Abstract
Gliomas, which generally have a poor prognosis, are the most common primary malignant brain tumors in adults. Recent genome-wide association studies have demonstrated that inherited susceptibility plays a role in the development of glioma. Although first-degree relatives of patients exhibit a two-fold increased risk of glioma, the search for susceptibility loci in familial forms of the disease has been challenging because the disease is relatively rare, fatal, and heterogeneous, making it difficult to collect sufficient biosamples from families for statistical power. To address this challenge, the Genetic Epidemiology of Glioma International Consortium (Gliogene) was formed to collect DNA samples from families with two or more cases of histologically confirmed glioma. In this study, we present results obtained from 46 U.S. families in which multipoint linkage analyses were undertaken using nonparametric (model-free) methods. After removal of high linkage disequilibrium SNPs, we obtained a maximum nonparametric linkage score (NPL) of 3.39 (P=0.0005) at 17q12–21.32 and the Z-score of 4.20 (P=0.000007). To replicate our findings, we genotyped 29 independent U.S. families and obtained a maximum NPL score of 1.26 (P=0.008) and the Z-score of 1.47 (P=0.035). Accounting for the genetic heterogeneity using the ordered subset analysis approach, the combined analyses of 75 families resulted in a maximum NPL score of 3.81 (P=0.00001). The genomic regions we have implicated in this study may offer novel insights into glioma susceptibility, focusing future work to identify genes that cause familial glioma.
Keywords: Glioma, family studies, linkage, haplotype pattern, NPL
INTRODUCTION
Gliomas account for ~40% of all primary malignant brain tumors (PBT) and are responsible for ~13,000 cancer related deaths in the US each year. Irrespective of treatment, most gliomas are associated with a poor prognosis with the most common type of glioma, glioblastoma (GBM), having a median overall survival of only 10–15 months (1).
Evidence strongly suggests that inherited susceptibility plays a role in the development of glioma, as first-degree relatives of patients with glioma have a two-fold increased risk of glioma (2–4). Hereditary genetic disorders such as neurofibromatosis type I and II, and Li-Fraumeni and Turcot’s syndromes are known to predispose to glioma (5;6). These syndromes are, however, rare and collectively make only a minor contribution to the familial risk of glioma.
Families segregating glioma outside the context of these syndromes provide a strong rationale for seeking to identify moderate-high risk susceptibility loci for glioma through genome-wide linkage scans. To date only one genome-wide linkage scan of glioma has been conducted, which was based on the analysis of four Finnish families. Evidence of linkage of glioma to 15q23–26 was provided; however, no genetic mutation was identified in this region (7).
To facilitate the collection of glioma families informative for linkage analysis, we established the “Genetic Epidemiology of Glioma International Consortium” (GLIOGENE-Linkage) in 2006 (8). GLIOGENE now includes 15 institutions in the United States, United Kingdom, Sweden, Denmark, and Israel. We have undertaken a genome-wide scan of 75 glioma families in the U.S. ascertained through GLIOGENE. This search was conducted using high-density single-nucleotide polymorphism (SNP) arrays, thereby allowing us to maximize the power to identify a disease-causing locus. Here we report evidence for Mendelian predisposition to glioma and strong evidence for a disease locus at 17q12–21.32.
MATERIALS AND METHODS
Ascertainment and collection of families
Forty six glioma families with at least two biologically related family members diagnosed with a histologically-confirmed glioma (International Classification of Disease – Oncology codes: low grade glioma (WHO grade I, II): juvenile pilocytic astrocytoma (9421/3), fibrillary astrocytoma (9420/3), protoplasmic astrocytoma (9410/3), gemistocytic astrocytoma (9411/3), diffuse astrocytoma (9400/3), oligodendroglioma (9450/3), oligoastrocytoma (9382/3); ependymoma (9391/3), high grade glioma (WHO grade III and IV): anaplastic astrocytoma (9401/3), anaplastic oligodendroglioma (9451/3), anaplastic oligoastrocytoma (9382/3), anaplastic ependymoma (9392/3), gliosarcoma (9442/3), gliomatosis cerebri (9381/3), and glioblastoma multiforme (9440/3)) were ascertained through the GLIOGENE consortium. We excluded all families with reported or confirmed diagnosis of neurofibromatosis 1, neurofibromatosis 2, Turcot’s syndrome or tuberous sclerosis. The scheme for recruitment, and data collection of families has been previously described (8). These forty-nine families were ascertained and genotyped in years 2004–2009, and are referred as the first stage families. An additional twenty-nine families were collected using the same sampling criterion in 2010 and genotyped in 2011. These twenty-nine families are referred to in this manuscript as the second stage or the replication families. All members of the included families were self-reported to be non-Hispanic white. Blood or saliva samples were obtained from both the offspring and spouse of deceased affected family members wherever possible to facilitate the genotype reconstruction of deceased family members. DNA was extracted from EDTA-venous blood samples and saliva samples that were collected using the Oragene kits (DNA Genotek). Biosamples and clinico-pathological information from patients and family members was collected with informed consent according to protocols approved by each center’s Institutional Review Board in accordance with the tenets of the Declarationof Helsinki.
