Abstract
Specific phosphorylation of the human ventricular cardiac myosin regulatory light chain (MYL2) modifies the protein at S15. This modification affects MYL2 secondary structure and modulates the Ca2+ sensitivity of contraction in cardiac tissue. Smooth muscle myosin light chain kinase (smMLCK) is a ubiquitous kinase prevalent in uterus and present in other contracting tissues including cardiac muscle. The recombinant 130 kDa (short) smMLCK phosphorylated S15 in MYL2 in vitro. Specific modification of S15 was verified using the direct detection of the phospho group on S15 with mass spectrometry. SmMLCK also specifically phosphorylated myosin regulatory light chain S15 in porcine ventricular myosin and chicken gizzard smooth muscle myosin (S20 in smooth muscle) but failed to phosphorylate the myosin regulatory light chain in rabbit skeletal myosin. Phosphorylation kinetics, measured using a novel fluorescence method eliminating the use of radioactive isotopes, indicates similar Michaelis-Menten Vmax and KM for regulatory light chain S15 phosphorylation rates in MYL2, porcine ventricular myosin, and chicken gizzard myosin. These data demonstrate that smMLCK is a specific and efficient kinase for the in vitro phosphorylation of MYL2, cardiac, and smooth muscle myosin. Whether smMLCK plays a role in cardiac muscle regulation or response to a disease causing stimulus is unclear but it should be considered a potentially significant kinase in cardiac tissue on the basis of its specificity, kinetics, and tissue expression.
Keywords: Cardiac myosin contractility regulation, myosin regulatory light chain phosphorylation, smooth muscle myosin light chain kinase, mass spectroscopy, phosphorylation specificity, phosphorylation kinetics
INTRODUCTION
Myosin II is the energy transducer in skeletal and cardiac muscle. It impels actin against resistive force using the ATP hydrolysis free energy. The myosin heavy chain has a globular N-terminus, called subfragment 1 (S1) where ATP and actin bind, and a tail portion that associates with other myosin tails in the sarcomere to form the thick filament. Within S1, the longest αhelix is a lever-arm that rotates to perform the linear translation of actin for work production. Two myosin light chains associate with the lever-arm, the 18.8 kDa regulatory light chain (RLC) and the 21.9 kDa essential light chain (ELC). Both light chains stabilize the lever-arm to facilitate momentum transfer to actin and RLC also participates in contraction regulation. Smooth muscle contraction is regulated by RLC phosphorylation in the presence of Ca2+. In striated muscle, Ca2+ regulates contraction through the actin filament regulatory proteins but RLC phosphorylation modulates Ca2+ sensitivity. Several RLC mutations in human cardiac tissue are implicated in heart disease and are thought to affect Ca2+ binding and RLC phosphorylation leading to changes in the Ca2+ sensitivity of contraction and in vitro motility [1]. Human ventricular myosin S1 has the heavy chain gene, MYH7, essential light chain, MYL3, and regulatory light chain, MYL2.
In muscle tissue, a Ca2+ and calmodulin dependent myosin light chain kinase (MLCK) phosphorylates RLC [2]. The N-terminal region of the RLC contains the MLCK-specific phosphorylation site, S15 and S20 in cardiac and smooth muscle, respectively [3]. MLCK has different isoforms expressed in different muscle tissues. Smooth muscle MLCK (smMLCK) is ubiquitous in many adult tissues and represents a major MLCK detectable in the cardiac muscle, in contrast to skeletal and cardiac MLCK which are probably tissue specific [4]. The smMLCK is better conserved among different species than the cardiac and skeletal muscle forms [5]. A short form (130 kDa) smMLCK was expressed in the heart at lower levels compared to those detected in smooth muscle-rich organs [6]. Skeletal muscle MLCK (skMLCK) was also reported to be present in the heart but at an abundance too low to maintain basal RLC phosphorylation [7]. A cardiac MLCK (cMLCK) has been proposed to be the predominant protein kinase that maintains the basal RLC phosphorylation required for normal physiological cardiac performance in vivo [8]. There are reports suggesting that cardiac RLC is not a good substrate for the smMLCK present in the cardiac myocytes [9; 10].
We show here that the 130 kDa smMLCK specifically and efficiently phosphorylates the isolated human ventricular myosin regulatory light chain (MYL2) at S15 in vitro. Identical specificity and similar efficiency for RLC phosphorylation by smMLCK was also obtained for RLC in porcine ventricular myosin and gizzard smooth muscle myosin. These results establish smMLCK as a useful reagent for phosphorylation of various RLCs including MYL2 and opens the possibility that cardiac phosphorylation in vivo could be dependent on activity of the 130 kDa smMLCK.
