (F) Colocalization of the autophagic marker GFP-LC3 and Lysotracker Red in SiHa cells treated without or with 50 µM DHA. The degree of colocalization was determined by ImageJ software as described in the Materials and Methods. *p < 0.05. (G) SiHa cells treated with 50 µM DHA in the presence or absence of 5 µM CQ for 24 h were analyzed by protein gel blotting with antibodies against LC3 and actin.
Since autophagic vesicles also accumulate when autophagic vesicle turnover (autophagosome-lysosome fusion or/and downstream cargo degradation) is inefficient,21 we asked whether DHA might induce autophagic vacuolization by impairing autophagic vesicle turnover. We therefore first assessed the mRNA level of LC3 (a reliable marker of autophagy induction) and the amount of p62 protein, which is associated with the completed autophagosomes and whose decrease reflects the lysosomal degradation of autophagosomes6,21 in response to DHA. As shown in Figure 1C, LC3 mRNA expression was increased after treatment with DHA and the increase was significant when compared with the controls. In contrast, DHA induced a dose- and time-dependent reduction in the expression level of p62 (Fig. 1D). These findings imply that autophagic vesicle turnover is not impaired by DHA treatment. To confirm this, we initially labeled the GFP-LC3 transfected cells with a pH-dependent dye that becomes fluorescent in acidic lysosomes,22 Lysotracker, to monitor autophagosome-lysosome fusion. Untreated SiHa cells demonstrated very few GFP-LC3 dots and Lysotracker-positive vesicles, whereas DHA-treated cells displayed a marked increase in the number of GFP-LC3 puncta and Lysotracker-positive vesicles (Fig. 1E). Importantly, we found that the colocalization of GFP-LC3 with Lysotracker markedly increased in DHA-treated cells (Fig. 1F), thus suggesting that the fusion between autophagosomes and lysosomes is enhanced in response to DHA. To assess autophagic vesicle turnover more precisely, SiHa cells were treated with DHA in the presence of chloroquine (CQ), which inhibits autophagic cargo degradation and leads to the accumulation of ineffective autophagosomes.22 CQ further promoted the DHA-triggered induction of LC3-II (Fig. 1G), in accordance with the idea that DHA stimulates autophagy without impairing autophagic vesicle turnover. Together, these results strongly indicate that DHA-induced autophagic vacuolization is a consequence of autophagic flux activation.
Decreased expression of p53 in response to DHA.
Given that SiHa cells possess the wild-type tumor suppressor p53 and that recent reports have suggested the involvement of p53 in the autophagic pathway,14,23,24 we elucidated the underlying mechanism of DHA induced-autophagy by examining the levels of p53 and LC3-II protein. In addition, cleaved poly(ADPribose) polymerase (PARP), a widely used apoptotic marker,1 was also examined to confirm the ability of DHA to induce apoptosis.
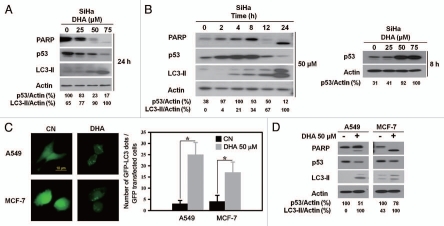
Protein gel blot assay revealed that DHA treatment caused dose- and time-dependent increases in LC3-II and cleaved PARP compared with control cells (Fig. 2A and B, left). These results are consistent with our previous finding that DHA induced not only apoptosis but also autophagy and suggest that DHA enhances apoptosis and autophagy simultaneously in SiHa cells. Along with the upregulation of autophagy as indicated by LC3-II accumulation, a dose-dependent decrease in p53 expression was noted after 24 h of DHA treatment (Fig. 2A). It was surprising, however, that in the time-course experiments, a transient increase in p53 activity was detected at 8 h after treatment, which declined to a barely detectable level after 24 h (Fig. 2B and left). We performed additional dose-response experiments and confirmed this upregulation of p53 at 8 h after DHA treatment (Fig. 2B and right), which is probably explained by nonspecific stress caused by DHA treatment.
Figure 2.
Decreased expression of p53 in response to DHA. (A) Protein gel blot analysis of the expression levels of p53, autophagic protein LC3, apoptotic protein PARP and actin in SiHa cells treated with the indicated dose of DHA for 24 h. (B) Left, protein gel blot analysis of p53, LC3, PARP and actin in SiHa cells treated with 50 µM DHA for the indicated time periods; right, protein gel blot analysis of p53 in SiHa cells treated with the indicated dose of DHA for 8 h. (C) A549 and MCF-7 cells were transfected with GFP-LC3 plasmids for 18 h and then incubated with 50 µM DHA for another 24 h before analysis by fluorescence microscopy. Left, representative images of GFP-LC3 staining in A549 and MCF-7 cells with or without DHA treatment (scale bar, 10 µm); right, quantification of the number of GFP-LC3 dots per transfected cells. Each bar represents the mean of 10 determinations repeated in three separate experiments. *p < 0.05. (D) Protein gel blot analysis of p53, LC3, PARP and actin levels in A549 and MCF-7 cells treated with or without 50 µM DHA for 24 h. The blots shown are representative of two independent experiments with similar results.
To confirm our observation, we tested the degree of autophagy induction and p53 expression after DHA treatment in MCF-7 and A549 cancer cells, which both express wild-type p53.23 DHA induced increases in GFP-LC3 dots in both MCF-7 and A549 GFP-LC3-transfected cells (Fig. 2C). Importantly, protein gel blot analysis showed that DHA also increased LC3-II protein levels and diminished p53 expression in both cell lines (Fig. 2D). These results are in agreement with our previous observations in SiHa cells, indicating that p53 downregulation is involved in DHA-induced autophagy.
p53 inactivation triggers autophagy and prevention of p53 degradation attenuates DHA-induced autophagy.