Genotyping
Prior to genotyping all DNA samples were quantified by PicoGreen (Invitrogen, Carlsbad, CA). Genotyping was conducted using Illumina Hap370K BeadChip arrays according to manufacturer’s protocol. DNA with GenCall scores <0.25 at any locus were considered ‘no-calls’. A SNP was considered to have failed if <95% of DNA samples generated a genotype at the locus. Cluster plots were manually inspected to resolve any ambiguities. We excluded SNPs from the analyses if the Hardy-Weinberg proportion P-value was <0.0001. We also removed SNPs for which the minor allele frequency was <5%.
Data processing and error checking
The pedigree relationship testing program Pedcheck software, version 1.1 (November 24, 1998) (9) was implemented to check for pedigree errors. Non-Mendelian error checking of genotypes and generation of linkage format files from Illumina array files was performed using in-house generated scripts. The genotyping calls were processed using the BeadStudio Genotyping (BSGT) software, version 3.2.32. The map order and distances between SNP markers was based on the NCBI GenomeBuild version 36.
Linkage analysis
We conducted multipoint non-parametric linkage analysis using Allegro software, version 2.0f (10). We used an equal family weighting scheme in the exponential model with a scoring function Spairs (11), which is optimal over a range of disease models (12).
Multipoint linkage assumes that markers are in linkage equilibrium. However, for closely spaced markers, this is not always the case thus increasing false-positive evidence for linkage (13). Pairwise LD (as measured by r2 value) between SNPs was calculated using Plink (v1.06) (14). Then, we obtained LD blocks based on the LD value criteria of r2 ≥ 0.01 and r2 ≥ 0.004. For each block, we retained the SNP from each set with the highest information content, defined by entropy information measure IE (15). Then, we performed linkage analysis using three different criteria: retaining all SNPs, excluding all SNPs with r2 >0.01, and excluding all SNPs with r2 value >0.004. For some pedigrees, individual members not informative for linkage were removed to accommodate Allegro pedigree size limitations.
We performed haplotype analyses using SimWalk2 (version 2.83) (16–18). SimWalk2 uses Markov chain Monte Carlo methods and simulated annealing algorithms to perform multipoint haplotype analysis, which estimates the most likely set of fully-typed maternal and paternal haplotypes of the marker loci for each individual in the pedigree. The input files to SimWalk2 were automatically generated from the linkage analysis files using a utility program Mega2 (Manipulation Engine for Genetic Analysis), version 4.3.1 (19). The options for running Mega2 were set with “SimWalk2” for analysis option and “Haplotype analysis” for analysis sub-option.
Genetic heterogeneity is an important feature of complex disease etiology such as glioma. We performed the ordered subset analysis (OSA) using trait related covariates that provide maximal evidence for linkage (20;21). In the OSA method, the family members’ covariate information is used to assign a covariate score to each family. Subsequently, families are ordered by family-specific covariate score and linkage analyses are performed on all subsets ranked by covariate scores. We used a computer program, FLOSS (version 1.4.1 for Windows), that implemented the OSA method (21). We used age of onset (minimum, maximum, mean, and range) and number of affected individuals per family as covariates in the FLOSS analyses.
RESULTS
Details of the 75 families: 46 from the first stage of analyses and 29 from the replication study are summarized in Table 1. For the first stage families, the average family size was 24.6 individuals, with the smallest family having eight individuals and the largest having 44 individuals. A total of 127 individuals (11.2%) were affected with a diagnosis of glioma, 835 (73.8%) were unaffected, and 169 (15.0%) were coded as “unknown affection status.” Of the 127 affected individuals, 61 (48%) were male compared to 50.1% of the total individuals. We genotyped 415 individuals (36.7%), with a range of 4 to 20 individuals genotyped per family. Twenty-eight pedigrees had two affected relatives per pedigree, twelve pedigrees had three affected relatives per pedigree, one pedigree had four affected relatives, three pedigrees had five affected relatives per pedigree, and two pedigrees had six affected relatives per pedigree. The average age of diagnosis for the probands was 46.9 years (standard deviation (sd)=13.4) and the average age of diagnosis for the other affected members in the pedigrees (excluding proband) was 46.9 years (sd=19.6) (Table 1).
Table 1.
