Abstract
Experimental work using human cancer cell lines often does not translate to the clinic. We posit this is because some cells undergo changes in vitro that no longer make them representative of human tumors. Here we describe a novel alignment method named SRCCM (Spearman’s Rank Correlation Classification Method) that measures similarity between cancer cell lines and human tumors via gene expression profiles, for the purpose of selecting lines that are biologically relevant. To demonstrate utility, we used SRCCM to assess similarity of 36 bladder cancer lines with 10 epithelial human tumor types (N=1630 samples) and with bladder tumors of different stages and grades (N=144 samples). While 34/36 lines aligned to bladder tumors rather than other histologies, only 16/28 had SRCCM assigned grades identical to that of their original source tumors. To evaluate the clinical relevance of this approach, we demonstrate that gene expression profiles of aligned cell lines stratify survival in an independent cohort of 87 bladder patients (HR = 3.41, logrank p = 0.0077), while unaligned cell lines using original tumor grades did not. We repeated this process on 22 colorectal cell lines and found that gene expression profiles of 17 lines aligning to colorectal tumors and selected based on their similarity with 55 human tumors stratified survival in an independent cohort of 177 colorectal cancer patients (HR = 2.35, logrank p = 0.0019). By selecting cell lines that reflect human tumors, our technique promises to improve the clinical translation of laboratory investigations in cancer.
Keywords: cell lines, tissue of origin, tumor grade, survival analysis, DNA microarray
INTRODUCTION
Cell lines derived from human tumors serve as model systems that have greatly increased our understanding of cancer biology. These are routinely used to characterize molecular mechanisms of disease (1–3) and therapeutic agents (4–6). High throughput cell line screening programs have been used to characterize the efficacy of anti-cancer agents (7) and to identify novel multi-agent therapies (8). Nevertheless, the validity of a cell line model depends on how representative it is of the tumor type under investigation. Notably, cell lines can lose molecular features that drive clinically relevant characteristics as they adapt to culture conditions (9). An analysis of NCI-60 cell lines found that the proportion of cell lines most similar to their presumed tissue of origin could be as low as 57%, and that the molecular profiles of patient tumors are more similar to normal tissues of the same type than their derived cell lines (10). Cross-contamination of cell lines is well-documented and also contributes to reduce the number of relevant cancer cell line models for research (11–13).
Stage and grade are cornerstone prognostic and predictive factors of tumor aggressiveness and disease outcome. In the case of bladder cancer, high grade cancers are more likely to metastasize than low grade ones and up to 30% of non-muscle invasive tumors progress to muscle invasion (14). Metastatic risk correlates with stage, with up to 50% of patients harboring muscle-invasive cancers developing metastatic disease during follow-up (15). Furthermore, tumor sensitivity to anti-cancer agents depends on stage and grade, with resistance more common in poorly differentiated tumors (16, 17). Lastly, the evaluation of chemotherapy and radiation treatments often occurs in subsets of cancer patients having tumors of a specific stage or grade (18–20). For all of these reasons, the choice of an appropriate cell line model that aligns with the specific human tumor characteristics being investigated is critical if results from cell line experiments are to have clinical relevance.
Here we develop a novel framework for classifying or aligning human cancer cell lines as a function of their similarity to several important phenotypes of human tumors via gene expression profiles. We then use the gene expression profiles of these aligned cancer cells as biomarkers, to determine which cells predict clinical outcomes in human cancer patients (i.e. are clinically relevant). As a proof of principle, we used bladder and colorectal cancer cell line panels and patient tumor cohorts to demonstrate that gene expression signatures of cancer cell lines selected on the basis of their alignments to human cancer phenotypes predict patient survival outcomes while the signatures of unaligned cells do not. Our practical approach can guide the selection of “clinically relevant” cell lines for use in experimental studies which may enhance the likelihood of clinical translation of laboratory investigations. Since this methodology is applicable to any cancer cell line and measurable phenotype (e.g. grade, invasiveness and metastasis), it has broad applicability in cancer research.
MATERIALS AND METHODS
The bladder and colorectal cell line panels
Gene expression profiles (CEL files) for 36 bladder cancer cell lines (BLA-36) are available from the Gene Expression Omnibus (GEO) (21), accession #GSE5845, Ref #22. The database contains expression profiles for 40 cell lines, including several cell lines of the same lineage. In our analysis, we chose to analyze unique cell lines or related cell lines that were derived in separate laboratories. As a result, we removed the cell lines FL3, SLT4, T24T (all derived from T24) and 253J-BV (derived from 253J) prior to our analysis. Information about the BLA-36 cell lines along with our alignments are provided in Supplementary Table S1. Gene expression profiles (CEL files) for 22 colorectal cell lines (CO-22) profiled in triplicate from GlaxoSmithKline are available through the National Cancer Institute’s cancer Bioinformatics Grid™ (caBIG), Ref #23.
The Spearman’s Rank Correlation Classification Method (SRCCM)
The SRCCM algorithm classifies a test sample based on the Spearman’s rank correlation between its gene expression profile and the gene expression profile of a set of training samples with known phenotypes (Figure 1A). The gene expression profile is based on a gene signature that is unique to each phenotype (Figure 1B–C). The tissue of origin gene signature consists of 12,402 “high fidelity” probes whose expression values are consistent across formalin fixed, paraffin embedded and fresh frozen preservation methods (24). The bladder stage and grade gene signatures have been previously published and independently validated (25). The bladder disease-specific survival (DSS) gene signature consists of the 181 probes with univariate Cox proportional hazard model logrank p-values < 0.01 in the training cohort from Memorial Sloan-Kettering Cancer Center (MSKCC, Ref # 26). The colorectal DSS gene signature is a previously published and validated signature consisting of 34 genes that correlate with recurrence and survival (27).
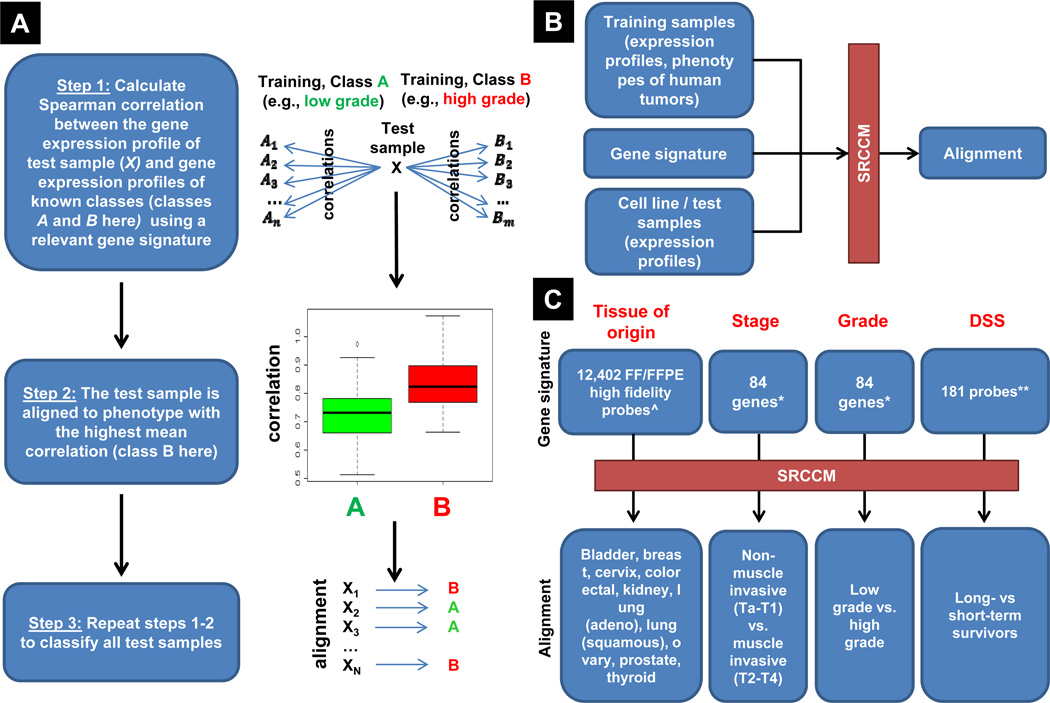
Figure 1.