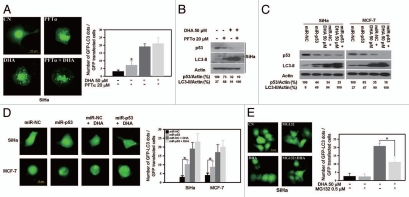
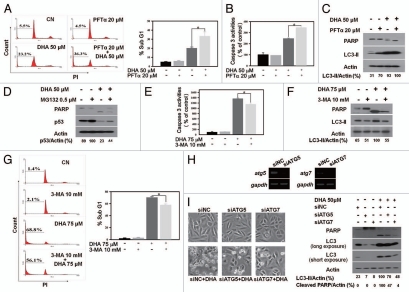
As DHA treatment downregulated p53, we investigated whether loss of p53 might cause DHA-induced autophagy using SiHa and MCF-7 cells. GFP-LC3-transfected cells showed a significant accumulation of GFP-LC3 puncta when p53 was inhibited with pifithrin-α (PFTα), a pharmacological p53 inhibitor25 (Fig. 3A and PFTα), or knocked down with a specific p53 microRNA (Fig. 3D and miR-p53). More importantly, p53 inhibition or knockdown followed by DHA treatment augmented the number of GFP-LC3 dots (Fig. 3A and PFTα + DHA and Fig. 3D, miR-p53 + DHA) and the level of LC3-II (Fig. 3B and C) compared with that observed with DHA treatment alone, though the increase in the number of GFP-LC3 dots and LC3-II expression level was not remarkable, which indicates that autophagy may reach a maximal level and/or that p53 inhibition-mediated autophagy induced by PFTα and DHA may act on a similar signaling. Next, since the p53 inactivation-triggered autophagy is mediated by proteasomes,14 to further address the issue of whether DHA-induced autophagy is regulated through p53 downregulation, GFP-LC3-transfected SiHa cells were treated with DHA in the presence or absence of MG132, a protease inhibitor to prevent p53 degradation. As shown in Figure 3E, MG132 indeed significantly inhibited the accumulation of GFP-LC3 puncta induced by DHA. Collectively, these results show that p53 loss is able to initiate autophagy and that p53 downregulation is responsible, at least partially if not completely, for DHA-induced autophagy in cancer cells harboring wild-type p53.
Figure 3.
p53 inhibition/knockdown triggers autophagy and prevention of p53 degradation attenuates DHA-induced autophagy. (A) SiHa cells were transfected with GFP-LC3 plasmids for 18 h, incubated for 2 h in the presence or absence of 20 µM PFTα, and then incubated for an additional 24 h with 50 µM DHA. Left, representative images of GFP-LC3 staining in SiHa cells (scale bar, 10 µm); right, the number of GFP-LC3 dots per transfected cells was scored after analysis by fluorescence microscopy. Each bar represents the mean of 10 determinations repeated in three separate experiments. *p < 0.05. (B) SiHa cells were incubated for 2 h in the presence or absence 20 µM PFTα and then treated for 24 h with 50 µM DHA. After cell lysis, the levels of p53 and LC3 were analyzed by protein gel blotting. (C) p53 knockdown with the miR RNAi expression system. SiHa and MCF-7 cells were infected with adenovirus expressing microRNA negative control (miR-NC) or p53 microRNA (miR-p53) and then treated with 50 µM DHA. LC3 and p53 levels were analyzed by protein gel blot analysis. (D) SiHa and MCF-7 cells were first transfected with GFP-LC3 plasmids for 18 h and then infected with adenovirus harboring miR-NC or miR-p53, followed by 24 h incubation with 50 µM DHA. Left, representative images of GFP-LC3 staining in SiHa and MCF-7 cells with or without treatment (scale bar, 10 µm); right, number of GFP-LC3 dots per transfected cells. Each bar represents the mean of 10 determinations repeated in three separate experiments. *p < 0.05. (E) SiHa cells were transfected with GFP-LC3 plasmids for 18 h and then incubated with 0.5 µM MG132 for 1 h before 50 µM DHA was added as described in the Materials and Methods. Left, representative images of GFP-LC3 staining in DHA-treated SiHa cells in the presence or absence of MG132 (scale bar, 10 µm); right, quantification of the number of GFP-LC3 dots per transfected cells. Each bar represents the mean of ten determinations repeated in three separate experiments. *p < 0.05.
DHA induces autophagy through p53-mediated AMPK/mTOR signaling.
A pivotal role in the control of autophagy is played by mTOR, which integrates input information from multiple upstream signal transduction pathways and negatively regulates autophagy.6 More recently, AMPK, which is a protein complex that responds to change in cellular AMP/ATP ratios, was found to stimulate autophagy via inhibition of mTOR.11 In addition, Tasdemir et al.14 have revealed that p53 mediates autophagy through an AMPK/mTOR-dependent pathway, leading us to hypothesize that AMPK/mTOR signaling would be involved in p53 loss and autophagy activation induced by DHA in cancer cells.
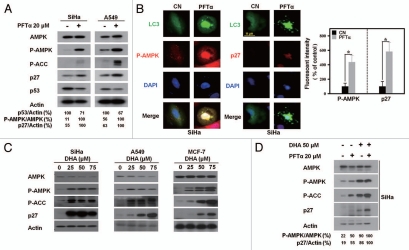
To test our hypothesis, alterations in AMPK/mTOR signaling molecules were first examined after p53 inactivation. As shown in Figure 4A, in both SiHa and A549 cells, after p53 inhibition with PFTα, AMPK and acetyl coA carboxylase (ACC), an AMPK substrate, were phosphorylated. The level of the cyclin-dependent kinase inhibitor p27, whose upregulation indicates mTOR inhibition,26,27 was enhanced. We also performed immunofluorescence staining experiments to verify the association between AMPK/mTOR signaling and p53-controlled autophagy. After PFTα treatment, GFP-LC3-transfected SiHa cells were stained with anti-phospho-AMPK or anti-p27 antibodies. Figure 4B clearly shows that the staining intensity of phospho-AMPK and p27 increased and became more distinct within the nucleus of the autophagic cells that exhibited LC3 aggregation in response to p53 inhibitor. These results strongly suggest that p53 also regulates autophagy via AMPK/mTOR signaling under our experimental conditions.