Descriptive statistics and characteristics of glioma pedigrees from US used for linkage analyses
| No. of affected individuals in a pedigree | No. of affected individuals in generation(s) | First Stage (n=46) | Second Stage (n=29) | Combined (n=75) |
|---|---|---|---|---|
| 2 | 1 | 14 | 8 | 22 |
| 2 | 13 | 10 | 23 | |
| 3 | 1 | 3 | 4 | |
| 3 | 1 | 7 | -- | 7 |
| 2 | 3 | 5 | 8 | |
| 3 | 2 | 1 | 3 | |
| 4 | 1 | -- | -- | -- |
| 2 | 1 | -- | 1 | |
| 3 | -- | -- | -- | |
| 4 | -- | 1 | 1 | |
| 5 | 1,2 | -- | -- | -- |
| 3 | 3 | -- | 3 | |
| 6 | 1,2 | -- | -- | -- |
| 3 | 2 | 1 | 3 |
| No. of affected male (%) | 61 (48.0) | 41 (58.6) | 102 (51.8) |
| No. of affected female (%) | 66 (52.0) | 29 (41.4) | 95 (48.2) |
| Avg. age (yrs) at dx, proband (SD) | 46.9 (13.4) | 47.6 (14.8) | 45.0 (14.4) |
| Age range at dx, proband | 6–69 | 22–75 | 6–75 |
| Avg. age (yrs) at dx, family (SD) | 46.9 (19.6) | 46.6 (20.5) | 45.3 (19.1) |
| Age range at dx, all affected individuals | 1–86 | 1–85 | 1–86 |
| Total No. Individuals | 1131 | 552 | 1683 |
| Avg. family size | 24.6 | 19.0 | 22.4 |
| No. genotyped (%) | 415 (36.7) | 309 (56.0) | 724 (43.0) |
Data quality
A number of parameters were usedthroughout the study to determine data quality. The average SNP call rate per array was >96%. Three pedigrees from the stage 1 samples had excessive Mendelian inheritance errors, which were likely due to misspecified relationships or sample switching. These three pedigrees were removed from the analyses. On average, Mendelian inconsistency was found in < 0.4% of the SNPs. These error SNPs were randomly distributed across the genome and were removed from further analyses.
Linkage analysis
The linkage analyses based on the SNPs retained using the three LD criterions (see the methods section) resulted in LOD score peaks at identical locations (data not shown). The first two criterions based on retaining all the SNPs and excluding all SNPs with r2 >0.01 gave higher LOD scores than the criterion based on excluding all SNPs with r2 >0.004. Conservatively, we report linkage analysis results based on the last criterion (i.e. excluding all SNPs with r2 value >0.004) which is based on a total of 23,476 SNPs across the genome.
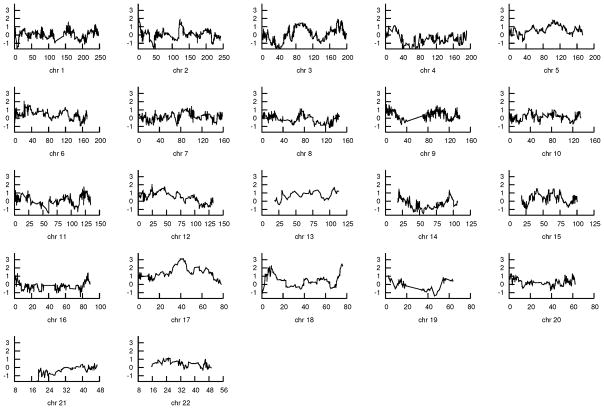
For the initial 46 families, the most significant genome-wide linkage (NPL) score of 3.39 was obtained at physical location 42,504,408 on chromosome 17q12–21.32 (P=0.0005) (Table 2 and Figure 1). The corresponding Z-score in this region was 4.20 (P=0.000007). In addition, three other chromosomal regions had maximum NPL scores exceeding 2.0. These regions were chromosome 6 (max NPL=2.10, P=0.02 at physical location 22,393,991, 6p22.3), chromosome 12 (max NPL=2.07, P=0.02 at physical location 24,344,720, 12p13.33–12.1), and chromosome 18 (max NPL=2.45, P=0.008 at physical location 75,238,190, 18q23).
Table 2.
The NPL scores and exact p-values for US glioma pedigrees after the removal of all SNPs with r2 value > 0.004 and minor allele frequency below 0.05.