Overview and use of the Spearman’s Rank Correlation Classification Method (SRCCM). A, overview of the SRCCM algorithm for cell line / test sample alignment. A test sample is aligned to the phenotype with the highest mean correlation between test and training gene expression profiles using a relevant gene signature. B, alignment of a test sample with a clinical phenotype using SRCCM is based on the gene expression profiles of the training and test samples and a relevant gene signature for the desired phenotype. C, published and derived gene signatures and phenotypes used by SRCCM for tissue of origin, and bladder cancer stage, grade, and disease specific survival (DSS) alignment. ^ signatures from Ref #24; * stage and grade signatures from Ref #25 are different, despite the same number of genes; ** univariate Cox proportional hazards model, logrank p-value < 0.01 in training cohort from Ref #26, see Materials and Methods for details.
Abbreviations: FF, fresh frozen; FFPE, formalin fixed and paraffin embedded.
The test sample is aligned to (i.e., classified as or assigned) the phenotype with the highest average correlation. Formally, let P = (P1, …, Pk) be the set of training phenotypes of interest with known gene expression profiles. In Figure 1A, k = 2 and P = (low grade, high grade). Let t be the gene expression profile of the test sample and be the gene expression profile of the ith training sample of phenotype p, i = 1, …, np. Let be the Spearman’s rank correlation between gene expression profiles t and , which is the standard Pearson product moment correlation calculated using the ranks of the data. Finally, let be the average Spearman’s rank correlation between the test sample’s gene expression profile and the gene expression profiles of phenotype p. Then the classification of the test sample is the phenotype p ∈ P that maximizes r̄(p). For purposes of ranking the test samples when k = 2 phenotypes, we use a correlation score = r(P2) − r(P1), where P = (non-muscle invasive, muscle invasive),(low grade, high grade), and (long-term survivor, short-term survivor), for stage, grade, and DSS alignment, respectively. Software for the SRCCM is available to the scientific community through the Comprehensive R Archive Network (28) as part of the correlation classification method (CCM) contributed package.
Bladder stage and grade alignments
The SRCCM algorithm was used along with separate published and validated stage and grade gene signatures (25) to classify tumors as muscle invasive (stages T2–T4) or non-muscle invasive (stages Ta-T1), and as high grade (G3–G4) or low grade (G1–G2). Each gene signature consists of 100 genes, with 54 genes common to both signatures. For each signature, we used the 84 genes common to the training (Lindgren) (25), validation (MSKCC), and BLA-36 datasets, with 46 genes common to both signatures. The two signatures were further validated using the Chungbuk National University Hospital (CNUH) cohort (29) and the Stransky cohort of patients from the Henri Mondor Hospital in France (30) using the genes common to all datasets (72 for stage and 70 for grade). For visualization purposes, the correlation score (described above) was used to rank the cell lines, with higher scores corresponding to muscle-invasive and high grade tumors, respectfully.
Bladder and colorectal disease specific survival alignments
We use DSS in all of our analyses with survival time equal to the time from diagnoses to time of death or last follow-up. All individuals not dead from disease are censored at the time of last follow-up or time of death from another cause. For DSS alignment, the “median cut” labeling method was used to identify long-term survivors (i.e., low risk individuals) and short-term survivors (i.e., high risk individuals) (31). Patients without censored survival times are assigned to the long-term survivor group if the survival time exceeds the median survival time of all patients in the training cohort, and assigned to the short-term survivor group otherwise. For a patient with an unobserved survival time of T that is censored at C, we calculate
where P(T > Median survival time) and P(T > C)are estimated using the Kaplan-Meier survival curve for all patients (32). The patient is then assigned to the long-term survivor group if P(long-term survivor) > 0.5, and assigned to the short-term survivor group otherwise.
RESULTS
A new method that aligns cell lines to human tumors based on gene expression similarity
We developed a new classification method, herein referred to as the Spearman’s Rank Correlation Classification Method (SRCCM) and first applied it to 36 bladder cancer cell lines (BLA-36) (Supplementary Table S1). These lines were assigned to the following tumor phenotypes: tissue of origin (from 10 epithelial cancers), bladder cancer stage (invasive vs. non-muscle invasive), grade (low vs. high), and disease specific survival (DSS, long-term vs. short-term survivors). SRCCM (Figure 1A) aligns cell lines to human tumor phenotypes based on the gene expression similarity between cell lines and human tumors calculated using gene signatures, described below and in Materials and Methods, that consists of a subset of all genes profiled in both groups (cell lines and human tumors) (Figure 1B–C). The SRCCM consists of three steps: 1) for each test sample, calculate the rank correlation between its gene expression profile and the gene expression profiles of each training sample with known phenotype (e.g., grade), 2) assign the phenotype with the highest mean correlation to the test sample, producing the alignment, 3) repeat the procedure for all test samples (Figure 1A). Therefore, each test sample (e.g., cell line in a set) is aligned to the human tumor phenotype it is most similar to, based on the similarity between gene expression profiles.
For each tumor phenotype, the SRCCM algorithm is validated on at least one independent test dataset and then applied to the BLA-36 cell lines. For each test sample, the primary output of the SRCCM is its molecular alignment (i.e., a qualitative phenotype) while the secondary output is a quantification of the relative strength of one alignment (e.g., to high grade) to another (e.g., to low grade) by subtracting the corresponding mean correlations, producing a correlation score. For stage, grade, and disease specific survival alignments, a higher correlation score indicates greater similarity with muscle invasive, high grade, and short-term survivors, respectively (see Materials and Methods).
Our SRCCM algorithm is based on the classification method of Wang et al. (33) but is modified in three important ways. The first modification is that we use Spearman's rank correlation rather than Pearson product moment correlation to measure gene expression similarity. Our choice of a rank-based classifier is motivated by the fact that the rank value of a probe’s expression level within a sample is insensitive to standard within-sample preprocessing methods and is not directly affected by other probes or samples on the array, whereas the expression value is highly dependent on the pre-processing and normalization methods used and can be affected by additional probes and samples (e.g., when robust multichip average (RMA) normalization is used). The second modification is that instead of measuring similarity based on a sample’s global gene expression profile (i.e., the expression levels of all common microarray probes) as used previously (33), we measure similarity based on gene signatures appropriate for the microarray data sets under study and the phenotype of interest (Figure 1C). For tissue alignment, because bladder samples (N=350) were both formalin fixed, paraffin embedded (FFPE) and fresh frozen (FF) (Supplementary Table S2), the gene signature used consists of 12,402 probes whose expression values were preserved across FF and FFPE (24). For stage and grade alignment, two previously published and independently validated gene signatures are used (25) (Supplementary Tables S3–S4). For disease specific survival alignment, the gene signature consists of the 181 probes that significantly correlate with survival in the training dataset (MSKCC, Supplementary Table S5) with univariate Cox proportional hazards model logrank p-values < 0.01. The final modification is that for each phenotype, we ensure that the SRCCM molecular alignment is accurate through validation on at least one independent dataset. References for all datasets are provided in Supplementary Tables S1–S5 and datasets are processed as described in Supplementary Materials. The SRCCM algorithm is comparable with common classification methods, including support vector machines, nearest centroid classification, and k-nearest neighbor but performs slightly better in terms of overall accuracy and consistency across multiple datasets (described in Supplemental Information and Supplementary Tables S6–S7).