Figure 4.
DHA induces autophagy through p53-mediated AMPK/mTOR signaling. (A) Protein gel blot analysis of the AMPK/mTOR signaling molecules in SiHa and A549 cells in the presence or absence of PFTα. Phosphorylation of AMPK (P-AMPK) and ACC (P-ACC) and upregulated p27 expression were detected in cells exposed to 20 µM PFTα for 24 h, compared with that in the nontreated control cells. The blots depicted are representative of three independent experiments with similar results. (B) Left, representative images of immunofluorescence staining with anti-P-AMPK and anti-p27 antibodies in GFP-LC3-transfected SiHa cells with or without p53 inhibitor treatment. SiHa cells were transiently transfected with GFP-LC3 plasmids prior to treatment with 20 µM PFTα for 24 h and then immunofluorescently stained as described in the Materials and Methods section (scale bar, 10 µm); right, relative fluorescent intensity analysis of P-AMPK and p27 staining in PFTα-treated SiHa cells compared with control cells, as described in Materials and Methods. (C) Elevated AMPK and blocked mTOR signaling in cancer cells treated with DHA. SiHa (left), A549 (middle) and MCF-7 (right) cells were treated with the indicated dose of DHA for 24 h before the protein lysates were normalized and immunoblotted with anti-AMPK, anti-P-AMPK, anti-P-ACC and anti-p27 antibodies. (D) SiHa cells were incubated for 2 h in the presence or absence 20 µM PFTα and then treated for 24 h with 50 µM DHA. After cell lysis, the levels of AMPK/mTOR signaling molecules were analyzed by protein gel blotting. Actin levels were analyzed as a control for equal loading. Blots are representative of two independent experiments with similar results.
Accordingly, in view of our above findings, we sought to determine whether DHA treatment enhances AMPK activity, which in turn inhibits mTOR. As expected, SiHa cells treated with DHA exhibited increased phospho-AMPK and phospho-ACC expression and upregulated p27 expression (Fig. 4C and left). Similarly, in A549 and MCF-7 cells, DHA treatment correlated with elevated AMPK activity and decreased mTOR activity (Fig. 4C and middle and right). This observation was also confirmed by the time-course experiments in A549 cells treated with DHA (Fig. S2A).
To obtain direct evidence for the interconnection between alterations in AMPK/mTOR signaling and the p53-mediated autophagic process in cells treated with DHA, SiHa cells were preincubated in the presence or absence of the p53 inhibitor PFTα and then subjected to DHA treatment. As shown in Figure 4D, DHA enhanced AMPK phosphorylation and attenuated mTOR activity, as indicated by p27 upregulation in the PFTα-primed cells as compared with that in the nonprimed cells. Similar results were also observed in A549 cell lines (Fig. S2B). Together with our above-mentioned observation that p53 regulated DHA-induced autophagy (Fig. 3), these correlative data indicate that DHA shares the similar signaling with PFTα and induces autophagy through p53-mediated AMPK/mTOR signaling.
Autophagy enhances DHA-induced apoptosis and inhibition of autophagy partially prevents DHA-induced apoptotic cell death.
Taken together, the results showed that DHA triggered apoptotic cell death and simultaneously activated the p53/AMPK/mTOR autophagic pathway in cancer cells. We were interested in investigating whether autophagy affected DHA-induced apoptotic cell death. Having found that p53 inactivation led to autophagy, SiHa and A549 cells were first treated with DHA in the presence or absence of PFTα and the contribution of autophagy to the DHA-induced apoptosis was examined. Pharmacological inhibition of p53 with DHA treatment resulted in a higher percentage of cells in the Sub G1 phase (Figs. 5A and S2C) and increased levels of caspase-3 activity (Fig. 5B) and cleaved PARP (Fig. 5C) compared with either treatment alone, suggesting that p53 loss-mediated autophagy contributes to apoptosis in cancer cells treated with DHA. To further ascertain this, SiHa cells were primed with MG132 to inhibit p53 loss-induced autophagy and the degree of apoptosis induced by DHA was determined by protein gel blot analysis. As shown in Figure 5D, MG132 remarkably attenuated DHA-induced apoptosis, as evidenced by a clear reduction in cleaved PARP. In addition, as our results implicated p53/AMPK-mediated mTOR inactivation as a mechanism underlying PFTα- and DHA-induced autophagy, one would expect that, by analogy with the effect of PFTα, mTOR inhibition would also strengthen DHA-induced apoptosis. Indeed, rapamycin, an mTOR inhibitor which is also widely used to induce autophagy,21 significantly augmented DHA-triggered apoptotic cell death as indicated by an increase in the Sub G1 cell population (Fig. S2D) and cleaved PARP formation (Fig. S2E) compared with that observed in A549 cells treated with DHA only. On the basis of these findings, we conclude that autophagic response promotes DHA-induced apoptosis.
Figure 5.