| chr | physical location | SNP | max NPL | NPL exact p-value | left physical location | right physical location |
|---|---|---|---|---|---|---|
| 1 | 158,235,632 | rs6676862 | 1.6120 | 0.0556 | 157,007,286 | 158,871,097 |
| 2 | 1,657,741 | rs7560004 | 1.9369 | 0.0283 | 1,443,268 | 4,151,604 |
| 3 | 178,984,865 | rs6764952 | 1.9355 | 0.0284 | 175,719,120 | 180,045,647 |
| 4 | 7,375,903 | rs4689705 | 1.1218 | 0.1320 | 6,691,863 | 7,852,481 |
| 5 | 103,137,679 | rs320654 | 1.8552 | 0.0338 | 89,093,506 | 124,487,446 |
| 6 | 22,393,991 | rs1205961 | 2.0998 | 0.0195 | 22,382,745 | 23,359,338 |
| 7 | 100,639,893 | rs1468358 | 1.4495 | 0.0755 | 100,151,356 | 101,310,585 |
| 8 | 72,212,959 | rs1481812 | 1.1769 | 0.1209 | 72,179,731 | 78,848,596 |
| 9 | 1,901,291 | rs10811102 | 1.6935 | 0.0473 | 1,381,887 | 2,049,341 |
| 10 | 37,456,382 | rs1878249 | 1.2281 | 0.1111 | 1,050,218 | 33,053,017 |
| 11 | 122,434,697 | rs11551598 | 1.7056 | 0.0461 | 122,356,773 | 123,153,543 |
| 12 | 24,344,720 | rs565934 | 2.0733 | 0.0208 | 2,152,566 | 25,405,864 |
| 13 | 105,045,125 | rs10508158 | 1.5536 | 0.0622 | 101,097,159 | 114,108,121 |
| 14 | 19,946,093 | rs1760897 | 1.5150 | 0.0669 | 19,536,664 | 93,349,018 |
| 15 | 41,144,698 | rs12050604 | 1.5787 | 0.0593 | 37,598,478 | 41,154,373 |
| 16 | 86,488,782 | rs8058389 | 1.4061 | 0.0817 | 84,489,852 | 87,500,055 |
| 17 | 42,504,408 | rs4074249 | 3.3913 | 0.0005 | 33,196,340 | 45,510,424 |
| 18 | 75,238,190 | rs1110784 | 2.4501 | 0.0082 | 73,651,628 | 76,085,595 |
| 19 | 2,561,822 | rs8104096 | 1.0822 | 0.1404 | 5,495,085 | 54,286,819 |
| 20 | 2,051,264 | rs6075668 | 1.3404 | 0.0918 | 16,749 | 3,398,635 |
| 21 | 45,595,912 | rs2838891 | 0.5032 | 0.3048 | 42,200,186 | 46,852,308 |
| 22 | 26,003,211 | rs134806 | 1.1607 | 0.1240 | 15,873,708 | 33,555,460 |
NOTE: chr, chromosome; physical location, the physical location at which maximum NPL score was observed; SNP, rs number of the SNP at which maximum NPL score was observed; max NPL, maximum NPL score obtained; NPL exact p-value, the exact p-value associated with maximum NPL score from Allegro; left physical location and right physical location, boundaries based on 1-LOD drop (approximating 95% confidence interval for the location of peak.
Figure 1. NPL scores for US glioma pedigrees across each chromosome.
Each plot shows NPL scores obtained after excluding all SNPs with r2 value >0.004. The y-axis is NPL score and x-axis is physical location.
We further investigated the contribution of each family to the LOD scores on chromosome 17. We investigated the age of onset of the affected members of these families linked to the region on chromosome 17. Eleven families showed maximal evidence of linkage at chromosome 17. Each of these families had individual NPL score greater than 0.59 (equivalent to a point-wise significance of 0.05). The age of onset for all individuals affected with glioma in the families linked to the chromosome 17 region was lower than the age of onset for all individuals affected with glioma coming from unlinked families (36.8 versus 43.9 years); however, the difference was not statistically significant (p-value=0.17). Most of the families linked to chromosome 17 have affected siblings with glioma (8 out of 11 families). No significant glioma histological differences were noted between families.
To better understand the 17q region and to identify risk haplotypes shared by affected individuals from the eleven families linked to this region, we genotyped five highly polymorphic microsatellite markers in 17q region (D17S932, D17S950, D17S791, D17S943, and D17S1869). The average information content value for all the markers was 0.56 to 0.70. Haplotype analyses revealed that the risk haplotype segregated among affected relatives. However, we also observed that a number of unaffected individuals were carriers of the disease haplotype implying decreased penetrance. The highest NPL score of 4.02 (P=0.000009) was obtained at the marker D17S950 using microsatellite-based multipoint linkage analyses.
Replication and Joint Analyses
To replicate the 17q12–21.32 region, we genotyped 29 independent glioma families ascertained using the same criteria as families in the first stage. The average family size was 19 individuals, with the smallest family having nine individuals and the largest had 32 individuals. A total of 70 individuals (12.6%) were affected with a confirmed diagnosis of glioma. Of the 70 affected individuals, 41 (58.6%) were male which is a higher percentage than the percentage of affected males in the first stage families; however, the difference is statistically not significant. We genotyped a total 309 individuals (56.0%) which is a higher percentage than the number of individuals genotyped in the first stage families. We were able to get DNA samples on more individuals in the second stage which could be due to the fact that stage 2 families were more recently ascertained and recruitment staff was more experienced. The majority of the families had only two affected relatives (n=21), and the other eight families had three to six affected relatives. The average age of diagnosis for the probands was 47.6 years (sd=14.8), and the average age of diagnosis for the affected members (excluding proband) was 46.6 years (sd=20.5) (Table 1) which was very similar to the average age of diagnosis for the affected individuals in the first stage families.
Using Allegro, we obtained NPL score of 1.26 (P=0.008) at 17q12–21.32 region for the Stage 2 families. The corresponding Z-score in this region was 1.47 (P=0.035).
To account for hypothesized genetic heterogeneity, using the FLOSS program, we obtained an ordered subset analyses-based NPL score of 2.16 from the 29 replication families (P=0.0008) with the number of affected individuals per family as significant family-specific covariate. Even though, the NPL score based on stage 2 families did not reach the LOD score threshold of 3.0, it should be noted that we are not performing genomewide analyses with these replication families. Therefore, the point-wise p-value of 0.0008 should be considered as a replication of the genetic region identified using the first stage families.