MATERIALS AND METHODS
Expression and purification of wild-type MYL2
The cDNA of MYL2 was a generous gift from Dr. D. Szczesna-Cordary, University of Miami. MYL2 was cloned into the pET-3d plasmid vector (EMD Chemicals, NJ) and transformed into Escherichia coli BL21 (DE3) competent cells (Agilent Technologies, Santa Clara, CA). Protein expression was induced with isopropyl β-D-1-thiogalactopyranoside (IPTG) (Roche, Mannheim, Germany) at a final concentration of 1 mM. After induction the cells continued to grow for 17–18 hours and then collected, pelleted, washed with PBS, pelleted and frozen in lysis buffer (20 mM Tris pH 7.5, 2 M urea, 0.1 mM PMSF, 1 μg/mL leupeptin and 1 mM dithiothreitol (DTT)) at −20°C . The cells were thawed and lysed by sonication and pelleted. The protein extract was incubated with SP Sepharose beads (Sigma, St. Louis, MO) for 3 hours with continuous rotation at 4°C. MYL2 was eluted from the beads in lysis buffer containing 50–500 mM KCl. Protein purity was checked using a 12% SDS-PAGE gel and stained with Coomassie Blue. Fractions containing pure protein were combined and protein concentration was determined by using the Bradford assay with bovine serum albumin as the protein standard [11]. Protein samples were stored at −20°C.
Cloning, expression and purification of MLCK and smooth muscle HMMs
The short smooth muscle variant of MLCK (isoform 5) was obtained from a human MLCK full-length cDNA clone (a generous gift from Dr. A. Bresnick, Albert Einstein College of Medicine, New York, NY). The recombinant short (130 kDa) smMLCK was constructed with six N-terminal histidine residues inserted between M1 and D2 (Swiss-Prot # Q15746 isoform 5). Expression and purification of smMLCK was described previously [12].
A pFastBac construct plasmid containing cDNA of recombinant WT chicken gizzard smooth muscle heavy meromyosin (HMM) heavy chain and cDNA of the WT chicken gizzard smooth muscle RLC and ELC were generous gifts from Dr. Hirofumi Onishi (Mayo Clinic Rochester). The recombinant HMM heavy chain has a myc-tag at its C-terminus and a His-tag at its N-terminus as described previously [13]. Expression and purification of the smooth muscle HMM, including both light chains, was described [12].
Tissue purified myosins
Cardiac myosin was prepared from porcine heart ventriculum as described earlier for bovine cardiac myosin [14]. Rabbit skeletal myosin was prepared from back and leg muscles by the method of Tonomura et al. [15].
Phosphorylation of regulatory light chains
MYL2 (55 μM) was phophorylated with 1.2 μM smMLCK in a low ionic strength buffer containing 30 mM KCl, 25 mM Tris-HCl buffer pH 7.6, 12 mM MgCl2, 0.2 mM CaCl2, 2 mM DTT, 0.1 mM PMSF, 1 μg/mL leupeptin, 5 mM ATP, and 1.0 μM calmodulin. The reaction was carried out at room temperature for 10 minutes. 3 mM EGTA was added to stop the reaction and protein was precipitated with ice cold acetone. Precipitated MYL2 was dissolved in 8 M urea sample buffer and run on a 10% Tris-Glycine polyacrylamide gel (Invitrogen, Carlsbad, CA). The gel was first treated with Pro-Q Diamond phosphoprotein stain to detect phosphoproteins and then with SYPRO Ruby stain to detect total protein in the sample (Invitrogen).
MYL2, cardiac or skeletal myosins or smooth muscle HMM at various concentrations were phosphorylated with 1.2 μM smMLCK at room temperature in a low ionic strength buffer containing 30 mM KCl, 25 mM Tris-HCl buffer pH 7.6, 12 mM MgCl2, 0.2 mM CaCl2, 2 mM DTT, 0.1 mM PMSF, 1 μg/mL leupeptin, 5 mM ATP, and 1.0 μM calmodulin. MYL2 and HMM are soluble but myosin forms filaments in these conditions. Small aliquots were taken during the incubation to assess time-dependence of the phosphorylation. The phosphorylation reaction was terminated by adding 3 mM EGTA, aliquot proteins precipitated with ice cold acetone, and precipitates dissolved in a sample buffer containing 8 M urea then subjected to 10% Tris-Glycine polyacrylamide gel electrophoresis [16]. Protein bands were stained with SYPRO-Ruby and the fluorescence quantitated with ImageJ (NIH, USA).
We analyzed phosphorylation kinetics using a Michaelis-Menten scheme:
Scheme 1.