Autophagy enhances DHA-induced apoptosis and inhibition of autophagy partially prevents DHA-induced apoptotic cell death. (A–C) Induction of autophagy by PFTα enhances DHA-induced apoptosis. SiHa cells were incubated for 2 h with or without 20 µM PFTα before incubation for 24 h with 50 µM DHA. The percentage of cells in the Sub G1 phase (A) and caspase-3 activity (B) were measured as described in the Materials and Methods section. Each bar represents the mean of more than four determinations repeated in three separate experiments. *p < 0.05. The levels of PARP, LC3 and actin were assessed by protein gel blotting (C). (D) Inhibition of autophagy by MG132 attenuates DHA-induced apoptosis. SiHa cells were incubated for 1 h with or without 0.5 µM MG132 before incubation for 24 h with 50 µM DHA. The levels of PARP, p53 and actin were assessed by protein gel blotting. (E–G) Inhibition of autophagy by 3-MA prevents DHA-induced apoptosis. SiHa cells were pretreated with 10 mM 3-MA for 1 h and then incubated with 75 µM DHA for 5 h. Caspase-3 activity (E) and the percentage of Sub G1 fraction (G) were analyzed as described in the Materials and Methods section. Each bar represents the mean of more than four determinations repeated in three separate experiments. *p < 0.05. The levels of PARP, LC3 and actin were assessed with a protein gel blot assay (F). (H) SiHa cells were seeded in six-well plates. After 24 h, the cells were treated with nontargeting control siRNA (siNC), ATG5 siRNA (siATG5) and ATG7 siRNA (siATG7). At 36 h after transfection, ATG5 and ATG7 mRNA expression levels were examined by RT-PCR analysis. The data shown are representative of three similar ones analyzed by agarose gel electrophoresis after RT-PCR. (I) SiHa cells were seeded in six-well plates. After 24 h, the cells were treated with siNC, siATG5 and siATG7. At 36 h after transfection, cells were treated for 24 h with 50 µM DHA. Light microscope images were captured (left) and cells were harvested and protein gel analysis was performed using the following antibodies: PARP, LC3 and actin as a loading control (right). The data shown are representative of two independent experiments with similar results.
To confirm the role of autophagy in DHA-induced apoptotic cell death, apoptotic levels were measured in SiHa cells treated with DHA in the presence or absence of 3-methyladenine (3-MA), a widely used inhibitor of autophagy.28 Inhibition of autophagy by 3-MA partially blocked DHA-induced apoptosis, as determined by the decreased levels of caspase-3 activity (Fig. 5E) and cleaved PARP (Fig. 5F) and the lower percentage of cells in the Sub G1 phase (Fig. 5G). Since 3-MA might induce nonspecific effects, SiHa were also subjected to small interfering RNA (siRNA) specific for two essential genes for autophagy, ATG5 and ATG7,6 to determine the effect of autophagy inhibition on DHA-induced apoptosis. While siATG5 or siATG7 had no major effects on cellular morphology and cleaved PARP formation after transfection, they strongly reduced DHA-induced autophagy and apoptosis as assessed by the level of LC3-II and cleaved PARP (Fig. 5H and I), respectively. Thus, these findings reveal that autophagy inhibition diminishes the DHA-dependent induction of apoptotic cell death.
Discussion
DHA predominantly exerts its anticancer effect by triggering the apoptotic cell death process.1,18 Here, we report for the first time that DHA also induces autophagy and this autophagic process contributes to the cytotoxic activity of DHA against wild-type p53 harbored cancer cells by enhancing apoptosis. As a process of consuming cellular components and generating energy, autophagy engages in a complex interconnection with apoptosis according to the nature of stimulus and cell type. It suppresses apoptosis by eliminating damaged organelles under cellular stress response to cancer therapy, or it sensitizes cells to apoptosis, which is ATP-dependent, by acting as an energy source.29–32 We showed that autophagy and apoptosis occurred simultaneously and acted cooperatively to induce cell death in DHA-treated cancer cells, as DHA treatment increased both LC3-II and cleaved PARP in dose- and time-dependent manners (Fig. 2A and B, left). In addition, the fact that morphological features of both autophagy and apoptosis were observed in the same cells treated with DHA (Fig. 1B) confirms the above results.
The finding that DHA induces both autophagy and apoptosis prompted us to closely investigate the interconnection between these two cellular processes. The evidence that further autophagy induction promoted apoptosis in cells treated with DHA (Figs. 5A–C and S2C) indicates that autophagy contributes to the apoptosis caused by DHA. In this respect, autophagy seems to facilitate the occurrence of apoptosis. This notion is supported by the observation that autophagy inhibition by MG132, 3-MA or ATG siRNA decreased cell sensitivity to DHA-mediated apoptosis (Fig. 5D–I). Furthermore, the form of cell death initiated by autophagy (autophagic cell death) is caspase independent.33 In our system, we observed significant changes in caspase-3 activity in cells preincubated with autophagy inducer or inhibitor (Fig. 5B and E) and subsequently treated with DHA, which also implies that autophagy, rather than directly causing cell death, assists DHA-induced apoptotic cell death. Indeed, it has been reported that autophagy can function upstream of apoptosis and participate in the process of membrane blebbing, one of the characteristics of apoptotic cell death, by maintaining cellular ATP levels.32,34,35 Whether this is the role played by autophagy in DHA-induced apoptosis needs to be addressed by further investigation. Additionally, although further autophagy induction by PFTα and rapamycin resulted in a statistically significant enhancement of apoptosis in DHA-treated cells, the increase was modest, which is not unexpected since these autophagic inducers share the same signaling with DHA to trigger autophagy and, compared with DHA, they have less potent effect on autophagic induction. Meanwhile, it is also worth noting that autophagy inhibition and knockdown failed to completely block apoptosis in cancer cells treated with DHA, suggesting that DHA-induced apoptosis is not exclusively based on mTOR-mediated autophgay activation and that other signaling pathways may participate in the apoptotic process caused by DHA treatment. At least the mechanisms involving accumulation of reactive oxygen species (ROS) and β-catenin have been reported.1–3