We further performed joint OSA on 75 families (46 from stage 1 and 29 from stage 2). Using the FLOSS, the combined analyses gave us an NPL score of 3.81 (P=0.00001) with the number of affected individuals per family as significant family-specific covariate. When we re-analysed the data excluding those families deemed unlinked from the FLOSS analysis, the LOD score increased to 4.24 (P=0.000005), further supporting linkage to this region.
We also performed joint OSA on 75 families at the three chromosomal regions 6p22.3, 12p13.33–12.1, and 18q23 that were suggestive of linkage based on the first stage families. Using the FLOSS, the combined analyses gave respective NPL scores 2.34, 1.75, and 2.04 with the number of affected individuals per family as significant family-specific covariate.
DISCUSSION
Here we provide evidence for a moderate-high penetrance susceptibility to glioma mapping to 17q12–21.32. We searched the National Center for Biotechnology Information (NCBI) Hapmap database and identified potentially important target genes related to cancers in this area. WNT9B (17q21), mapping at 44,941,702 bps, belongs to the WNT gene family, is associated with basal cell carcinoma. The WNT gene family consists of several structurally related genes that encode signalling proteins that have been implicated in oncogenesis and in several developmental processes, including the regulation of cell fate and patterning during embryogenesis (22;23). Another gene that resides in this region is CDC27, at location 17q12–23.2. The protein encoded by this gene is a component of the anaphase-promoting complex (APC), an E3 ubiquitin ligase that controls the cell cycle. The DNA damage response mediator MDC1 directly interacts with this protein (24). A third gene in this region is NPEPPS, at location 17q21. This protein-coding gene encodes an enzyme that degrades enkaphalins in the brain, and mouse studies suggest that it is also involved in regulating the cell cycle. Another candidate gene mapping to this region is NSF, located at 17q21, which has been shown to be associated with schizophrenia (25) and neurotransmission and neurodevelopment (26). Finally, the ITGB3 gene located at 17q21.32 has been reported to be is involved in brain ischemia (27).
We also identified three additional regions, on chromosomes 6p, 12p, and 18q, which are suggestive for linkage using data from first stage families. The associated allele-sharing LOD scores also exceeded threshold for suggestive evidence of linkage. The exact p-values for NPL scores in these regions were between 0.01 and 0.02. However, using the joint OSA on 75 families, only 6p22.3 and 18q23 regions remained suggestive for linkage.
The contribution of individual families to the genetic region implicated in this study varied, suggesting that gliomas have strong genetic heterogeneity. This provides justification for the further study of larger number of families at high risk for this relatively rare cancer.
Recently, our group conducted three independent genome-wide association studies (28–30) which identified seven risk loci for glioma at 5p15.33 (TERT rs2736100), 7p11.2 (EGFR rs11979158 and rs2252586), 8q24.21 (CCDC26 rs4295627), 9p21.3 (CDKN2A-CDKN2B rs4977756), 11q23.3 (PHLDB1 rs498872), and 20q13.33 (RTEL1 rs6010620). Collectively these loci accounted for only 7–14% of the excess familial risk of glioma (28). The linkage peak for the GWAS hits was not significant in the families analyzed herein, underscoring that the genes associated with sporadic glioma have little contribution to familial glioma. Thus, additional studies are needed to discover additional risk loci for familial glioma.
In summary, our analysis provides evidence for disease locus for glioma susceptibility at 17q12–21.3. The next step for this study is to sequence this region in the affected and unaffected members of families that showed high linkage and determine if there is a pathogenic mutation in the germline. We will look for deleterious mutations (frame shift truncating or nonsense mutations, missense variants, insertions or deletions), that could be cancer promoting genes. If we identify specific genes from the sequencing analysis, we will then screen for these mutations in the linking relatives. To date, this is the largest genetic linkage study of glioma. The regions implicated in this study underscore glioma heterogeneity especially since they are different from those identified in previous GWAS. Further studies could potentially provide novel insights into the biological mechanisms and may ultimately lead to the identification of a causal gene or genes in these regions and to guidelines for genetic counseling.