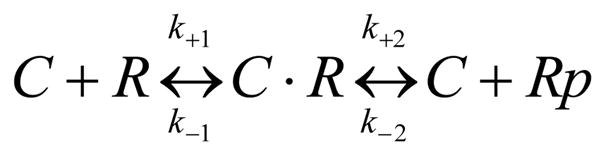
where C is MLCK, R the RLC (either on myosin heavy chain or free in solution), and Rp the phosphorylated RLC. ATP binding is saturated in our conditions. Rates k+1 and k−1 with equilibrium constant K1 represent the MLCK/RLC binding step and rates k+2 and k−2 catalysis. We assume catalysis is irreversible (Michaelis-Menten assumption) hence k−2 ≈ 0 and that K1 ≈ 1/KM (Michaels-Menten KM) because k−1 ≫ k2. The latter implies the reasonable assumptions that the binding step is rapidly reversible while catalysis is slow compared to other steps in Scheme 1.
Mass Spectrometry
Phosphorylated proteins were subjected to gel electrophoresis [17], followed by staining with Sypro Ruby. The PAGE gel bands were destained with 15 mM potassium ferricyanide and 50 mM sodium thiosulfate in water until clear and rinsed with water several times to remove all the color [18]. The bands were reduced with 50 mM TCEP/50 mM Tris pH 8.1 at 55ºC for 40 min and alkylated with 40 mM iodoacetamide at room temperature for 40 min in the dark. Proteins were digested in-situ with 30 μl (0.005μg/μl) trypsin (Promega Corporation, Madison, WI) in 25 mM Tris pH 8.1 / 0.0002% Zwittergent 3–16, at 37°C overnight followed by peptide extraction with 10 μl 2% trifluoroacetic acid, followed by 60 μl of acetonitrile. The peptide extracts was concentrated to less than 5 μl on a SpeedVac spinning concentrator (Savant Instruments, Holbrook NY) and then brought up in 0.15% formic acid/0.05% trifluoroacetic acid for protein identification by nano-flow liquid chromatography electrospray tandem mass spectrometry (nanoLC-ESI-MS/MS) using a ThermoFinnigan LTQ Orbitrap Hybrid Mass Spectrometer (Thermo Fisher Scientific, Bremen, Germany) coupled to an Eksigent nanoLC-2D HPLC system (Eksigent, Dublin, CA). The digested peptide mixture was loaded onto a 250 nl OPTI-PAK trap (Optimize Technologies, Oregon City, OR) custom packed with Michrom Magic C8 solid phase (Michrom Bioresources, Auburn, CA). Chromatography was performed using 0.2% formic acid in both the A solvent (98%water/2%acetonitrile) and B solvent (80% acetonitrile/10% isopropanol/10% water), and 5%B to 50%B gradient over 60 min at 325 nl/min through a hand packed PicoFrit (New Objective, Woburn, MA) 75 μm × 200 mm column (Michrom Magic C18 3μm). The LTQ Orbitrap mass spectrometer experiment was set to perform a Fourier Transform (FT) full scan from 375–1600 m/z with resolution set at 60,000 (at 400m/z), followed by linear ion trap MS/MS scans on the top five ions. Dynamic exclusion was set to 1 and selected ions were placed on an exclusion list for 15 sec. The lock-mass option was enabled for the FT full scans using the ambient air polydimethylcyclosiloxane (PCM) ion of m/z = 445.120024 or a common phthalate ion m/z = 391.284286 for real time internal calibration [19].
Database Searching
Tandem mass spectra were extracted by BioWorks version 3.2. All MS/MS samples were analyzed using Mascot (Matrix Science, London, UK; version 2.2.04) set up to search the Swiss-Prot database (699052 entries) assuming full trypsin digestion with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 10.0 PPM. Oxidation of methionine, iodoacetamide derivative of cysteine, and phosphoserine, phosphothreonine, and phosphotyrosine were specified as variable modifications. Any phosphopeptide identifications were manually validated. For MYL2, post tryptic digestion “AGGANSNVFSMFEQTQIQEFK” peptide was the only phosphopeptide match found from the data base search. Related phosphopeptides were likewise identified for RLC in porcine ventricular myosin and chicken gizzard myosin. No trypsin fragment of phosphorylated skeletal RLC was identified.
RESULTS
In vitro smMLCK phosphorylation of MYL2
Phosphorylated and unphosphorylated MYL2 were separated on a urea-PAGE gel where mobility varies with charge. Figures 1B and 1A compares the same gel area successively treated with Pro-Q Diamond phosphoprotein and Sypro-Ruby total protein stains. Lane 1 contains unphosphorylated MYL2 while lane 2 contains phosphorylated MYL2. Phosphorylated MYL2 has the added negative charge of the acidic phosphate group attached to the serine and migrates faster than the nonphosphorylated form (Figure 1A). Pro-Q allows direct, in-gel detection of phosphate groups attached to protein thereby confirming that the MYL2 protein was phosphorylated (Figure 1B).