Another question we addressed in this study is the mechanism underlying DHA-induced autophagy. Prompted by the observation of p53 loss in cancer cells treated with DHA (Fig. 2), we further evaluated the specific involvement of p53 in the autophagic process induced by DHA. Based on the finding that p53 inhibition or knockdown led to autophagy and prevention of p53 degradation decreased DHA-induced autophagy (Fig. 3), we therefore propose that p53 may regulate the onset of autophagy induced by DHA in tumor cells harboring wild-type p53. Although p53 is best known as a transcriptional factor that controls cell cycle and apoptosis, increasing evidence suggests that p53 inactivation also triggers autophagy in normal cells, transformed cells and cancer cells.13,14 Despite obtaining similar results in our experimental systems (SiHa, A549 and MCF-7 cancer cells), the effect of p53 on autophagy is not a general phenomenon, since p53 knockdown fails to enhance autophagy in skeletal muscle cells.14,24 One possible explanation for this is the different genetic background of the cell models. Likewise, considering that the DHA-induced autophagy is mediated by p53 and that some cancer cells are p53 deficient/mutant, it is reasonable to assume that the p53-mediated autophagy induced by DHA is dependent on cell type as well. Indeed, when we treated PC3 (p53 null) and DU145 (p53 mutant) prostate cancer cells23 with DHA, an increase in LC3-II was also detected (Fig. S3A). In addition, PFTα pretreatment neither influenced the basal level of LC3-II nor the increased LC3-II level induced by DHA in both cell lines (Fig. S3B). These results indicate that p53 loss-mediated autophagy induced by DHA is relatively important and not essential in some cell types and that besides the p53 pathway, DHA may also trigger autophagy through other mechanisms. However, although p53 is not required for autophagy induction, cancer cells harboring wild-type p53 may have an advantage to undergo autophagy via p53 inactivation after DHA treatment and in p53 deficient/mutant cells, DHA may still be able to induce autophagy by directly targeting downstream signaling events involved in p53-regulated autophagy, such as mTOR. The evidence supporting this hypothesis is that DHA, a highly polyunsaturated fatty acid susceptible to peroxidation, can accumulate ROS in cancer cells1,18 and ROS has been shown to induce autophagy by inhibiting mTOR directly.6,36 It is possible that DHA may induce autophagy through inhibiting mTOR directly by producing ROS in p53 deficient/mutant cells. Studies are currently underway to test this hypothesis.
The signaling by which p53 induces autophagy has been suggested to be a direct action of mTOR through AMPK activation.14,15,37 As a central signal integrator, mTOR receives signals arising from nutrients, growth factors and many cellular kinases including AMPK.26 Phosphorylation of AMPK activates downstream signaling that leads to mTOR inhibition and triggers autophagy,38 which is consistent with the AMPK function of initiating catabolic processes.11 Our data are in line with a recent finding by Tasdemir and colleagues14 that inactivation of p53 inhibits mTOR activity via AMPK activation. Additionally, we showed that treatment with the combination of p53 inhibitor and DHA had a stronger effect on the activation of AMPK/mTOR signaling than treatment with either drug alone, indicating that p53/AMPK/mTOR signaling is involved in DHA-induced autophagy. It is worth noting that, downstream of mTOR, p27 was assessed to indicate mTOR activity, since mTOR phosphorylation does not imply its activation.26 p27 inhibits cyclin-dependent kinase, resulting in cell cycle arrest.39 It has also been shown that p27 is sufficient to induce autophagy.40 Therefore, our observation of a marked increase in p27 in cells treated with DHA suggests that p27 may somewhat correlate with the autophagic activation governed by p53 loss. This is supported by the fact that p27 arrests cell cycle in the G1 and S phase41 and that p53 loss induces autophagy in a cell cycle-dependent fashion, with a preference for the G1 and S phases.42 Since both p27 and ATGs are located downstream of mTOR,26 it is possible that the autophagy and the cell cycle arrest induced by p53 attenuation in cancer cells treated with DHA occur in parallel via mTOR inactivation. Nevertheless, considering that only p27 was used to reflect mTOR activity, we could not exclude the possibility that p27 activation may also account for a substantial portion of autophagy induced by DHA.
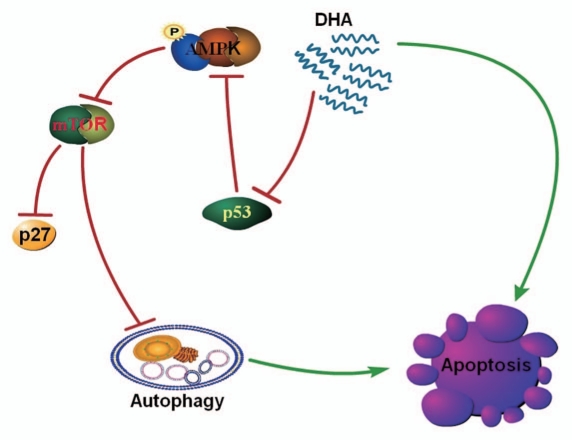
Collectively, this study highlights the significance of autophagy in DHA-induced cancer cell death. DHA initially triggers a p53-mediated autophagic process via AMPK/mTOR signaling and the DHA-induced autophagy, alongside other signaling sensitizes tumor cells to apoptosis (Fig. 6). Our finding that DHA-induced autophagy contributes to cancer cell death may have important implications in developing future strategies to use DHA in cancer prevention and therapy.
Figure 6.
Proposed mechanism of DHA-induced autophagy and apoptosis in human cancer cells harboring wild-type p53. DHA-induced autophagic activation is mediated by the p53/AMPK/mTOR pathway and contributes to apoptotic cell death. DHA causes the phosphorylation of AMPK at Thr172 and activates AMPK by decreasing the expression of the tumor suppressor protein p53. This leads to inhibition of mTOR signaling, which negatively regulates autophagy, a process defined by the sequestration of bulk cytoplasm and organelles in autophagosomes and their subsequent lysosomal degradation. This p53-controlled autophagy induced by DHA decreases the viability of cancer cells harboring wild-type p53 by promoting apoptosis. Of note, other signaling cascades may also contribute to the apoptosis induced by DHA.
Materials and Methods
Cells and cell culture.