Acknowledgments
The authors acknowledge the contributions of the following individuals to the overall brain tumor research programs MD Anderson Cancer Center: Phyllis Adatto, Fabian Morice, Sam Payen, Lacey McQuinn, Rebecca McGaha, Sandra Guerra, Leslie Paith, Katherine Roth, Dong Zeng, Hui Zhang, Dr. Alfred Yung, Dr. Howard Colman, Dr. Charles Conrad, Dr. John de Groot, Dr. Arthur Forman, Dr. Morris Groves, Dr. Victor Levin, Dr. Monica Loghin, Dr. Vinay Puduvalli, Dr. Raymond Sawaya, Dr. Amy Heimberger, Dr. Frederick Lang, Dr. Nicholas Levine, Lori Tolentino; Brigham and Women’s Hospital: Kate Saunders, Donna Dello Iacono; Case Western Reserve University: Dr. Stanton Gerson, Dr. Warren Selman, Dr. Robert Maciunas, Dr. Nicholas Bambakidis, Dr. David Hart, Dr. Jonathan Miller, Dr. Alan Hoffer, Dr. Mark Cohen, Dr Lisa Rogers, Dr. Charles J Nock, Wendi Barrett, Anita Merriam, Quinn Ostrom, Sarah Robbins, Perica Davitkov, Dr. Michael Vogelbaum, Dr. Robert Weil, Dr. Manmeet Ahluwalia, Dr. David Peereboom, Dr. Edward Benzel, Dr. Susan Staugaitis, Cathy Schilero, Cathy Brewer, Kathy Smolenski, Diane Fabec, Theresa Naska, Jennifer Hornacek-Guadalupe; Columbia University Medical Center: Dr. Steven Rosenfeld; Israel: Dr. Zvi Ram, Dr. Deborah T Blumenthal, Dr. Felix Bokstein (Tel-Aviv Sourasky Medical Center), Dr. Felix Umansky (Hadassah Hebrew University Medical Center, Henry Ford Hospital), Dr. Menashe Zaaroor (Rambam Health Care Campus) Dr. Avi Cohen (Soroka University Medical Center, Chaim Sheba Medical Center), Dr. Tzeela Tzuk-Shina (Rambam Medical Center and Faculty of Medicine, Technion-Israel Institute of Technology); Denmark: Dr. Bo Voldby (Aarhus University Hospital), Dr. Rene Laursen M.D. (Aalborg University Hospital), Dr. Claus Andersen (Odense University Hospital), Dr. Jannick Brennum (Glostrup University Hospital), Matilde Bille Henriksen (Institute of Cancer Epidemiology, the Danish Cancer Society); Memorial Sloan-Kettering Cancer Center: Maya Marzouk, Mary Elizabeth Davis, Eamon Boland, Marcel Smith, Ogechukwu Eze, Mahalia Way; NorthShore University HealthSystem: Pat Lada, Nancy Miedzianowski, Michelle Frechette, Dr. Nina Paleologos; Sweden: Gudrun Bystrom, Sara Huggert, Mikael Kimdal and Monica Sandstrom (Umea University); University of California, San Francisco: Dr. Tarik Tihan, Dr. Shichun Zheng, Dr. Mitchel Berger, Dr. Nicholas Butowski, Dr. Susan Chang, Dr. Jennifer Clarke, Dr. Michael Prados, Terri Rice, Jeannette Sison, Valerie Kivett, Xiaoqin Duo, Helen Hansen, George Hsuang, Rosito Lamela, Christian Ramos, Joe Patoka, Katherine Wagenman, Mi Zhou, Adam Klein, Nora McGee, Jon Pfefferle, Callie Wilson, Pagan Morris, Mary Hughes, Marlin Britt-Williams, Jessica Foft, Julia Madsen, Csaba Polony; University of Illinois at Chicago: Candice Zahora, Dr. John Villano, Dr. Herbert Engelhard.
The authors acknowledge the input of the Gliogene External Advisory Committee: Dr. Ake Borg (Department of Oncology, Lund University, Lund, Sweden), Dr. Stephen K Chanock (National Cancer Institute, U. S. National Institutes of Health), Dr. Peter Collins (University of Cambridge, United Kingdom), Dr. Robert Elston (Department of Epidemiology and Biostatistics, Case Western Reserve University), Dr. Paul Kleihues (Department of Pathology, University Hospital, Zurich, Switzerland), Carol Kruchko (Central Brain Tumor Registry of the United States), Dr. Gloria Petersen (Health Sciences Research, Mayo Clinic), Dr. Sharon Plon (Baylor Cancer Genetics Clinic, Baylor College of Medicine).
Finally, we would like to thank the patients and their families for participating in this research.
FUNDING
This work was supported by grants from the National Institutes of Health, Bethesda, Maryland (5R01 CA119215, 5R01 CA070917, R01CA52689, P50097257, R01CA126831). Additional support was provided by the American Brain Tumor Association, The National Brain Tumor Society, and the Tug McGraw Foundation. For more information about the Gliogene Consortium, refer to the following Web site: http://www.gliogene.org.