Molecular alignment of human bladder cell lines as a function of tumor tissue of origin
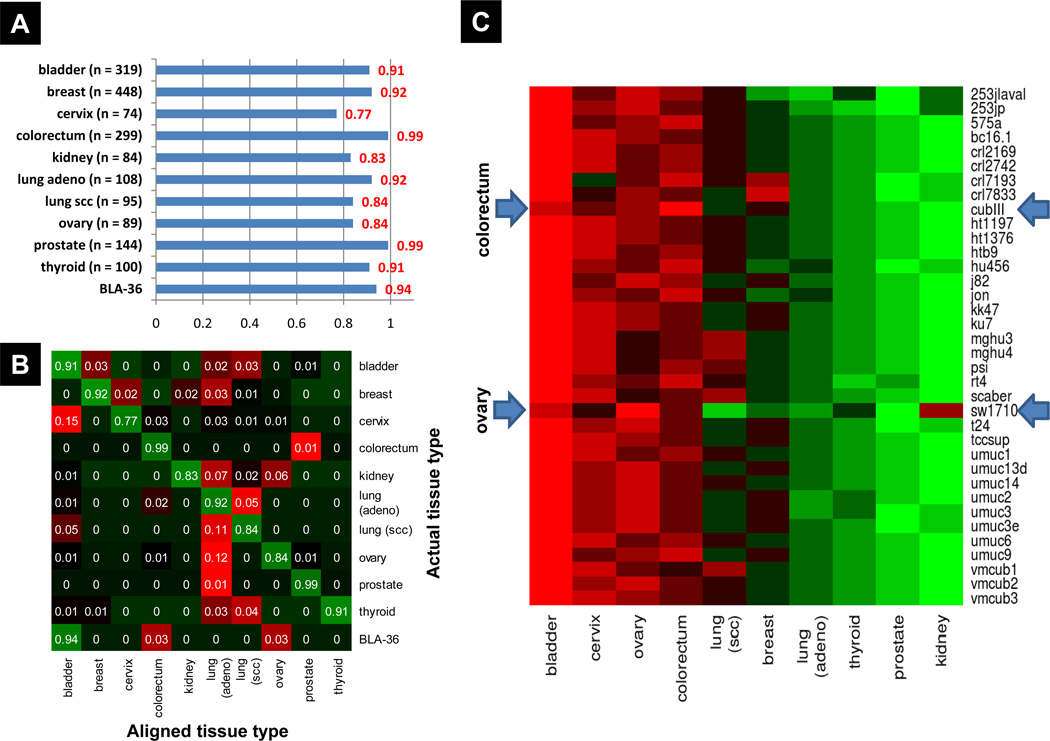
We first determined whether the BLA-36 cell lines had gene expression profiles that were more similar to bladder cancer or other common carcinomas of cervical, prostate, ovarian, breast, thyroid, kidney, colorectal, and lung origin. These tumor types were selected based on the following criteria 1) epithelial histology, 2) availability of at least one public expression microarray dataset containing ≥ 30 tumor samples, and 3) profiled on an Affymetrix platform, the platform used for BLA-36 profiling. Because distinct molecular profiles for lung cancer subtypes have been well characterized (34), we considered lung adenocarcinoma (adeno) and lung squamous cell carcinoma (scc) separately. A total of 3320 tumor samples were used in this analysis with 1630 used in training and 1690 in validation of the SRCCM algorithm (Supplementary Table S2). When applied on the 1690 validation samples from 10 tumor types, the SRCCM algorithm had a mean accuracy of 89%, with tumor-specific accuracy ranging from 77% for cervical tumors to 99% for both colorectal and prostate tumors (Figure 2A). The overall accuracy for bladder cancer was 91% (Figure 2A). Interestingly, the majority of misaligned samples are aligned with lung (adeno), including 12% of all ovary samples and 11% of all lung (scc) samples (Figure 2B). The majority of misaligned cervical samples are aligned with bladder, which comprise 15% of all cervical samples. Importantly, when we applied the SRCCM to the BLA-36 panel, 34 of 36 (94%) cells align with tumors of bladder origin. Of the two not predicted to be of bladder tumor origin, one (CubIII) is predicted to be colorectal and the other (SW1710) ovarian (Figure 2C).
Figure 2.
Independent validation and BLA-36 cell line alignment to tissue of origin using the SRCCM algorithm. Test samples and BLA-36 cell lines are aligned to 10 epithelial cancers from a training dataset including bladder, breast, cervix, colorectum, kidney, lung adenocarcinoma (adeno), lung squamous cell carcinoma (scc), ovary, prostate, and thyroid samples (N = 1630). A, tissue specific accuracy of SRCCM alignment algorithm on independent datasets (N= 1690). B, confusion matrix for independent validation presented as a heatmap, with green indicating correct alignment and red indicating incorrect alignment. For each tissue type (i.e., row of the matrix), the proportion of samples aligned with each tissue type is reported. C, BLA-36 tissue of origin alignment heatmap. For each cell line, the color represents the average correlation with each tissue type, with red indicating strong positive correlation and green indicating weak positive correlation. All cell lines are most strongly correlated (i.e., aligned) with bladder, with the exception of CubIII and SW1710 (arrows) which are aligned with colorectum and ovary, respectively.
Molecular alignment of human bladder cell lines as a function of tumor stage and grade
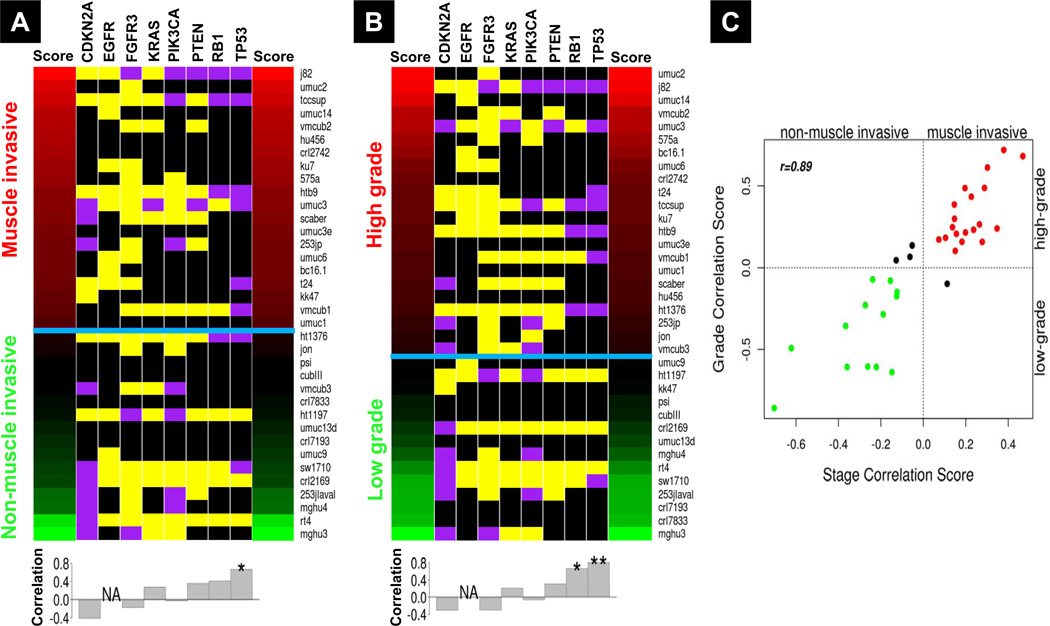
We next used the SRCCM to align the 36 bladder cancer cell lines as a function of human bladder tumor stage and grade, using previously published and validated stage and grade gene signatures (25) (Figure 1C). Stage alignment is to non-muscle or muscle invasive (Ta-T1 or T2–T4) tumors and grade alignment is to high or low grade (G3–G4 or G1–G2) tumors. For both stage and grade alignments, we use the Lindgren cohort (25) for training and leave-one-out cross-validation (LOOCV), and use three additional datasets for independent validation: patient profiles from the Memorial Sloan-Kettering Cancer Center (MSKCC) (26), the Chungbuk National University Hospital (CNUH) (29), and the Stransky cohort of patients from the Henri Mondor Hospital in France (30) (Tables S3–S4). The SRCCM performs well in LOOCV and independent validation for stage and grade (Table 1). The high area under the curve (AUC) in LOOCV (0.93~0.94) and independent validation (0.81~0.92) indicates the SRCCM algorithm accurately separates stage and grade subgroups in tumor samples. When the SRCCM was applied to the 36 cell lines, 20 aligned with muscle-invasive and 16 with non-muscle-invasive tumors (Figure 3A). For grade, 22 and 14 aligned with high-grade and low-grade tumors, respectively (Figure 3B). The rankings of the cell lines in stage and grade alignment are highly correlated (Spearman r = 0.89, p < 0.0001), with 32/36 cell lines resembling the muscle-invasive high-grade and non-muscle-invasive low-grade tumor types (Figure 3C) commonly seen in patients (25, 35, 36).