Human cervical cancer SiHa cells, human lung cancer A549 cells and human breast cancer MCF-7 cells were purchased from American Type Culture Collection (HTB-35, CCL-185 and HTB-22). Cells were maintained in Dulbecco's modified Eagle's medium (GIBCO, 31600-034) supplemented with 10% heat-inactivated fetal bovine serum (GIBCO, 10082147), penicillin and streptomycin (GIBCO, 15140-122). The cells were cultured in a humidified 5% CO2 atmosphere at 37°C.
Chemical treatment and antibodies.
DHA (Cayman Chemical, 90310) dissolved in absolute ethanol (MERCK, 1009831011), PFTα (Sigma, P4359) dissolved in dimethyl sulfoxide (DMSO, Sigma, D2650), CQ (Sigma, C6228) dissolved in phosphate-buffered saline (PBS, Sigma, D8537), rapamycin (Tocris, 1292) dissolved in DMSO, MG132 (Tocris, 1748) dissolved in DMSO and 3-MA (Sigma, M9281) dissolved in PBS were added to the medium to final concentrations as described in each experiment. Cells grown to 60% confluency were switched to serum-free medium and the culture was allowed to expand for 24 h before giving any treatment. For PFTα plus DHA treatment, cells were first incubated with 20 µM PFTα for 2 h; then DHA was added and the cells were incubated for 24 h. For 3-MA plus DHA treatment, cells were pretreated with 10 mM 3-MA for 1 h in Hanks' balanced salt solution (Sigma, H8264) and then incubated with DHA for 5 h. For MG132 plus DHA treatment, cells were first incubated with 0.5 µM MG132 for 1 h; then DHA was added and the cells were incubated for 24 h. To label lysosomes, cells were treated with 50 µM DHA for 24 h followed by Lysotracker Red DND-99 (50 nM, Molecular Probes, L-7528) staining according to the manufacturer's instructions.
The antibodies used and their sources are as follows. LC3B, phospho-ACC (Ser79), AMPK and phospho-AMPK (Thr172) (Cell Signaling Technology, 2775, 3661, 2532 and 2535); anti-p53 (DO-1) (sc-126), anti-p27 (C-19) (sc-528) and anti-PARP (F-2) (sc-8007) (Santa Cruz Biotechnology); anti-actin (A5441) and anti-p62 (P0067) (Sigma); goat anti-rabbit (401315) and goat anti-mouse (401215) secondary antibodies (Calbiochem) and secondary antibodies conjugated with Texas red (goat anti-rabbit) (Santa Cruz Biotechnology, sc-2708).
Protein gel blot analysis.
Protein gel blot analysis was done as described previously in reference 43. Briefly, cell lysates (15–30 µg) were separated by 8–12% sodium dodecyl sulfate-PAGE and electroblotted onto a polyvinylidene difluoride membrane (Millipore, ISEQ00010). The blots were blocked in 5% (w/v) skimmed-milk protein and incubated with a 1:2,000 dilution of primary antibodies overnight at 4°C, followed by incubation with goat anti-rabbit or anti-mouse peroxidase-conjugated secondary antibody. The blots were then visualized by enhanced chemiluminescence (Millipore, WBKLS0100).
Electron microscopy.
Cells were harvested, pelleted, fixed in 2.5% glutaraldehyde (MP Biomedicals, 195199) in PBS for 24 h at 4°C, post-fixed in 1% osmium tetroxide (Sigma, 75632) and 0.5% potassium ferricyanide (Electron Microscopy Sciences, 20150) in cacodylate buffer (Electron Microscopy Sciences, 11650) for 1 h, and then embedded in straight resin (Agar Scientific, R1045). The solidified blocks were cut into 60-nm-thick sections, which were stained with uranyl acetate (Agar Scientific, R1260A) and lead citrate (Agar Scientific, R1210) and then examined under a Zeiss EM 900 transmission electron microscope (Carl Zeiss).
Transfection and transduction.
Transient transfection was performed with GFP-LC3 expression vector (a kind gift from Dr. Tamotsu Yoshimori, National Institute of Genetics, Mishima, Japan), using Lipofectamine 2000 reagent (Invitrogen, 11668-019) as recommended by the vendor. After 18 h of transfection, cells were subjected to different treatments. For knockdown experiments using microRNA, adenoviruses containing a microRNA expression cassette for p53, prepared with the BLOCK-iT miR RNAi expression system (a gift from professor Chang Deok Kim, Department of Dermatology and Research Institute for Medical Sciences, College of Medicine, Chungnam National University, Korea), were employed to infect the cells. After 12 h, the cells were treated with either PFTα or DHA, or both. The sequence used to knock down p53 is 5′-CGA TAT TGA ACA ATG GTT CAC TGA A-3′. For knockdown experiments using siRNA (Invitrogen), 100 nM siRNA was transfected into cells using Lipofectamine 2000 reagent for 36 h before cells were subjected to 50 µM DHA treatment for another 24 h. siRNA against the following human genes were used: ATG5 (5′AUC CCA UCC AGA GUU GCU UGU GAU C-3′) and ATG7 (5′-CCA AGG AUG GUG AAC CUC AGU GAA U-3′).
Apoptosis assays.
For the TUNEL assay, SiHa cells plated on glass coverslips were grown for 24 h and then incubated with or without 50 µM DHA for 24 h; the cells were stained using the DeadEnd™ Fluorometric TUNEL System (Promega, G3250), according to the manufacturer's instructions. Apoptosis was determined as the percentage of positive cells per 1,000 4′,6-diamidino-2-phenylindole (DAPI, Roche, 10236276001)-stained nuclei. These glass coverslips were then visualized under a fluorescence microscope (Olympus iX70). For the caspase-3 activity assay, protein lysates (15–30 µg) were added to a 200 µM reaction mixture containing 50 µmol/L fluorogenic Asp-Glu-Val-Asp (DEVD) substrate (Fluorogenic, C-1133) and the mixture was incubated for 1–4 h at 37°C as described previously in reference 44. Caspase-3 activity was obtained by measuring the fluorescence emission in a fluorometer (Fluoroscan Ascent Labsystems). For the flow cytometry assay, both floating and attached cells were collected after drug treatment, washed in PBS, fixed with 70% ethanol for 24 h, treated with 500 µg/mL RNase A (Sigma, R4642) and then stained with 50 µg/mL propidium iodide (PI, Sigma, P4170) for 10 min at 37°C. DNA staining with PI was analyzed with a FACSCalibur flow cytometer (BD Biosciences).