Figure 1.
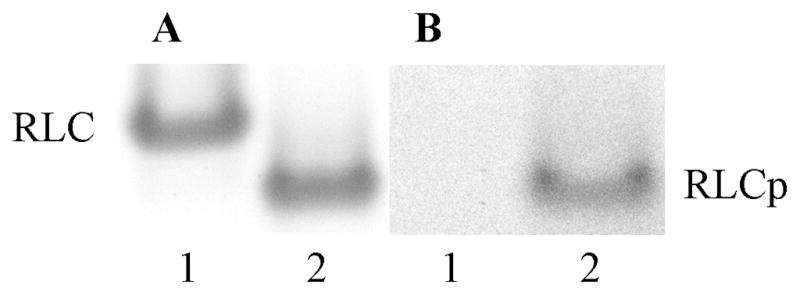
Phosphoprotein and total protein detected in unphosphorylated (RLC, lane 1) and phosphorylated (RLCp, lane 2) MYL2. Images are made from the same 10% polyacrylamide gel successively treated with Pro-Q Diamond phosphoprotein and Sypro-Ruby total protein stains. Panel A: SYPRO Ruby stain for total protein detection. Panel B: Pro-Q Diamond stain for phosphoprotein detection.
Specificity of MYL2 phosphorylation by smMLCK determined with mass spectrometry
MYL2 was phosphorylated by smMLCK in vitro. Trypsin digestion and nanoLC-tandem mass spectrometry with an Orbitrap was used for phosphopeptide analysis. The only phosphopeptide detected was a singly phosphorylated version of the sequence AGGANS15NVFS19MFEQTQIQEFK containing an oxidized methionine. This peptide contains two serines and a threonine that are at positions S15, S19, and T24 in the MYL2. Supplementary Figure S1, shows the corresponding [M+2H]+2 precursor ion of this peptide from the Orbitrap full scan with a delta mass within 1 ppm of the theoretical (theoretical mass is molecular mass divided by charge Z=2). Supplementary Figure S2, shows the ion trap collision induced dissociation (CID) MS/MS spectrum illustrating detection of the Y-ion series. In a phosphorylated species, CID causes loss of H3PO4 (including O from the host residue) resulting in a 98 Da mass reduction. This was not observed in S19 or T24. The y16 ion mass corresponding to phosphorylation at S15 minus the 98 Da from loss of H3PO4, 1961 Da, is present confirming phosphorylation of S15. Supplementary Table S2, shows the theoretical y-ion series ions with the phosphorylation at three possible sites for this sequence. The S19 or T24 positions are not phosphorylated, as there are no corresponding y or y-H3PO4 ions present in the CID MS/MS spectra.
Phosphorylation kinetics of MYL2, porcine ventricular myosin, chicken gizzard HMM, and rabbit skeletal muscle myosin
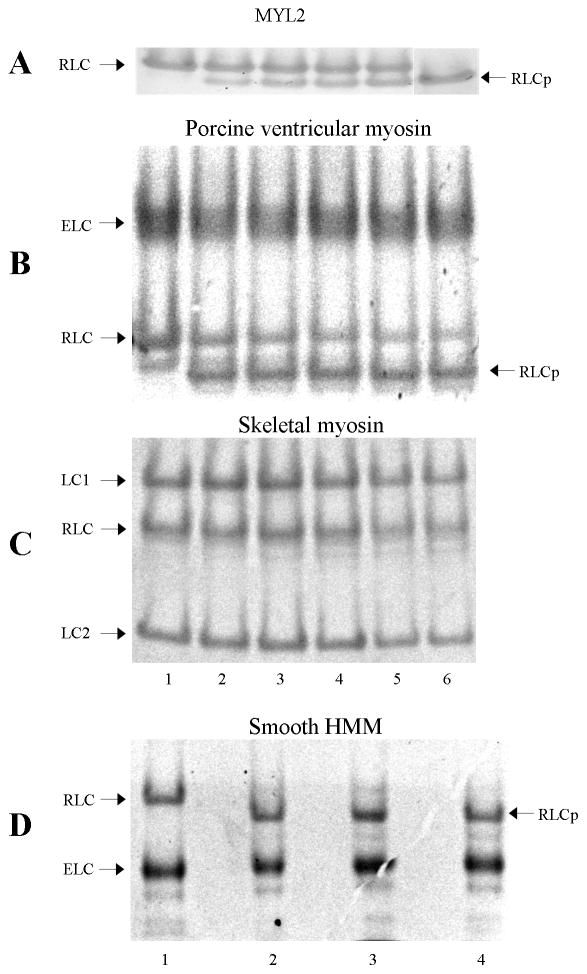
Figure 2 shows the time course for smMLCK phosphorylation of MYL2 (Panel A), porcine ventricular myosin (Panel B), rabbit skeletal myosin (Panel C), and chicken gizzard HMM (Panel D) on a urea-PAGE gel where mobility varies with charge. Mass spectrometry analysis on several of the gel bands shown in Figure 2 was identical to that performed on MYL2 as described in the previous section. The mass spectrometry indicated specific phosphorylation of S15 (S20 for smooth muscle) in the myosins tested except for skeletal myosin where no phosphorylation was detected. Mass spectrometry also indicated that the gel band appearing below the main RLC band in porcine ventricular myosin (Panel B) at the zero time point is an unphosphorylated RLC with altered charge due to methylation or other post translational modification.