The members of the Gliogene Consortium: Department of Epidemiology, The University of Texas MD Anderson Cancer Center, Houston, Texas (Melissa L. Bondy, Sanjay Shete, Christopher I Amos, Georgina N Armstrong, Yanhong Liu, Robert K Yu); Department of Pathology, The University of Texas MD Anderson Cancer Center, Houston, Texas (Kenneth D Aldape); Department of Neuro-Oncology, The University of Texas MD Anderson Cancer Center, Houston, Texas (Mark R Gilbert); Department of Neurosurgery, The University of Texas MD Anderson Cancer Center, Houston, Texas (Jeffrey Weinberg); Department of Pediatrics, Section of Hematology and Oncology, Dan L. Duncan Cancer Center, Baylor College of Medicine, Houston, Texas (Ching C. Lau, Eastwood Hon-chiu Leung, Caleb Davis, Rita Cheng, Chris Man, Rudy Guerra, Sivashankarappa Gurusiddappa, Michael E. Scheurer); Section of Cancer Genetics, Institute of Cancer Research, Sutton, Surrey, United Kingdom (Richard S. Houlston, Fay J Hosking, Lindsay Robertson, Elli Papaemmanuil); Department of Epidemiology and Public Health, Yale University School of Medicine, New Haven, Connecticut (Elizabeth B. Claus); Department of Neurosurgery, Brigham and Women’s Hospital, Boston, Massachusetts (Elizabeth B. Claus); Case Comprehensive Cancer Center, Case Western Reserve University School of Medicine, Cleveland, Ohio (Jill Barnholtz-Sloan, Andrew E. Sloan, Gene Barnett, Karen Devine, Yingli Wolinsky); The Neurological Institute of Columbia University, New York, New York (Rose Lai, Erika Florendo, Delcia Rivas, Christina Corpuz); Cancer Control and Prevention Program, Department of Community and Family Medicine, Duke University Medical Center, Durham, North Carolina (Dora Il’yasova, Joellen Schildkraut); Cancer and Radiation Epidemiology Unit, Gertner Institute, Chaim Sheba Medical Center, Tel Hashomer, Israel (Siegal Sadetzki, Galit Hirsh Yechezkel, Revital Bar-Sade Bruchim, Lili Aslanov); Sackler School of Medicine, Tel-Aviv University, Tel-Aviv, Israel (Siegal Sadetzki); Department of Neurology; Institute of Cancer Epidemiology, Danish Cancer Society, Copenhagen, Denmark (Christoffer Johansen, Hanne Bodtcher); Neurosurgery Department, Rigshospitalet, University Copenhagen (Michael Kosteljanetz), Neuropathology Department, Rigshospitalet, University Copenhagen (Helle Broholm); Department of Epidemiology and Biostatistics, Memorial Sloan-Kettering Cancer Center, New York, New York (Jonine L. Bernstein, Sara H. Olson, Erica Schubert), Department of Neurology, Memorial Sloan-Kettering Cancer Center, New York, New York (Lisa DeAngelis); Mayo Clinic Comprehensive Cancer Center, Mayo Clinic, Rochester, Minnesota (Robert B. Jenkins, Ping Yang, Amanda Rynearson); Department of Radiation Sciences Oncology, Umea University, Umea, Sweden (Beatrice S. Melin, Roger Henriksson, Ulrika Andersson), Department of Medical Biosciences, Umea University, Umea, Sweden (Thomas Brannstrom); Evanston Kellogg Cancer Care Center, NorthShore University HealthSystem, Evanston, Illinois (Nicholas A. Vick); Departments of Neurological Surgery and Epidemiology and Biostatistics (Margaret Wrensch, John Wiencke, Joe Wiemels, Lucie McCoy) Division of Epidemiology and Biostatistics, University of Illinois at Chicago, Chicago, Illinois (Bridget J McCarthy, Faith G Davis).
Footnotes
Conflict of Interest
The authors declare no competing financial interests.
The Danish (C. Johansen), Israeli (S. Sadetzki), and Swedish (B. Melin) sites recruit population-based participants nationwide.
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005 Mar 10;352(10):987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Malmer B, Gronberg H, Bergenheim AT, Lenner P, Henriksson R. Familial aggregation of astrocytoma in northern Sweden: an epidemiological cohort study. Int J Cancer. 1999 May 5;81(3):366–70. doi: 10.1002/(sici)1097-0215(19990505)81:3<366::aid-ijc9>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 3.Wrensch M, Lee M, Miike R, Newman B, Barger G, Davis R, et al. Familial and personal medical history of cancer and nervous system conditions among adults with glioma and controls. Am J Epidemiol. 1997 Apr 1;145(7):581–93. doi: 10.1093/oxfordjournals.aje.a009154. [DOI] [PubMed] [Google Scholar]
- 4.Hemminki K, Tretli S, Sundquist J, Johannesen TB, Granstrom C. Familial risks in nervous-system tumours: a histology-specific analysis from Sweden and Norway. Lancet Oncol. 2009 May;10(5):481–8. doi: 10.1016/S1470-2045(09)70076-2. [DOI] [PubMed] [Google Scholar]
- 5.Bondy ML, Scheurer ME, Malmer B, Barnholtz-Sloan JS, Davis FG, Il’yasova D, et al. Brain tumor epidemiology: consensus from the Brain Tumor Epidemiology Consortium. Cancer. 2008 Oct 1;113(7 Suppl):1953–68. doi: 10.1002/cncr.23741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kyritsis AP, Bondy ML, Rao JS, Sioka C. Inherited predisposition to glioma. Neuro Oncol. 2010 Jan;12(1):104–13. doi: 10.1093/neuonc/nop011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paunu N, Lahermo P, Onkamo P, Ollikainen V, Rantala I, Helen P, et al. A novel low-penetrance locus for familial glioma at 15q23-q26.3. Cancer Res. 2002 Jul 1;62(13):3798–802. [PubMed] [Google Scholar]