RT-PCR and quantitative real-time PCR.
Total RNA was isolated with Trizol reagent (Invitrogen, 10296-010) and 1 µg of the total RNA was reverse-transcribed into cDNA with M-MLV reverse transcriptase (ElpisBio, EBT-1501) in the presence of anchored oligo (dT) (ElpisBio, EBT-1523). For PCR, 1 µL of the cDNA was used for each 20 µL of HiPi PCR PreMix reaction system (ElpisBio, EBT-1202) for the amplification of different genes, following the instructions of the manufacturer. Varying cycles of PCR were performed to determine the linear ranges of PCR products. PCR products were analyzed by electrophoresis on a 2% agarose gel (Genosapiens, 21500) with ethidium bromide staining and photographed under UV transillumination (Spectroline). Real-time PCR was performed in triplicate with SYBR Green master mix (Invitrogen, 4367659) for 15 min at 95°C for initial denaturation, followed by 40 cycles of segments of 95°C for 30 sec and 60°C for 30 sec in the 7000 Applied Biosystems Sequence Detection System (Applied Biosystems). The primers used to amplify the LC3, ATG5 and ATG7 genes are 5′-ATG CCG TCG GAG AAG ACC TT-3′ (forward) and 5′-TTA CAC TGA CAA TTT CAT CCC G-3′ (reverse), 5′-CCT GAC CAG AAA CAC TTC GCT G-3′ (forward) and 5′-TGG AGG AAA GCA GAG GTG ATG C-3′ (reverse), 5′-GTC GTC TTC CTA TTG ATG GAC ACC-3′ (forward) and 5′-CAA AGC AGC ATT GAT GAC CAG C-3′ (reverse), respectively. The expression levels of the LC3, ATG5 and ATG7 genes were normalized against the expression levels of the housekeeping gene gapdh.
Immunocytochemistry.
After treatment, SiHa cells were fixed in 4% paraformaldehyde (Fisher, NC9245948), blocked with 1% bovine serum albumin (Bovogen, BSAL0.1) in PBS, stained with rabbit anti-p27 (1:300) or rabbit anti-phospho-AMPK (1:500) primary antibodies, visualized using an anti-rabbit IgG conjugated with Texas red and counterstained with DAPI. These fluorescently stained cells were then observed under a fluorescence microscope as previously described using DP Controller software (Olympus) for image acquisition. Final images were a noncontrast-adjusted merge of two or three channels. At least 15 fields from each coverslip were examined and more than two experiments were performed for each condition. To determine the percentage of colocalization, untransfected cells in each image were masked using ImageJ software prior to analysis. Then, the regions of interest within images from three experiments were automatically analyzed with ImageJ Colocalization Plugin and at least 50 cells were analyzed for each condition. For fluorescent intensity analysis, the average fluorescent intensity was quantified with ImageJ software to correct for the differences in cell number followed by the relative fluorescent intensity calculation.
Statistical analysis.
Multiple comparisons were analyzed using one-way analysis of variance and individual differences then tested by the Fisher's protected least-significant difference test after demonstrating the existence of significant intergroup difference by one-way analysis of variance. Two-group comparisons were performed using Student's t-test. Results are shown as mean ± SD values, with a minimum of two separate experiments for each issue addressed. p values of less than 0.05 were considered significant.
Acknowledgements
We are grateful to Dr. Tamotsu Yoshimori for the gift of GFP-LC3 constructs and to Professor Chang Deok Kim for the adenoviral vectors and packing cells. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Education, Science and Technology (2010-0016447, 2010-0001290 and 2011-0013263).