Table 1.
Performance of stage and grade classification using the SRCCM alignment algorithm
| Stage | Accuracy | AUC | ||
|---|---|---|---|---|
| Ta-T1 | T2–T4 | Overall | ||
| LOOCV in Lindgren* | 0.722 | 0.933 | 0.789 | 0.930 |
| Independent Validation in MSKCC† | 0.880 | 0.818 | 0.835 | 0.895 |
| Independent Validation in CNUH‡ | 0.702 | 0.721 | 0.709 | 0.813 |
| Independent Validation in Stransky§ | 0.919 | 0.750 | 0.817 | 0.920 |
| Grade | Accuracy | AUC | ||
| Low grade | High grade | Overall | ||
| LOOCV in Lindgren* | 0.861 | 0.875 | 0.868 | 0.942 |
| Independent Validation in MSKCC† | 0.722 | 0.836 | 0.813 | 0.814 |
| Independent Validation in CNUH‡ | 0.724 | 0.867 | 0.776 | 0.871 |
| Independent Validation in Stransky§ | 0.790 | 0.849 | 0.824 | 0.877 |
Abbreviation: LOOCV, leave-one-out cross validation; AUC, area under curve.
Profiled on two specialized platforms, Swegene Human 27K RAP UniGene 188 and SWEGENE H_v3.0.1. 35K arrays; available from the Gene Expression Omnibus (21)
Profiled on the HG-U133A microarray platform, available as supplementary material to publication (24).
Profiled on the Illumina human-6 v2.0 microarray platform, available from Gene Expression Omnibus at Accession #GSE13507
Profiled on the HG-U195 and HG-U195Av2 microarray platforms, available from Array Express (52) at Accession #E-TABM-147
Figure 3.
A, Stage and B, grade alignment heatmaps. Cell lines are ranked by their correlation scores (see Materials and Methods) from high (red) to low (green) and classified as either muscle invasive (MI, T2–T4) or non-muscle invasive (NMI, Ta-T1) for stage and high (G3–G4) or low (G1–G2) grade, separated by a blue horizontal line. The heatmap also contains the documented gene mutations according to the COSMIC database (53), with purple, yellow, and black indicating known mutation, wild-type, and unknown mutation status, respectively. The histograms below the heatmaps show correlations of mutations with each alignment. P-values of Fisher exact tests: *, 0.05≤p<0.1; **, p<0.05. C, Plot of correlation scores for SRCCM assigned stage and grade. Red points denote cell lines aligned with high grade MI tumors, green points denote cell lines aligned with low grade NMI tumors, and all other cell lines are plotted in black.
To determine the value of the stage and grade realignment, we evaluated the correlation between SRCCM assigned stages and grades and the status of several common gene mutations (see Supplementary Materials) known to be associated with biological phenotypes in human bladder cancer (Figure 3A,B). For assigned cell line stages, we observed positive correlations with TP53 (r = 0.671), RB1 (r = 0.408), PTEN (r = 0.354), and KRAS (r = 0.277) mutants, and negative correlations with CDKN2A (r = −0.408) and FGFR3 (r = −0.181) mutants, consistent with the role of the first three in aggressive disease and the association of the latter two with a non-muscle invasive phenotype (37–39). Notably, the correlation with TP53 reaches a marginal significance (Fisher’s exact p = 0.06), despite the fact that TP53 mutation status is known for just 11 cell lines. For assigned cell line grades, we observe positive correlations with TP53 (r = 0.810, Fisher’s exact p = 0.024), RB1 (r = 0.667, Fisher’s exact p = 0.076), PTEN (r = 0.304), and KRAS (r = 0.207) mutants, and negative correlations with CDKN2A (r = −0.310) and FGFR3 (r = −0.305) mutants.
Cell lines assigned their original tumor grades do not correlate with survival but correlation is restored using molecularly aligned cell line grades
We collected stage and grade information for the tumor samples used to derive the BLA-36 lines (Supplementary Table S1) and compared the original tumor information with the SRCCM assigned grades described above. Only 8 cell lines had stage information from their original tumors, while 28 cell lines had grade information so only the latter was used in the analysis below. Of the 9 cell lines originating from low grade tumors, 4 (44%) aligned with low grade tumors in the Lindgren bladder tumor set (N=144, Table S4) and 12 (63%) of the 19 cell lines from high grade tumors are aligned with high grade lesions. Strikingly, 43% (12/28) of the cell lines don’t resemble their tumor origins with respect to grade gene signatures.
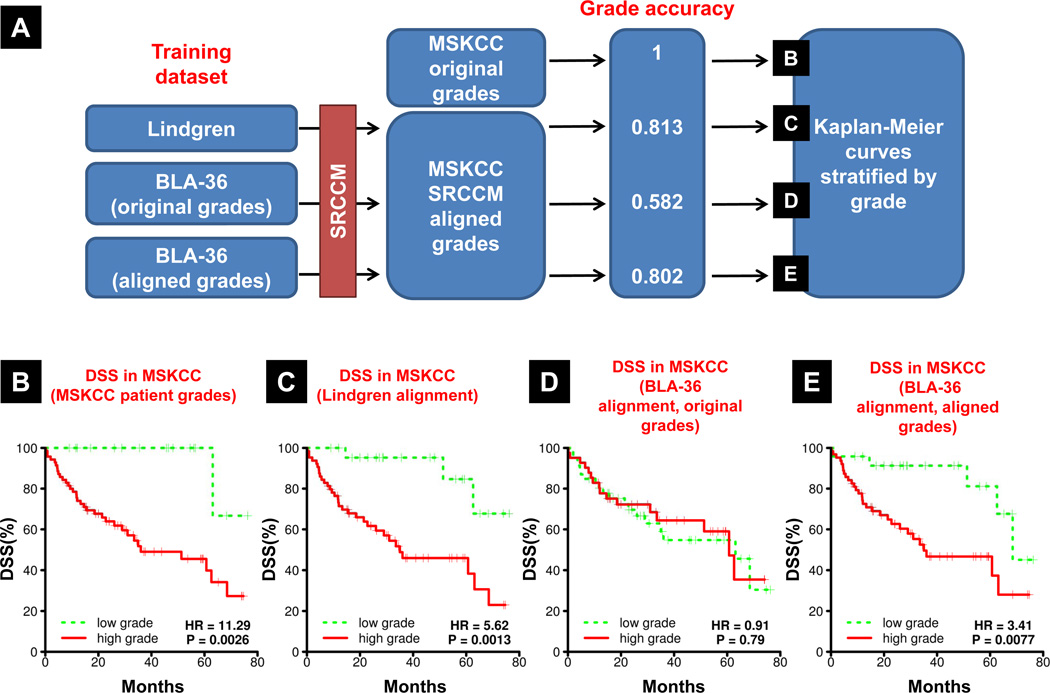
Because of the disagreement between BLA-36 original tumor grade and SRCCM assigned grade, and because grade is a strong indicator of patient survival (40), we next investigated the clinical relevance of this alignment (Figure 4A). Specifically, we asked whether the grades of the original tumors from which the cell lines were derived or the grades assigned via SRCCM were more predictive of DSS. To evaluate this, we aligned 87 tumor samples from an independent patient cohort (MSKCC, Supplementary Table S4) to either a patient dataset (Lindgren, Supplementary Table S4) or the BLA-36 cell lines, using their original or reassigned grades, and then generated Kaplan-Meier (KM) DSS curves for the assigned low and high grade samples. In the MSKCC cohort, patient grade is a very strong predictor of survival, producing KM survival curves with a hazard ratio (HR) of 11.29 and a logrank p-value of 0.0026 (Figure 4B). Use of the Lindgren cohort establishes the efficacy of the SRCCM algorithm to predict MSKCC grade and then generate KM survival curves since this resulted in excellent stratification (HR = 5.62, logrank p = 0.0013, Figure 4C). Using the grades of the original tumors from where BLA-36 were derived, the relationship between grade and survival is lost (HR = 0.91, logrank p = 0.79, Figure 4D). In contrast, when the MSKCC samples are aligned with SRCCM assigned cell line grades, the relationship between grade and survival is restored (HR = 3.41, logrank p = 0.0077, Figure 4E). Together, these results indicate that cell lines no longer resemble the tumors they were derived from and cell lines selected by their original grades no longer correlate with survival. In contrast, the SRCCM is effective in identifying lines that correlate with clinical outcome.