Figure 2.
Time course for smMLCK phosphorylation of RLC from various myosin isoforms. Unphosphorylated (RLC) and phosphorylated (RLCp) forms of the regulatory light chain are indicated. ELC, LC1, and LC2 are the essential light chains. Panel A: MYL2. Time points shown for lanes 1–6 are 0, 0.25, 0.5, 0.75, 1, and 10 min. Time points 1. and 10 minutes are separated on the gel by time points 2, 2.5, 3, 4, 5 min (not shown). Panel B: Porcine ventricular myosin. Time points shown for lanes 1–6 are 0, 10, 20, 40, 60, and 120 min. Panel C: Rabbit skeletal myosin. Time points identical to Panel B. Panel D: Chicken gizzard (smooth muscle) HMM. Time points shown for lanes 1–4 are 0, 5, 15, and 25 min. Running conditions are 10% polyacrylamide and 8M urea. Panels B–D show only the section of the gel with myosin ELC and RLC. The myosin heavy chain does not enter into the 10% polyacrylamide-urea gel. Phosphorylation rates are roughly similar for all samples tested except for skeletal myosin where smMLCK is unable to phosphorylate the RLC (see Table 1).
Panel B containing porcine ventricular myosin shows a residual slowly phosphorylated component of ~38% of the total RLC content (comparing RLCp to the sum of RLC and RLCp band intensities). MYL2 and smooth muscle HMM do not have a slowly phosphorylated component. The slowly phosphorylated RLC component apparently contains a less accessible S15 to MLCK. In porcine ventricular myosin, synthetic filament formation probably sterically inhibits MLCK from the vicinity of some of the S15’s. Overnight (24 hr) RLC phosphorylation at 4 °C eliminates the unphosphorylated component. We assume the 0–30 min time points correspond to 100% phosphorylation of S15 accessible RLC when fitting the time dependent curves as described below.
Figure 3 shows the MYL2 phosphorylation time course for three substrate concentrations [MYL2]0 = 55, 25, and 12.5 μM. Time point used in the analysis are: t = 0.25, 0.5, 0.75, 1, 1.5, 2, 2.5, 3, and 10 min after mixing smMLCK and MYL2. At t = 0.25 min an anomalously large phosphorylated RLC component was detected probably due to the finite time needed to stop the reaction (detection dead-time). We renormalized data by taking the t = 0.25 min point as the reaction initiation and subtracting the phosphorylated band density at t = 0.25 min from all phosphorylated band densities. We adjusted [MYL2]0 to 36, 16, and 8 μM to remove the substrate already converted to product (RLCp) at t = 0.25 min. The Scheme 1 model generates the best fitting curves shown giving KM = 1.5 μM, k+2 = 5.8 min−1, and Vmax = k+2[MLCK]0 = 7.0 μM/min (Table 1).
Figure 3.
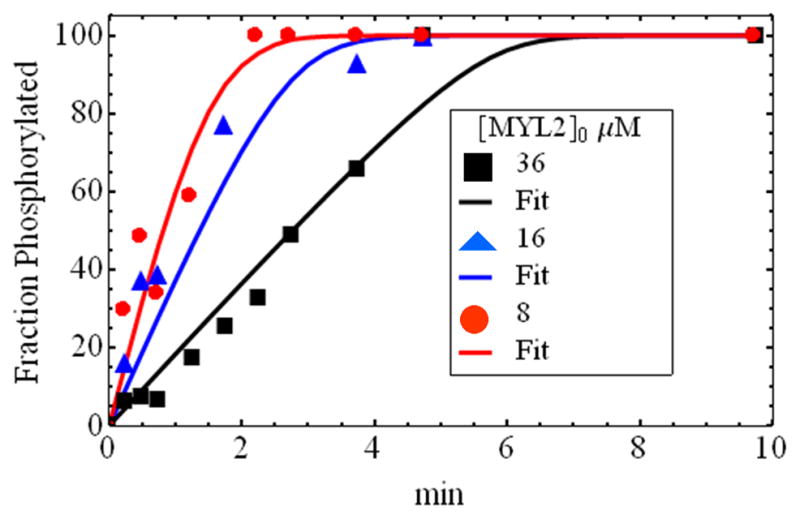
Time course for MYL2 phosphorylation by smMLCK at 3 MYL2 initial concentrations. Fitted curves were derived using Scheme 1. Initial concentrations for MYL2 given in the legend are after renormalization to account for detection dead-time. Original MYL2 concentrations are 12.5, 25, and 55 μM that correspond to 8, 16, and 36 μM, respectively, after renormalization. Kinetic constants are summarized in Table 1.