- 8.Malmer B, Adatto P, Armstrong G, Barnholtz-Sloan J, Bernstein JL, Claus E, et al. GLIOGENE an International Consortium to Understand Familial Glioma. Cancer Epidemiol Biomarkers Prev. 2007 Sep;16(9):1730–4. doi: 10.1158/1055-9965.EPI-07-0081. [DOI] [PubMed] [Google Scholar]
- 9.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998 Jul;63(1):259–66. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2000 May;25(1):12–3. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- 11.Whittemore AS, Halpern J. A class of tests for linkage using affected pedigree members. Biometrics. 1994 Mar;50(1):118–27. [PubMed] [Google Scholar]
- 12.McPeek MS. Optimal allele-sharing statistics for genetic mapping using affected relatives. Genet Epidemiol. 1999;16(3):225–49. doi: 10.1002/(SICI)1098-2272(1999)16:3<225::AID-GEPI1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 13.Huang Q, Shete S, Amos CI. Ignoring linkage disequilibrium among tightly linked markers induces false-positive evidence of linkage for affected sib pair analysis. Am J Hum Genet. 2004 Dec;75(6):1106–12. doi: 10.1086/426000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996 Jun;58(6):1347–63. [PMC free article] [PubMed] [Google Scholar]
- 16.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996 Jun;58(6):1323–37. [PMC free article] [PubMed] [Google Scholar]
- 17.Sobel E, Sengul H, Weeks DE. Multipoint estimation of identity-by-descent probabilities at arbitrary positions among marker loci on general pedigrees. Hum Hered. 2001;52(3):121–31. doi: 10.1159/000053366. [DOI] [PubMed] [Google Scholar]
- 18.Sobel E, Papp JC, Lange K. Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet. 2002 Feb;70(2):496–508. doi: 10.1086/338920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukhopadhyay N, Almasy L, Schroeder M, Mulvihill WP, Weeks DE. Mega2: data-handling for facilitating genetic linkage and association analyses. Bioinformatics. 2005 May 15;21(10):2556–7. doi: 10.1093/bioinformatics/bti364. [DOI] [PubMed] [Google Scholar]
- 20.Hauser ER, Watanabe RM, Duren WL, Bass MP, Langefeld CD, Boehnke M. Ordered subset analysis in genetic linkage mapping of complex traits. Genet Epidemiol. 2004 Jul;27(1):53–63. doi: 10.1002/gepi.20000. [DOI] [PubMed] [Google Scholar]
- 21.Browning BL. FLOSS: flexible ordered subset analysis for linkage mapping of complex traits. Bioinformatics. 2006 Feb 15;22(4):512–3. doi: 10.1093/bioinformatics/btk012. [DOI] [PubMed] [Google Scholar]
- 22.Kirikoshi H, Inoue S, Sekihara H, Katoh M. Expression of WNT10A in human cancer. Int J Oncol. 2001 Nov;19(5):997–1001. doi: 10.3892/ijo.19.5.997. [DOI] [PubMed] [Google Scholar]
- 23.Smolich BD, McMahon JA, McMahon AP, Papkoff J. Wnt family proteins are secreted and associated with the cell surface. Mol Biol Cell. 1993 Dec;4(12):1267–75. doi: 10.1091/mbc.4.12.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coster G, Hayouka Z, Argaman L, Strauss C, Friedler A, Brandeis M, et al. The DNA damage response mediator MDC1 directly interacts with the anaphase-promoting complex/cyclosome. J Biol Chem. 2007 Nov 2;282(44):32053–64. doi: 10.1074/jbc.M705890200. [DOI] [PubMed] [Google Scholar]
- 25.Lencz T, Lambert C, DeRosse P, Burdick KE, Morgan TV, Kane JM, et al. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc Natl Acad Sci U S A. 2007 Dec 11;104(50):19942–7. doi: 10.1073/pnas.0710021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gratacos M, Costas J, de CR, Bayes M, Gonzalez JR, Baca-Garcia E, et al. Identification of new putative susceptibility genes for several psychiatric disorders by association analysis of regulatory and non-synonymous SNPs of 306 genes involved in neurotransmission and neurodevelopment. Am J Med Genet B Neuropsychiatr Genet. 2009 Sep 5;150B(6):808–16. doi: 10.1002/ajmg.b.30902. [DOI] [PubMed] [Google Scholar]
- 27.Lorger M, Krueger JS, O’Neal M, Staflin K, Felding-Habermann B. Activation of tumor cell integrin alphavbeta3 controls angiogenesis and metastatic growth in the brain. Proc Natl Acad Sci U S A. 2009 Jun 30;106(26):10666–71. doi: 10.1073/pnas.0903035106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009 Aug;41(8):899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009 Aug;41(8):905–8. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanson M, Hosking FJ, Shete S, Zelenika D, Dobbins SE, Ma Y, et al. Chromosome 7p11.2 (EGFR) variation influences glioma risk. Hum Mol Genet. 2011 May 16;20(14):2897–904. doi: 10.1093/hmg/ddr192. [DOI] [PMC free article] [PubMed] [Google Scholar]