Abstract
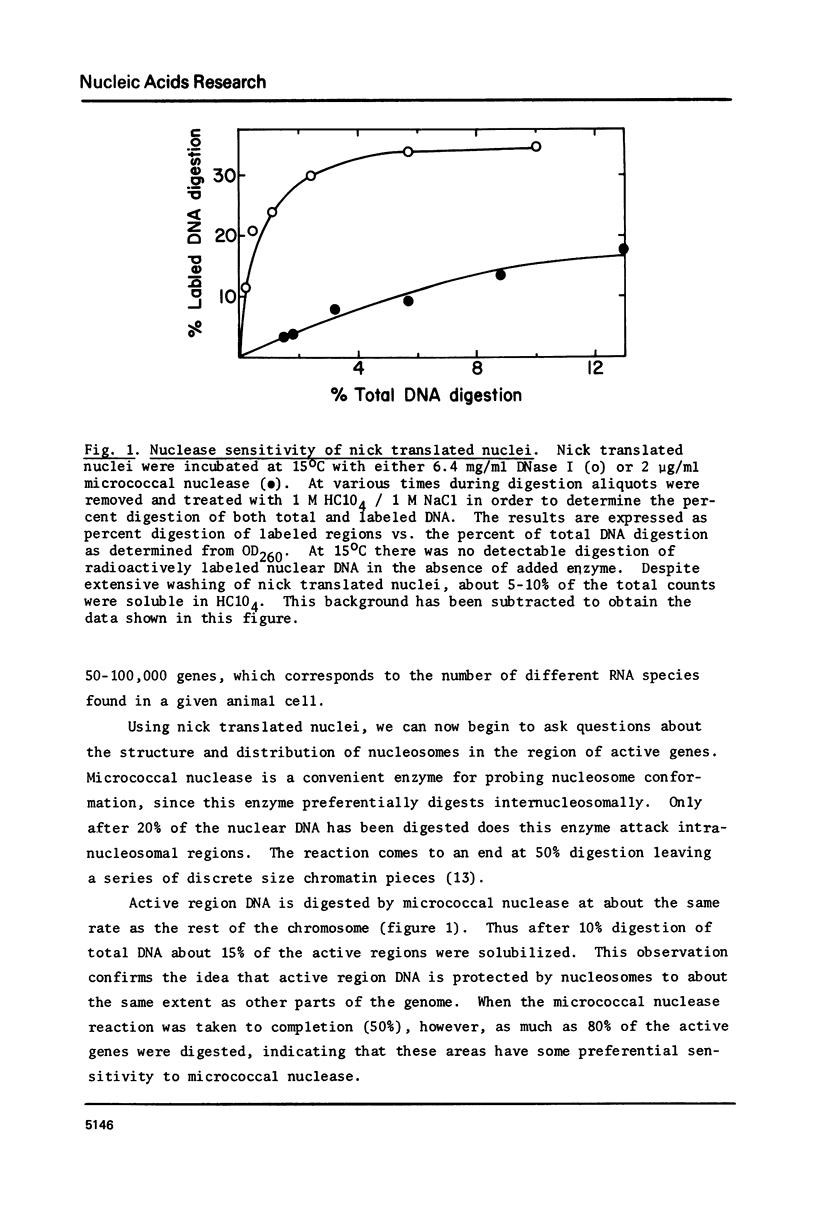
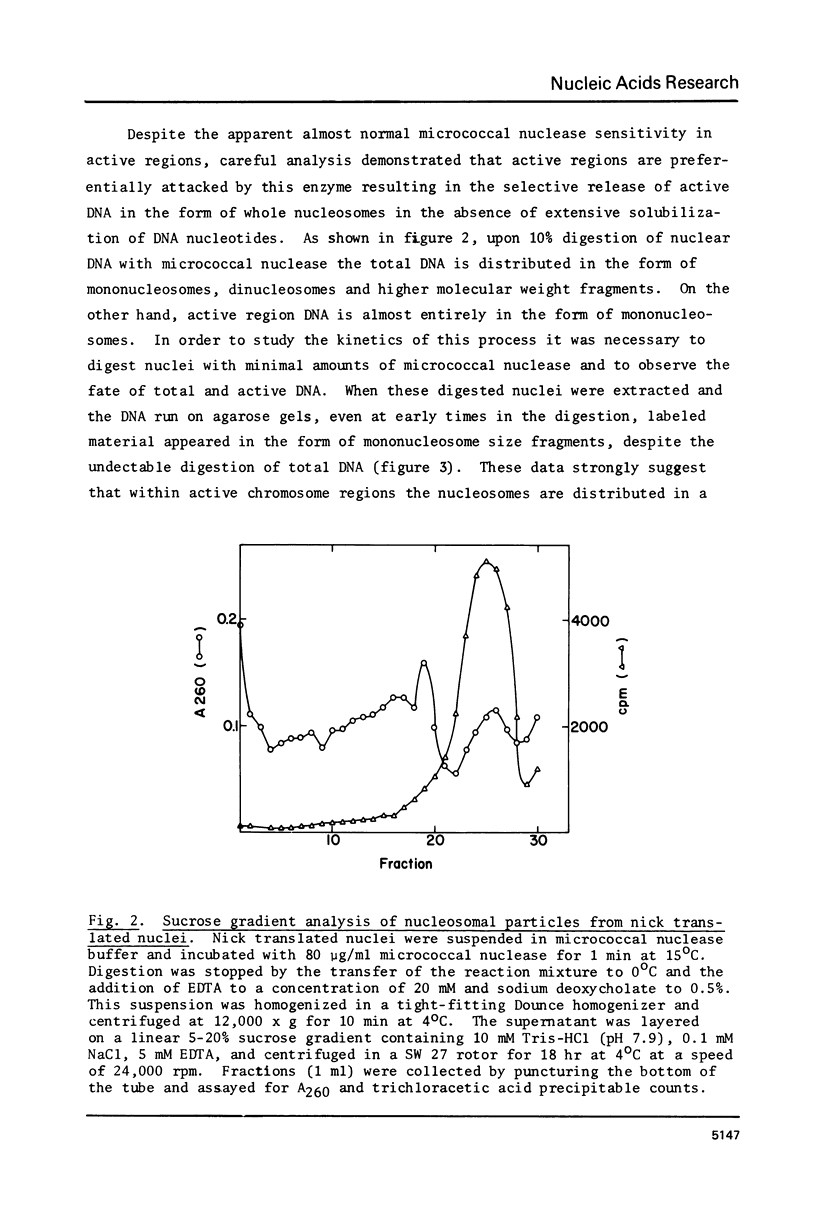
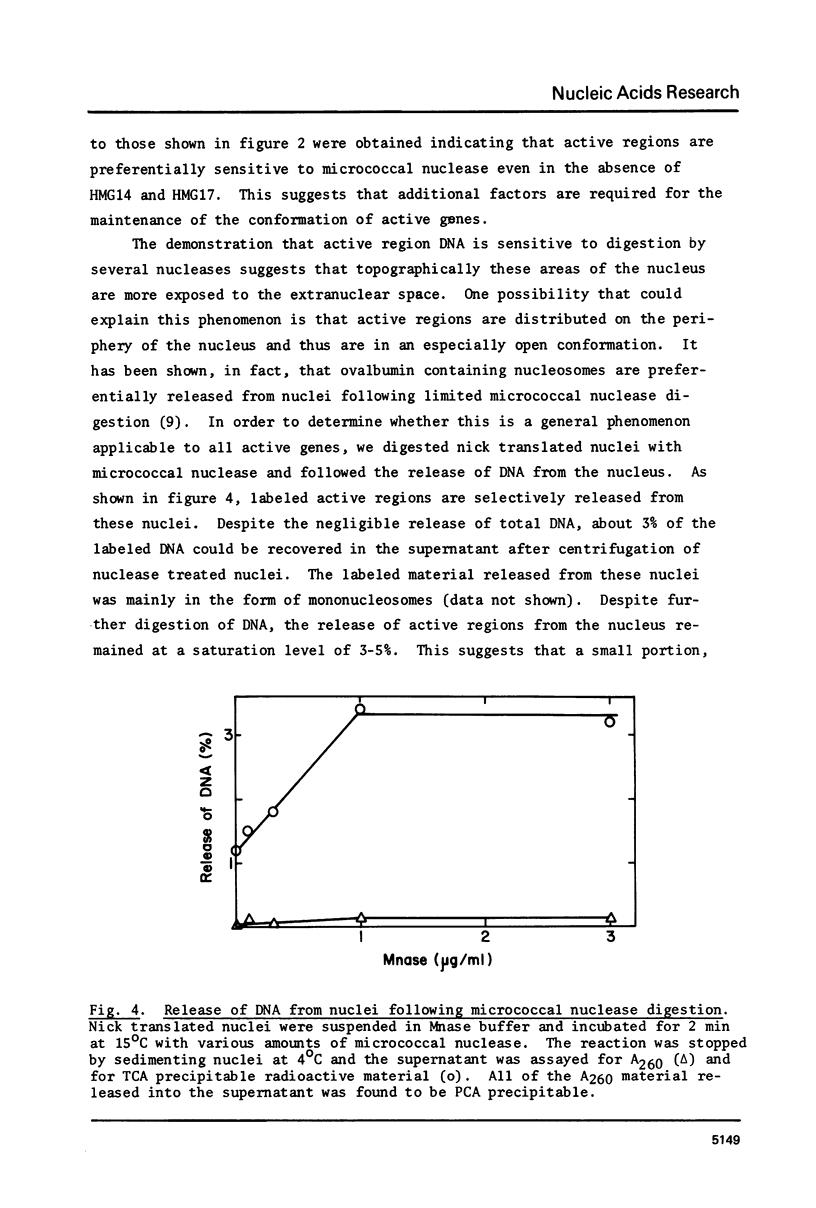
The active regions of chicken erythrocyte nuclei were labeled using the standard DNase I directed nick translation reaction. These nuclei were then used to study the characteristics and, in particular, the nuclease sensitivity of active genes. Although DNase I specifically attacks active genes, micrococcal nuclease solubilizes these regions to about the same degree as the total DNA. On the other hand micrococcal nuclease does selectively cut the internucleosomal regions of