Table 1.
Michaelis-Menten kinetic constants for MLCK and RLC1
| k+2 (min−1) | KM (μM) | Vmax (μM/min) | |
|---|---|---|---|
| smMLCK(hu)/MYL22 | 5.8 ± 0.8 | 1.5± 1 | 7.0± 1 |
| smMLCK(hu)/smRLC(ch)3 | 2.5 | 5.2 | 3.0 |
| smMLCK(hu)/cRLC(pig)4 | 1.5 | 1.5 | 1.2 |
| cMLCK(hu)/ MYL25 | 0.041 | 4.3 | 0.05 |
Abbreviations used: human (hu), chicken gizzard (ch), smooth muscle (sm), and cardiac (c).
Calculated from data in Figure 3. Standard errors are indicated.
Consistent with k+2 = 2.7 min−1 and KM = 5.2 μM measured in [20].
Consistent with data in Figure 3.
From [27] and for our MLCK concentration of 1.2 μM.
We also measured RLC phosphorylation time course for smooth muscle HMM and porcine ventricular myosin at single substrate concentrations. Smooth muscle myosin data (Figure 2 Panel D) is consistent with k+2 = 2.5 min—1, KM = 5.2 μM, and Vmax = 3.0 μM/min in agreement with k+2 = 2.7 min−1 and KM = 5.2 μM measured previously for this system [20]. Porcine ventricular myosin data (Figure 2 Panel B) is consistent with k+2 = 1.0 min−1, KM = 1.5 μM, and Vmax = 1.2 μM/min. Hence the myosin bound porcine RLC is somewhat less active than MYL2 probably because the porcine substrate is bound to the myosin lever arm and aggregated into filaments. These comparisons show that smMLCK is a specific and efficient kinase of both smooth muscle and cardiac myosin light chains.
DISCUSSION
MLCK is a family of Ca2+/calmodulin dependent protein kinases present in smooth [21], skeletal [22], and cardiac muscle [10], and nonmuscle cells [23]. In smooth muscle, MLCK-mediated phosphorylation of RLC is responsible for regulating contraction; in skeletal and cardiac muscle, its role is more subtle and less understood but likely affecting the Ca2+ sensitivity of contraction in addition to the Ca2+ dependent regulation exerted by the actin filament regulatory proteins [5; 24]. In the present work we have used the 130 kDa smMLCK.
Reports of the identity and amount of the RLC kinases present in the heart are variable although smMLCK has been consistently detected in the heart. Tissue distribution of sk-, c-, and smMLCK expression showed specificity for the sk- and c-MLCK isoforms but ubiquitous expression (including the heart) of smMLCK [9; 25]. Immunoblotting using a specific antibody against chicken gizzard MLCK and indirect immunofluorescent microscopy found the 130 kDa smMLCK in various chicken muscle and nonmuscle tissues including the I-band region of isolated cardiac and skeletal myofibrils [26].
It was proposed that the cardiac RLC was not effectively phosphorylated by smMLCK and that the R17 residue preceding S20 in smooth muscle RLC (G12 and S15 in MYL2) was a key determinant for smMLCK specificity [8]. Using mass spectrometry we demonstrated conclusively that smMLCK specifically phosphorylates S15 in MYL2, porcine cardiac myosin, and gizzard myosin (MYL2 mass spectrometry data shown in Supplementary Figures S1 and S2, and Supplementary Table S3). Phosphorylation kinetics, measured using a novel fluorescence method eliminating radioactive isotopes, indicates smMLCK phosphorylates MYL2 at a rate exceeding even smooth muscle RLC (Table 1). In cardiac RLC, R17 is replaced by glycine conserving neither the side chain size nor charge, nevertheless, smMLCK is similarly effective at phosphorylating smooth muscle or cardiac RLC. Sequence comparison suggests that the S15 proximity to A13 may promote smMLCK specificity, catalytic efficiency, or both for cardiac and smooth muscle RLC.