Figure 4.
Original cell line grades no longer correlate with disease specific survival but correlation is restored via SRCCM alignment to patient tumors. A, Overview of the methodology used. MSKCC tumors are aligned to the grades of Lindgren tumors or BLA-36 cell lines using the grades of the tumors they were derived from (original grades) or the assigned grades from alignment with the Lindgren patients (aligned grades). We report the accuracy of MSKCC grade assignment. We then generate KM survival curves based on B, the original patient grades in MSKCC (positive control), C, MSKCC grades based on alignment to Lindgren patient tumors (second positive control), D, MSKCC grades based on alignment to BLA-36 (original grades), and E, MSKCC grades based on alignment to BLA-36 (aligned grades).
The clinical impact of selecting human bladder cancer cell lines based on their similarity with human tumors
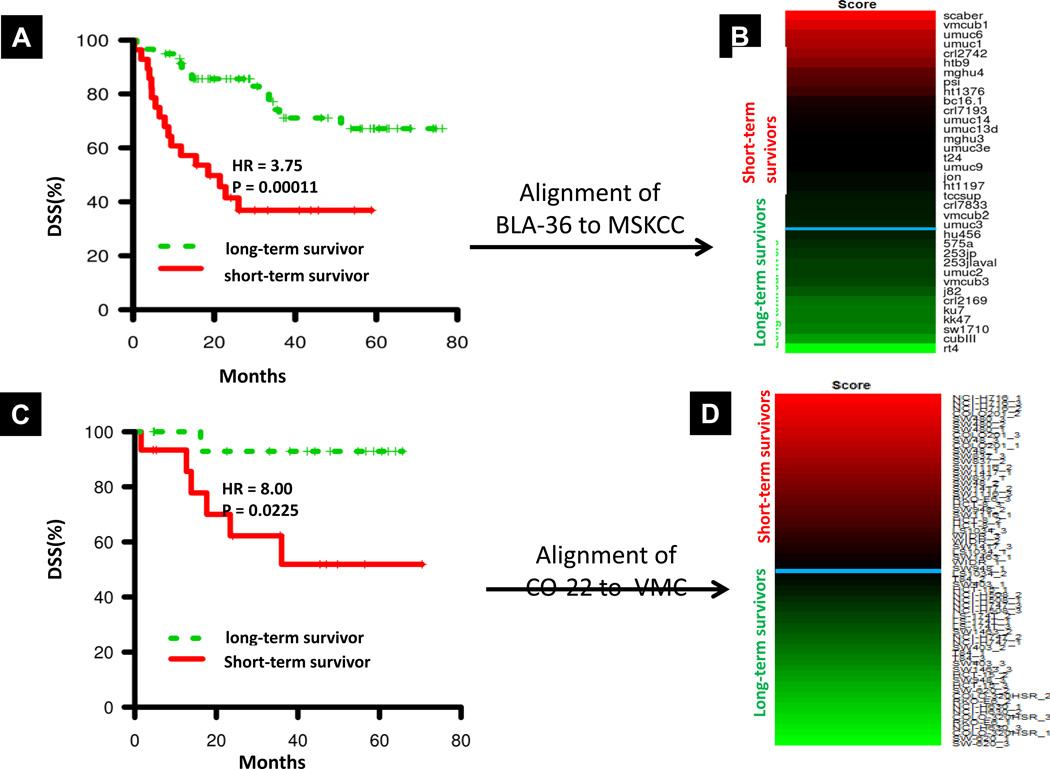
The overarching objective of this work is to identify the most clinically relevant cancer cell lines, those most strongly correlated with disease outcome. Above, we noted that the aligned grades were more clinically significant than the actual grades of the original tumors from which the cell lines were derived. To define the most clinically relevant cell lines, here we first align the BLA-36 cell lines directly with DSS, using the MSKCC cohort as the training dataset. The MSKCC cohort contains 87 tumor samples and has a median survival time of 26.0 months and a 5-year DSS rate of 56.3% (Supplementary Table S5). Patients were assigned to risk groups (i.e., long- and short-term survivors) using the “median cut” labeling method (31) as described in Materials and Methods. For SRCCM analysis, we use a gene signature consisting of all probes correlated with DSS in MSKCC (univariate Cox proportional hazards model, logrank p-value < 0.01), and applied the SRCCM algorithm to align the BLA-36 cell lines to long- and short-term survivors in the MSKCC cohort. In a LOOCV on MSKCC, the long- and short-term survivors produce survival curves with a hazard ratio of 3.75 and a logrank p-value = 0.00011 (Figure 5A). We then used the SRCCM to align the BLA-36 cell lines with long- and short-term survivors in MSKCC and ranked the cell lines according to their correlation score (Figure 5B).
Figure 5.
Disease specific survival (DSS) in BLA-36 and CO-22 as a function of long- and short-term survivors determined by “median cut” labeling (see Materials and Methods). A, KM survival curves for bladder cancer patients in the training MSKCC dataset obtained through SRCCM alignment and LOOCV. B, DSS alignment heatmap for BLA-36 cell lines. Cell lines are ranked from high (red) to low (green) by their correlation scores (see Materials and Methods) and classified as either long-term or short-term survivors, separated by a horizontal blue line, based on their alignment to long- and short-term survivors in MSKCC. C, KM survival curves for colorectal cancer patients in the training VMC dataset obtained through SRCCM alignment and LOOCV. D, DSS alignment heatmap for CO-22 cell lines. Cell lines are ranked from high (red) to low (green) by their correlation scores (see Materials and Methods) and classified as either long-term or short-term survivors, separated by a horizontal blue line, based on their alignment to long- and short-term survivors in VMC.