Abbreviation
- DHA
docosahexaenoic acid
- PFTα
pifithrin-α
- mTOR
mammalian target of rapamycin
- AMPK
AMP-activated protein kinase
- ACC
acetyl coA carboxylase
- LC3
microtubule-associated protein 1 light chain 3
- PARP
poly(ADP-ribose) polymerase
- Atg
autophagy-related
- 3-MA
3-methyladenine
- GFP
green fluorescent protein
- TUNEL
terminal deoxyribonucleotidyl transferase-mediated dUTP nick end labeling
- DAPI
4′,6-diamidino-2-phenylindole
- miR
microRNA
- siRNA
small interfering RNA
- CQ
chloroquine
- DMSO
dimethyl sulfoxide
- ROS
reactive oxygen species
- PI
propidium iodide
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Gleissman H, Yang R, Martinod K, Lindskog M, Serhan CN, Johnsen JI, et al. Docosahexaenoic acid metabolome in neural tumors: identification of cytotoxic intermediates. FASEB J. 2010;24:906–915. doi: 10.1096/fj.09-137919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim K, Han C, Dai Y, Shen M, Wu T. Omega-3 polyunsaturated fatty acids inhibit hepatocellular carcinoma cell growth through blocking beta-catenin and cyclooxygenase-2. Mol Cancer Ther. 2009;8:3046–3055. doi: 10.1158/1535-7163.MCT-09-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim K, Han C, Xu L, Isse K, Demetris AJ, Wu T. Cyclooxygenase-2-derived prostaglandin E2 activates beta-catenin in human cholangiocarcinoma cells: evidence for inhibition of these signaling pathways by omega 3 polyunsaturated fatty acids. Cancer Res. 2008;68:553–560. doi: 10.1158/0008-5472.CAN-07-2295. [DOI] [PubMed] [Google Scholar]
- 4.Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–2906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 5.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 8.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papandreou I, Lim AL, Laderoute K, Denko NC. Hypoxia signals autophagy in tumor cells via AMPK activity, independent of HIF-1, BNIP3 and BNIP3L. Cell Death Differ. 2008;15:1572–1581. doi: 10.1038/cdd.2008.84. [DOI] [PubMed] [Google Scholar]
- 10.Herrero-Martín G, Hoyer-Hansen M, Garcia-Garcia C, Fumarola C, Farkas T, Lopez-Rivas A, et al. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J. 2009;28:677–685. doi: 10.1038/emboj.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meley D, Bauvy C, Houben-Weerts JH, Dubbelhuis PF, Helmond MT, Codogno P, et al. AMP-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem. 2006;281:34870–34879. doi: 10.1074/jbc.M605488200. [DOI] [PubMed] [Google Scholar]
- 12.Maiuri MC, Tasdemir E, Criollo A, Morselli E, Vicencio JM, Carnuccio R, et al. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ. 2009;16:87–93. doi: 10.1038/cdd.2008.131. [DOI] [PubMed] [Google Scholar]
- 13.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 14.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 17.Scarlatti F, Granata R, Meijer AJ, Codogno P. Does autophagy have a license to kill mammalian cells? Cell Death Differ. 2009;16:12–20. doi: 10.1038/cdd.2008.101. [DOI] [PubMed] [Google Scholar]
- 18.Gleissman H, Johnsen JI, Kogner P. Omega-3 fatty acids in cancer, the protectors of good and the killers of evil? Exp Cell Res. 2010;316:1365–1373. doi: 10.1016/j.yexcr.2010.02.039. [DOI] [PubMed] [Google Scholar]
- 19.Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 21.Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 22.Maclean KH, Dorsey FC, Cleveland JL, Kastan MB. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest. 2008;118:79–88. doi: 10.1172/JCI33700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berglind H, Pawitan Y, Kato S, Ishioka C, Soussi T. Analysis of p53 mutation status in human cancer cell lines: a paradigm for cell line cross-contamination. Cancer Biol Ther. 2008;7:699–708. doi: 10.4161/cbt.7.5.5712. [DOI] [PubMed] [Google Scholar]
- 24.Tasdemir E, Chiara Maiuri M, Morselli E, Criollo A, D'Amelio M, Djavaheri-Mergny M, et al. A dual role of p53 in the control of autophagy. Autophagy. 2008;4:810–814. doi: 10.4161/auto.6486. [DOI] [PubMed] [Google Scholar]
- 25.Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science. 285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 26.Faivre S, Kroemer G, Raymond E. Current development of mTOR inhibitors as anticancer agents. Nat Rev Drug Discov. 2006;5:671–688. doi: 10.1038/nrd2062. [DOI] [PubMed] [Google Scholar]
- 27.Nourse J, Firpo E, Flanagan WM, Coats S, Polyak K, Lee MH, et al. Interleukin-2-mediated elimination of the p27Kip1 cyclin-dependent kinase inhibitor prevented by rapamycin. Nature. 1994;372:570–573. doi: 10.1038/372570a0. [DOI] [PubMed] [Google Scholar]
- 28.Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide-3-kinase. J Biol Chem. 2010;285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16:966–975. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- 30.Yokoyama T, Miyazawa K, Naito M, Toyotake J, Tauchi T, Itoh M, et al. Vitamin K2 induces autophagy and apoptosis simultaneously in leukemia cells. Autophagy. 2008;4:629–640. doi: 10.4161/auto.5941. [DOI] [PubMed] [Google Scholar]
- 31.Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14:500–510. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- 32.Yoshimori T. Autophagy: paying Charon's toll. Cell. 2007;128:833–836. doi: 10.1016/j.cell.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 33.Shao Y, Gao Z, Marks PA, Jiang X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2004;101:18030–18035. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sy LK, Yan SC, Lok CN, Man RY, Che CM. Timosaponin A-III induces autophagy preceding mitochondria-mediated apoptosis in HeLa cancer cells. Cancer Res. 2008;68:10229–10237. doi: 10.1158/0008-5472.CAN-08-1983. [DOI] [PubMed] [Google Scholar]
- 35.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 36.Dewaele M, Maes H, Agostinis P. ROS-mediated mechanisms of autophagy stimulation and their relevance in cancer therapy. Autophagy. 2010;6:838–854. doi: 10.4161/auto.6.7.12113. [DOI] [PubMed] [Google Scholar]
- 37.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaw RJ. Glucose metabolism and cancer. Curr Opin Cell Biol. 2006;18:598–608. doi: 10.1016/j.ceb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 39.Polyak K, Lee MH, Erdjument-Bromage H, Koff A, Roberts JM, Tempst P, et al. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78:59–66. doi: 10.1016/0092-8674(94)90572-X. [DOI] [PubMed] [Google Scholar]
- 40.Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- 41.Massagué J. G1 cell cycle control and cancer. Nature. 2004;432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 42.Tasdemir E, Maiuri MC, Orhon I, Kepp O, Morselli E, Criollo A, et al. p53 represses autophagy in a cell cycle-dependent fashion. Cell Cycle. 2008;7:3006–3011. doi: 10.4161/cc.7.19.6702. [DOI] [PubMed] [Google Scholar]
- 43.Nam E, Park C. Maspin suppresses survival of lung cancer cells through modulation of Akt pathway. Cancer Res Treat. 2010;42:42–47. doi: 10.4143/crt.2010.42.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee JY, Kim JH, Chae G, Lee BK, Ha KS, Kwon YG, et al. Cyclic AMP prolongs graft survival by suppressing apoptosis and inflammatory gene expression in acute cardiac allograft rejection. Exp Mol Med. 2010;42:69–79. doi: 10.3858/emm.2010.42.1.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.