Others have suggested that smMLCK could serve as a backup kinase in the heart mitigating age or disease related decline in cMLCK expression [27]. Our data is consistent with this role for smMLCK since we showed it efficiently phosphorylates MYL2 and because the lower expression level for smMLCK compared to cMLCK could be compensated by smMLCK’s more favorable kinetics [28]. It seems likely that smMLCK participates in cardiac muscle regulation or in cardiac muscle response to disease causing stimulus.
Supplementary Material
HIGHLIGHTS.
Cardiac myosin regulatory light chain (MYL2) is phosphorylated at S15
Smooth muscle myosin light chain kinase (smMLCK) is a ubiquitous kinase
It is a widely believed that MYL2 is a poor substrate for smMLCK
In fact, smMLCK efficiently and rapidly phosphorylates S15 in MYL2
Phosphorylation kinetics measured by novel fluorescence method without radioactivity
Acknowledgments
We thank Ben Madden and Dan McCormick from the Mayo Proteomics Research Center for the mass spectrometry, Ozgur Ogut for valuable advice, and Miriam F. Halstead for technical assistance. Bovine brain calmodulin was a generous gift from Whyte Owen, Mayo Clinic Rochester. This work was supported by NIH grants R01AR049277 and R01HL095572 and by the Mayo Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Greenberg MJ, Watt JD, Jones M, Kazmierczak K, Szczesna-Cordary D, Moore JR. Regulatory light chain mutations associated with cardiomyopathy affect myosin mechanics and kinetics. Journal of Molecular and Cellular Cardiology. 2009;46:108–115. doi: 10.1016/j.yjmcc.2008.09.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olsson MC, Patel JR, Fitzsimons DP, Walker JW, Moss RL. Basal myosin light chain phosphorylation is a determinant of Ca2+ sensitivity of force and activation dependence of the kinetics of myocardial force development. Am J Physiol Heart Circ Physiol. 2004;287:H2712–H2718. doi: 10.1152/ajpheart.01067.2003. [DOI] [PubMed] [Google Scholar]
- 3.Szczesna-Cordary D. Regulatory Light Chains of Striated Muscle Myosin. Structure, Function and Malfunction Current Drug Targets-Cardiovascular & Haematological Disorders. 2003;3:187–197. doi: 10.2174/1568006033481474. [DOI] [PubMed] [Google Scholar]
- 4.Herring BP, Dixon S, Gallagher PJ. Smooth muscle myosin light chain kinase expression in cardiac and skeletal muscle. American Journal of Physiology - Cell Physiology. 2000;279:C1656–C1664. doi: 10.1152/ajpcell.2000.279.5.C1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stull JT, Nunnally MH, Moore RL, Blumenthal DK. Myosin light chain kinases and myosin phosphorylation in skeletal muscle. Adv Enzyme Regulation. 1985;23:123–140. doi: 10.1016/0065-2571(85)90043-3. [DOI] [PubMed] [Google Scholar]
- 6.Blue EK, Goeckeler ZM, Jin Y, Hou L, Dixon SA, Herring BP, Wysolmerski RB, Gallagher PJ. 220- and 130-kDa MLCKs have distinct tissue distributions and intracellular localization patterns. American Journal of Physiology - Cell Physiology. 2002;282:C451–C460. doi: 10.1152/ajpcell.00333.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis JS, Hassanzadeh S, Winitsky S, Lin H, Satorius C, Vemuri R, Aletras AH, Wen H, Epstein ND. The overall pattern of cardiac contraction depends on a spatial gradient of myosin regulatory light chain phosphorylation. Cell. 2001;107:631–641. doi: 10.1016/s0092-8674(01)00586-4. [DOI] [PubMed] [Google Scholar]
- 8.Ding P, Huang J, Battiprolu PK, Hill JA, Kamin KE, Stull JT. Cardiac myosin light chain kinase is necessary for myosin regulatory light chain phosphorylation and cardiac performance in vivo. J Biol Chem. 2010;285:40819–40829. doi: 10.1074/jbc.M110.160499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhi G, Ryder JW, Huang J, Ding P, Chen Y, Zhao Y, Kamm KE, Stull JT. Myosin light chain kinase and myosin phosphorylation effect frequency-dependent potentiation of skeletal muscle contraction. Proc Natl Acad Sci USA. 2005;102:17519–17524. doi: 10.1073/pnas.0506846102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudnakova TV, Stepanova OV, Dergilev KV, Chadin AV, Shekhonin BV, Watterson DM, Shirinsky VP. Myosin light chain kinase colocalizes with nonmuscle myosin IIB in myofibril precursors and sarcomeric Z-lines of cardiomyocytes. Cell Motil Cytoskeleton. 2006;63:375–383. doi: 10.1002/cm.20127. [DOI] [PubMed] [Google Scholar]