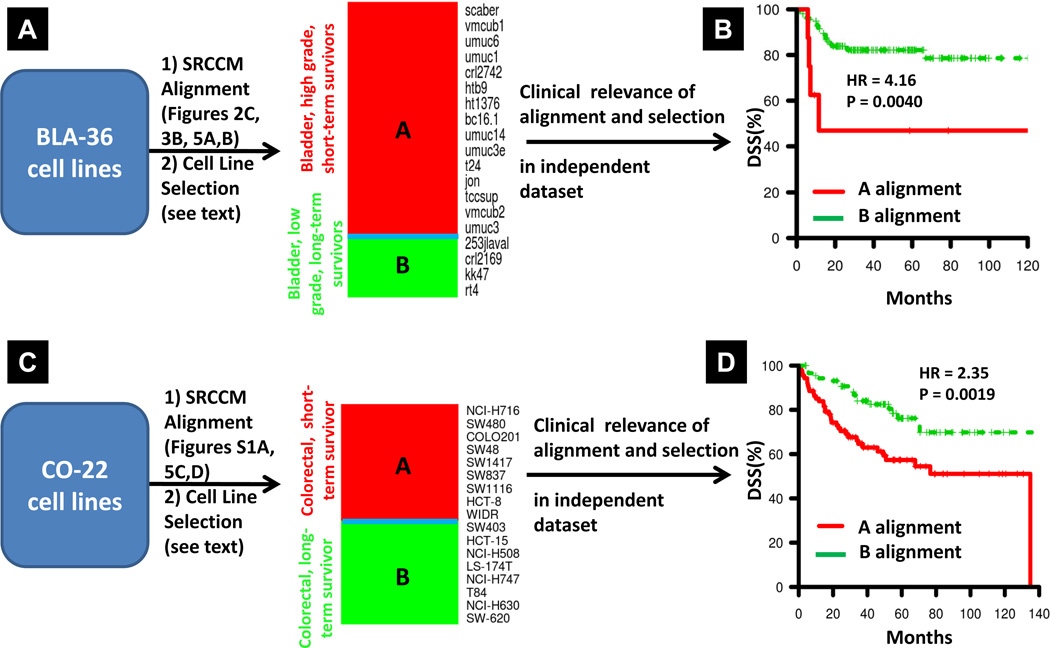
Next we compared this ranking to the tumor of origin alignment (Figure 2C) and to the grade ranking (Figure 3B) that was found clinically relevant (Figure 4E) and selected only cells aligning with bladder tumors that were either in high-grade/short-term survivors or low-grade/long-term survivor groups. This provides us with 15 and 4 cell lines in each group respectively (Figure 6A). We then evaluated the clinical relevance of this selection by using it to align an independent cohort of patients from CNUH (N=129, 5-year DSS rate = 80.5%, Ref #29, Supplementary Table S5). Patients aligned to these high-grade/short-term and low-grade/long-term survivors produced survival curves with a hazard ratio of 4.16 and a logrank p-value = 0.0040 (Figure 6B) indicating the selection of cell lines based on their tissue of origin, grade, and DSS alignments has biological significance.
Figure 6.
Selection and validation of clinically relevant BLA-36 and CO-22 cell lines. A, SRCCM alignment of BLA-36 cell lines to tissue of origin, grade, and DSS. Heatmap of the clinically relevant BLA-36 cell lines aligning with bladder, high grade, and short term-survivors or bladder, low grade, and long-term survivors. The cell lines are ranked by their DSS alignment score. B, validation of the clinical relevance of the selected cell lines in an independent dataset. KM survival curves are generated for patients in CNUH following alignment to the clinically relevant cell lines in (A). C, SRCCM alignment of CO-22 cell lines to tissue of origin and DSS. Heatmap of the clinically relevant CO-22 cell lines having all replicates aligning with colorectum and no ambiguous DSS alignments. The cell lines are ranked by their average DSS alignment score. D, validation of the clinical relevance of the selected cell lines in an independent dataset. KM survival curves are generated for patients in MCC following alignment to the clinically relevant cell lines in (C).
Selecting human colorectal cancer cell lines based on their similarity to human tumors leads to the identification of clinically relevant lines
To evaluate the generalizability of our SRCCM algorithm and cell line selection concept, we analyzed a panel of 22 colorectal cell lines (CO-22) profiled by GlaxoSmithKline. The CO-22 cell lines were first aligned to tissue of origin, where the SRCCM alignment accuracy for colorectal tumors in the independent test datasets was 99% (Figure 2A). All CO-22 cell line samples aligned with colorectum with the exception of COLO-320HSR, whose samples all aligned with uterus, and RKO-E6, where one sample aligned with bladder (Supplementary Figure S1A).
We next aligned the CO-22 cell lines directly to DSS, using a set of 55 patients with colorectal cancer from the Vanderbilt Medical Center as the training dataset (VMC cohort, Ref #27). The VMC cohort has a median survival time of 38.0 and a 5-year DSS rate of 73.4% (Supplementary Table S5). Patients were assigned to risk groups using the “median cut” labeling method as was done for the BLA-36 cell lines (see Materials and Methods). For SRCCM analysis, we used a previously published and validated gene signature consisting of 34 genes that correlates with recurrence and survival (27). In a LOOCV on VMC, the long- and short-term survivors produce survival curves with a hazard ratio of 8.00 and a logrank p-value = 0.0225 (Figure 5C). We then used the SRCCM to align the CO-22 cell lines with long- and short-term survivors in VMC, ranked the cell lines according to their correlation score, and used the median correlation score to separate long- and short-term survivors (Figure 5D).
For cell line selection, a cell line was deemed clinically relevant if all of its replicates aligned with colorectum and all replicates had the same DSS alignment (Figure 6C). Cell lines were removed if they had ambiguous DSS alignments defined as alignments with one or more replicates aligned with short-term survivors and at least one aligned with long-term survivors. Our selection resulted in the removal of the following lines: COLO-320HSR, which aligned with uterus; LS1034, SW1463, and SW948, which had ambiguous DSS alignments, and RKO-E6, which had an ambiguous DSS alignment and also had one replicate aligning with bladder.
The clinical relevance of the remaining 17 cell lines was then evaluated in an independent cohort of patients from the Moffitt Cancer Center (MCC, N = 177, 5 year DSS rate = 67.0%, Ref #27, Supplementary Table S5). Alignment of the MCC patients to the selected colorectal cell lines produces survival curves with a hazard ratio of 2.35 and a logrank p-value of 0.0019, establishing their clinical relevance (Figure 6D). Importantly, when all 22 colorectal cell lines were used in this analysis (i.e., without the selection step), the hazard ratio falls to 1.69 and the separation of survival curves is no longer statistically significant (logrank p-value = 0.054, Supplementary Figure S1B).
DISCUSSION
For a cell line model to have clinical utility it must be representative of the human tumor of interest. Importantly, cell line models selected based on the characteristics of the tumors from which they are derived, their xenograft behavior, or their technical convenience may not be clinically relevant and therefore lack clinical translatability. In this paper we describe a novel framework for the selection of the most clinically relevant cell lines. At the heart of this approach is the SRCCM alignment algorithm, with software we have made available to the scientific community, and which is applicable to any cancer cell line. Although we have selected clinically relevant cell lines for bladder and colorectal cancers, we caution that because gene expression profiles vary across culture conditions (42, 43) and with cell passage number, the cell lines selected here as most clinically relevant may not be so in another lab with the same lines. Instead, we recommend SRCCM alignment and cell line selection using gene expression profiles obtained in the specific lab. In time, we will determine whether this possible variation in the repertoire of the “most clinically relevant” lines does indeed occur.
In developing SRCCM, a variety of classification methods for aligning cell lines with clinical phenotypes could have been implemented, including standard machine learning algorithms such as nearest neighbor and support vector machines (44, 45). However, the SRCCM algorithm was developed after carefully considering its ease of interpretation and its applicability. Correlation is an intuitive measure of similarity, while the specific use of Spearman’s correlation ensures broad applicability to additional datasets and cell lines, as the choice of data normalization and processing methods will have relatively little impact on the output of the algorithm.
Independent validation of SRCCM was a critical feature of our work. Because classifiers developed from high dimensional microarray datasets are prone to over-fitting (46), and technical batch effects can correlate with biological outcomes of interest (47), independent validation is essential if reported classification accuracies are expected to be representative of performance in additional datasets. We validated the tissue of origin, stage, and grade SRCCM alignments on multiple independent datasets, and achieved excellent overall accuracies.. We validated the DSS SRCCM alignments on an independent dataset by first aligning the BLA-36 cell lines to long- and short-term survivors in the MSKCC training cohort and then aligning 129 patients in an independent cohort (CNUH) to the 19 clinically relevant cell lines to produce KM survival curves with clear stratification.
The SRCCM tissue alignment finds that 34/36 (94%) of the bladder cell lines and 20/22 (91%) of the colorectal cell lines resemble their tissue of origin more closely than the 9 additional carcinomas we considered. This is reassuring, as a previous tissue classification of the NCI-60 cell line panel suggested that as few as 57% of cell lines most closely resembled their presumed tissue of origin (10). In general, the BLA-36 cell lines correlate more strongly with tissue types where squamous cell histologies are common (e.g., cervix and ovary) than with tissues where squamous cell histologies are rare (e.g., breast, thyroid, prostate, and kidney) (Figure 2C), suggesting that it may be relatively difficult to distinguish between tissue types if their histologies are similar. In the independent validation on over 300 bladder samples, 9% were misaligned, and the fact that 2/36 (6%) of the cell lines are misaligned in the BLA-36 panel is within the expected margin of error. Further analysis of these two lines, CubIII and SW1710, is required to determine whether these are of bladder origin.
Most striking is our finding that the grade alignment of the BLA-36 cell lines is consistent with only 44% of the low grade tumors and 63% of the high grade tumors that the cell lines were derived from. One possible explanation for these results is that the alignment is inaccurate. However, our findings support the validity of the alignment and loss of relevance of the original tumor grade. For example, we found that TP53 and RB1 mutations are associated with aligned high grade muscle-invasive tumors while FGFR3 mutations are associated with aligned low grade, non-muscle invasive tumors, which is consistent with the prevalence of TP53, RB1, and FGFR3 mutations in patients (37–39). Since tumor grade is an important clinical parameter that correlates with both stage and survival in patients (40, 48), for a cell line model to have clinical relevance, it should properly represent these clinical phenotypes. Importantly, we have found that selection of cell line models based on clinical information from their original tumors will fail to properly represent these phenotypes for the cell lines we analyzed. In contrast, once aligned using SRCCM the new cell line grades relate to clinical outcomes (Figure 4D,E).
Cell line models have also been selected based on their xenograft behavior. However, animal models do not always recapitulate human physiology and behavior (49, 50). In a meta-analysis looking at 39 pharmacological agents with both xenograft data and results from Phase II clinical trials, Johnson et al. found that out of six xenograft histologies, only lung xenografts had the ability to predict clinical activity in the same human cancer histology, though interestingly several were predictive of clinical activity in other histologies (51). In contrast, xenografts derived from aligned cell lines validated for their ability to reflect human tumor biology, using alignment and selection process shown here (Figure 6), may provide a readout of drug activity in xenografts that parallel that seen in human tumors.
In conclusion, we describe a framework for aligning cell lines with the clinicopathologic phenotypes of human tumors such as tissue of origin, stage, grade, and DSS, that can be used to select the most clinically relevant cell line models. Our technique has the potential to reduce the discrepancy often encountered between in vitro and in vivo laboratory data and patient interventions. Although the approach is demonstrated in bladder and colorectal cancer cells, SRCCM provides a general framework that is applicable to all cancer types and is freely available to the scientific community.
Supplementary Material
Acknowledgements
This work was supported in part by National Institutes of Health grants CA075115 and CA143791 to DT.
Abbreviations
- adeno
adenocarcinoma
- scc
squamous cell carcinoma
- NMI
non-muscle invasive
- MI
muscle invasive
- SRCCM
Spearman’s rank correlation classification method
- KM
Kaplan-Meier
- HR
hazard ratio
- DSS
disease specific survival
Footnotes
Conflicts of Interest: none
REFERENCES
- 1.Marsit CJ, Houseman EA, Christensen BC, et al. Identification of methylated genes associated with aggressive bladder cancer. PLoS One. 2010;5:e12334. doi: 10.1371/journal.pone.0012334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inoue K, Slaton JW, Kim SJ, et al. Interleukin 8 expression regulates tumorigenicity and metastasis in human bladder cancer. Cancer Res. 2000;60:2290–2299. [PubMed] [Google Scholar]
- 3.Nutt JE, Mellon JK, Qureshi K, Lunec J. Matrix metalloproteinase-1 is induced by epidermal growth factor in human bladder tumour cell lines and is detectable in urine of patients with bladder tumours. Brit J Cancer. 1998;78:215–220. doi: 10.1038/bjc.1998.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saitoh H, Mori K, Kudoh S, Itoh H, Takahashi N, Suzuki T. BCG effects on telomerase activity in bladder cancer cell lines. Int J Clin Oncol. 2002;7:165–170. doi: 10.1007/s101470200024. [DOI] [PubMed] [Google Scholar]
- 5.Zhang YB, Niu HT, Chang JW, Dong GL, Ma XB. EZH2 silencing by RNA interference inhibits proliferation in bladder cancer cell lines. Eur J Cancer Care (Engl) 2011;20:106–112. doi: 10.1111/j.1365-2354.2009.01148.x. [DOI] [PubMed] [Google Scholar]
- 6.Krause-Heuer AM, Grunert R, Kuhne S, et al. Studies of the mechanism of action of platinum(II) complexes with potent cytotoxicity in human cancer cells. J Med Chem. 2009;52:5474–5484. doi: 10.1021/jm9007104. [DOI] [PubMed] [Google Scholar]
- 7.Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 2010;10:241–253. doi: 10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
- 8.Borisy AA, Elliott PJ, Hurst NW, et al. Systematic discovery of multicomponent therapeutics. Proc Natl Acad Sci U S A. 2003;100:7977–7982. doi: 10.1073/pnas.1337088100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masters JR. Human cancer cell lines: fact and fantasy. Nat Rev Mol Cell Biol. 2000;1:233–236. doi: 10.1038/35043102. [DOI] [PubMed] [Google Scholar]
- 10.Sandberg R, Ernberg I. Assessment of tumor characteristic gene expression in cell lines using a tissue similarity index (TSI) Proc Natl Acad Sci U S A. 2005;102:2052–2057. doi: 10.1073/pnas.0408105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cell line misidentification: the beginning of the end. Nat Rev Cancer. 2010;10:441–448. doi: 10.1038/nrc2852. [DOI] [PubMed] [Google Scholar]
- 12.MacLeod RAF, Dirks WG, Matsuo Y, Kaufmann M, Milch H, Drexler HG. Widespread intraspecies cross-contamination of human tumor cell lines arising at source. International Journal of Cancer. 1999;83:555–563. doi: 10.1002/(sici)1097-0215(19991112)83:4<555::aid-ijc19>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 13.Drexler HG, Dirks WG, Matsuo Y, MacLeod RA. False leukemia-lymphoma cell lines: an update on over 500 cell lines. Leukemia. 2003;17:416–426. doi: 10.1038/sj.leu.2402799. [DOI] [PubMed] [Google Scholar]