- 11.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 12.Ajtai K, Halstead MF, Nyitrai M, Penheiter AR, Zheng Y, Burghardt TP. The Myosin C-Loop Is an Allosteric Actin Contact Sensor in Actomyosin. Biochemistry. 2009;48:5263–5275. doi: 10.1021/bi900584q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kojima S, Fujiwara K, Onishi H. SH1 (Cysteine 717) of smooth muscle myosin: its role in motor function. Biochemistry. 1999;38:11670–11676. doi: 10.1021/bi990081f. [DOI] [PubMed] [Google Scholar]
- 14.Ajtai K, Garamszegi SP, Park S, Velazquez Dones AL, Burghardt TP. Structural characterization of β-cardiac myosin subfragment 1 in solution. Biochemistry. 2001;40:12078–12093. doi: 10.1021/bi0112098. [DOI] [PubMed] [Google Scholar]
- 15.Tonomura Y, Appel P, Morales M. On the molecular weight of myosin II. Biochemistry. 1966;5:515–521. doi: 10.1021/bi00866a017. [DOI] [PubMed] [Google Scholar]
- 16.Facemyer KC, Cremo CR. A new method to specifically label thiophosphorylatable proteins with extrinsic probes. labeling of Serine-19 of the regulatory light chain of smooth muscle myosin. Bioconjugate Chem. 1992;3:408–413. doi: 10.1021/bc00017a009. [DOI] [PubMed] [Google Scholar]
- 17.Han YS, Ogut O. Regulation of Fibre Contraction in a Rat Model of Myocardial Ischemia. Plos One. 2010;5:e9528. doi: 10.1371/journal.pone.0009528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: A method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999;20:601–605. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 19.Olsen JV, de Godoy LMF, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Parts per Million Mass Accuracy on an Orbitrap Mass Spectrometer via Lock Mass Injection into a C-trap. Molecular & Cellular Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Birukov GK, Csortos C, Marzilli L, Dudek S, Ma SF, Bresnick AR, Verin AD, Cotter RJ, Garcia JGN. Differential Regulation of Alternatively Spliced Endothelial Cell Myosin Light Chain Kinase Isoforms by p60Src. J Biol Chem. 2001;276:8567–8573. doi: 10.1074/jbc.M005270200. [DOI] [PubMed] [Google Scholar]
- 21.Hong F, Halderman BD, Jackson D, Carter M, Baker JE, Cremo CR. Biochemistry of smooth muscle myosin light chain kinase. Arch Biochem Biophys. 2011;510:135–146. doi: 10.1016/j.abb.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pires EMV, Perry V. Purification and Properties of Myosin Light-Chain Kinase from Fast Skeletal Muscle Biochemical. Journal. 1977;167:137–146. doi: 10.1042/bj1670137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hathaway DR, Adelstein RS. Human platelet myosin light chain kinase requires the calcium-binding protein calmodulin for activity. Proc Natl Acad Sci USA. 1979;76:1653–1657. doi: 10.1073/pnas.76.4.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh MP, Vallet B, Autric F, Demaille JG. Purification and characterization of bovine cardiac calmodulin-dependent myosin light chain kinase. J Biol Chem. 1979;254:12136–12144. [PubMed] [Google Scholar]
- 25.Takashima S. Phosphorylation of Myosin Regulatory Light Chain by Myosin Light Chain Kinase, and Muscle Contraction. Circulation Journal. 2009;73:208–213. doi: 10.1253/circj.cj-08-1041. [DOI] [PubMed] [Google Scholar]
- 26.Conti MA, Adelstein RS. Nonmuscle myosin II moves in new directions. J Cell Sci. 2008;121:11–18. doi: 10.1242/jcs.007112. [DOI] [PubMed] [Google Scholar]
- 27.Chan JY, Takeda M, Briggs LE, Graham ML, Lu JT, Horikoshi N, Weinberg EO, Aoki H, Sato N, Chien KR, Kasahara H. Identification of Cardiac-Specific Myosin Light Chain Kinase. Circ Res. 2008;102:571–580. doi: 10.1161/CIRCRESAHA.107.161687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seguchi O, Takashima S, Yamazaki S, Asakura M, Asano Y, Shintani Y, Wakeno M, Minamino T, Kondo H, Furukawa H, Nakamura K, Naito A, Takahashi T, Ohtsuka T, Kawakami K, Isomura T, Kitamura S, Tomoike H, Mochizuki N, Kitakaze M. A cardiac myosin light chain kinase regulates sarcomere assembly in the vertebrate heart. J Clin Invest. 2007;117:2812–2824. doi: 10.1172/JCI30804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.