- 14.Herr HW, Dotan Z, Donat SM, Bajorin DF. Defining optimal therapy for muscle invasive bladder cancer. J Urol. 2007;177:437–443. doi: 10.1016/j.juro.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 15.Stein JP, Lieskovsky G, Cote R, et al. Radical cystectomy in the treatment of invasive bladder cancer: long-term results in 1,054 patients. J Clin Oncol. 2001;19:666–675. doi: 10.1200/JCO.2001.19.3.666. [DOI] [PubMed] [Google Scholar]
- 16.Kemp CJ, Sun SL, Gurley KE. p53 induction and apoptosis in response to radio- and chemotherapy in vivo is tumor-type-dependent. Cancer Res. 2001;61:327–332. [PubMed] [Google Scholar]
- 17.Mizutani Y, Wada H, Fukushima M, et al. The significance of dihydropyrimidine dehydrogenase (DPD) activity in bladder cancer. Eur J Cancer. 2001;37:569–575. doi: 10.1016/s0959-8049(00)00440-8. [DOI] [PubMed] [Google Scholar]
- 18.Grossman HB, Natale RB, Tangen CM, et al. Neoadjuvant chemotherapy plus cystectomy compared with cystectomy alone for locally advanced bladder cancer. N Engl J Med. 2003;349:859–866. doi: 10.1056/NEJMoa022148. [DOI] [PubMed] [Google Scholar]
- 19.Sylvester RJ, Oosterlinck W, van der Meijden AP. A single immediate postoperative instillation of chemotherapy decreases the risk of recurrence in patients with stage Ta T1 bladder cancer: a meta-analysis of published results of randomized clinical trials. J Urol. 2004;171:2186–2190. doi: 10.1097/01.ju.0000125486.92260.b2. quiz 435. [DOI] [PubMed] [Google Scholar]
- 20.von der Maase H, Hansen SW, Roberts JT, et al. Gemcitabine and cisplatin versus methotrexate, vinblastine, doxorubicin, and cisplatin in advanced or metastatic bladder cancer: results of a large, randomized, multinational, multicenter, phase III study. J Clin Oncol. 2000;18:3068–3077. doi: 10.1200/JCO.2000.18.17.3068. [DOI] [PubMed] [Google Scholar]
- 21.Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for functional genomics data sets-10 years on. Nucleic Acids Research. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JK, Havaleshko DM, Cho H, et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci U S A. 2007;104:13086–13091. doi: 10.1073/pnas.0610292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wooster R, Greshock J, Bachman KE, et al. Molecular Target Class Is Predictive of In vitro Response Profile. Cancer Res. 2010;70:3677–3686. doi: 10.1158/0008-5472.CAN-09-3788. [DOI] [PubMed] [Google Scholar]
- 24.Smith SC, Baras AS, Dancik G, et al. A 20-gene model for molecular nodal staging of bladder cancer: development and prospective assessment. Lancet Oncol. 2011;12:137–143. doi: 10.1016/S1470-2045(10)70296-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindgren D, Frigyesi A, Gudjonsson S, et al. Combined gene expression and genomic profiling define two intrinsic molecular subtypes of urothelial carcinoma and gene signatures for molecular grading and outcome. Cancer Res. 2010;70:3463–3472. doi: 10.1158/0008-5472.CAN-09-4213. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 27.Smith JJ, Deane NG, Wu F, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R: A Language and Environment for Statistical Computing [Internet] Vienna (AU): R Development Core Team; [cited 2010 July 1]. Available from: http://www.R-project.org. [Google Scholar]
- 29.Kim WJ, Kim EJ, Kim SK, et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Mol Cancer. 2010;9:3. doi: 10.1186/1476-4598-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stransky N, Vallot C, Reyal F, et al. Regional copy number-independent deregulation of transcription in cancer. Nat Genet. 2006;38:1386–1396. doi: 10.1038/ng1923. [DOI] [PubMed] [Google Scholar]
- 31.Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2:E108. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Therneau TM, Grambsch PM. Modeling survival data : extending the Cox model. New York: Springer; 2000. [Google Scholar]
- 33.Wang H, Huang S, Shou J, et al. Comparative analysis and integrative classification of NCI60 cell lines and primary tumors using gene expression profiling data. BMC Genomics. 2006;7:166. doi: 10.1186/1471-2164-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garber ME, Troyanskaya OG, Schluens K, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dyrskjot L, Thykjaer T, Kruhoffer M, et al. Identifying distinct classes of bladder carcinoma using microarrays. Nat Genet. 2003;33:90–96. doi: 10.1038/ng1061. [DOI] [PubMed] [Google Scholar]
- 36.Blaveri E, Simko JP, Korkola JE, et al. Bladder cancer outcome and subtype classification by gene expression. Clin Cancer Res. 2005;11:4044–4055. doi: 10.1158/1078-0432.CCR-04-2409. [DOI] [PubMed] [Google Scholar]
- 37.Abdel-Fattah R, Challen C, Griffiths TR, Robinson MC, Neal DE, Lunec J. Alterations of TP53 in microdissected transitional cell carcinoma of the human urinary bladder: high frequency of TP53 accumulation in the absence of detected mutations is associated with poor prognosis. Br J Cancer. 1998;77:2230–2238. doi: 10.1038/bjc.1998.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bakkar AA, Wallerand H, Radvanyi F, et al. FGFR3 and TP53 gene mutations define two distinct pathways in urothelial cell carcinoma of the bladder. Cancer Res. 2003;63:8108–8112. [PubMed] [Google Scholar]
- 39.Wu XR. Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer. 2005;5:713–725. doi: 10.1038/nrc1697. [DOI] [PubMed] [Google Scholar]
- 40.Pagliaro LC, Sharma P. Review of metastatic bladder cancer. Minerva Urol Nefrol. 2006;58:53–71. [PubMed] [Google Scholar]
- 41.Jorissen RN, Gibbs P, Christie M, et al. Metastasis-Associated Gene Expression Changes Predict Poor Outcomes in Patients with Dukes Stage B and C Colorectal Cancer. Clin Cancer Res. 2009;15:7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kissel T, Behrens I. Do cell culture conditions influence the carrier-mediated transport of peptides in Caco-2 cell monolayers? Eur J Pharm Sci. 2003;19:433–442. doi: 10.1016/s0928-0987(03)00146-5. [DOI] [PubMed] [Google Scholar]
- 43.Lin CS, Lin GT, Chow S, Lin J, Wang G, Lue TF. Effect of cell passage and density on protein kinase G expression and activation in vascular smooth muscle cells. J Cell Biochem. 2004;92:104–112. doi: 10.1002/jcb.20043. [DOI] [PubMed] [Google Scholar]
- 44.Byvatov E, Schneider G. Support vector machine applications in bioinformatics. Appl Bioinformatics. 2003;2:67–77. [PubMed] [Google Scholar]
- 45.Thamaraiselvi G, Kaliammal A. Data Mining: Concepts and Techniques. SRELS Journal of Information Management. 2004;41:339–348. [Google Scholar]
- 46.Lee S. Mistakes in validating the accuracy of a prediction classifier in high-dimensional but small-sample microarray data. Stat Methods Med Res. 2008;17:635–642. doi: 10.1177/0962280207084839. [DOI] [PubMed] [Google Scholar]
- 47.Leek JT, Scharpf RB, Bravo HC, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 49.Peterson JK, Houghton PJ. Integrating pharmacology and in vivo cancer models in preclinical and clinical drug development. Eur J Cancer. 2004;40:837–844. doi: 10.1016/j.ejca.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 50.Voskoglou-Nomikos T, Pater JL, Seymour L. Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clin Cancer Res. 2003;9:4227–4239. [PubMed] [Google Scholar]
- 51.Johnson JI, Decker S, Zaharevitz D, et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Brit J Cancer. 2001;84:1424–1431. doi: 10.1054/bjoc.2001.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parkinson H, Sarkans U, Kolesnikov N, et al. ArrayExpress update-an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Research. 2011;39:D1002–D1004. doi: 10.1093/nar/gkq1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bamford S, Dawson E, Forbes S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.