active genes resulting in the appearance of mononucleosomal fraction which is enriched in active gene DNA. A small percentage of the active chromatin is also released from the nucleus by low speed centrifugation following micrococcal nuclease treatment. The factors which make active genes sensitive to DNase I were shown to reside on individual nucleosomes from these regions. This was established by showing that isolated active mononucleosomes were preferentially sensitive to DNase I digestion. Although the high mobility group proteins are essential for the maintenance of DNase I sensitivity in active regions, these proteins are not necessary for the formation of the conformation which makes these genes preferentially accessible to micrococcal nuclease. The techniques employed in this paper enable one to study the chromatin structure of the entire population of actively expressed genes. Previous studies have elucidated the structure of a few special highly prevalent genes such as ovalbumin and hemoglobin. The results of this paper show that this special conformation is a general feature of all active genes irregardless of the extent of expression.
Full text
PDF




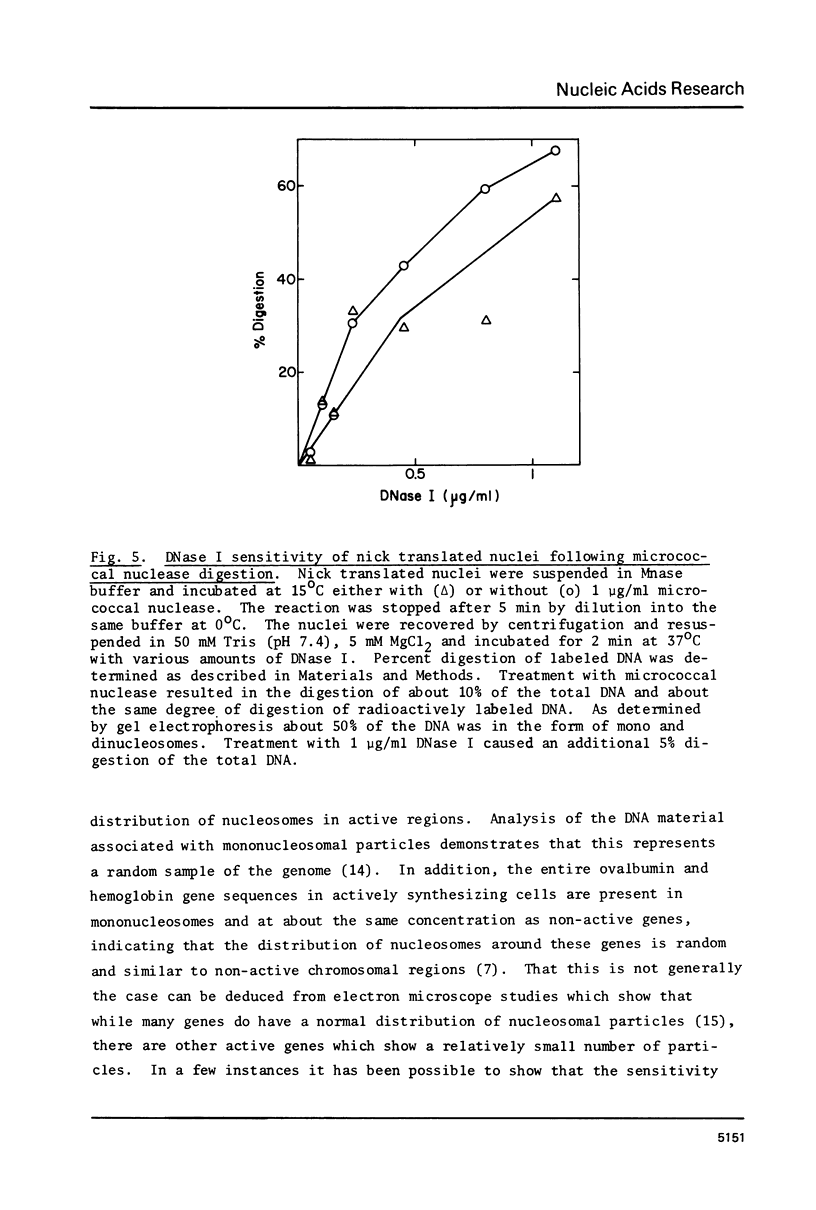
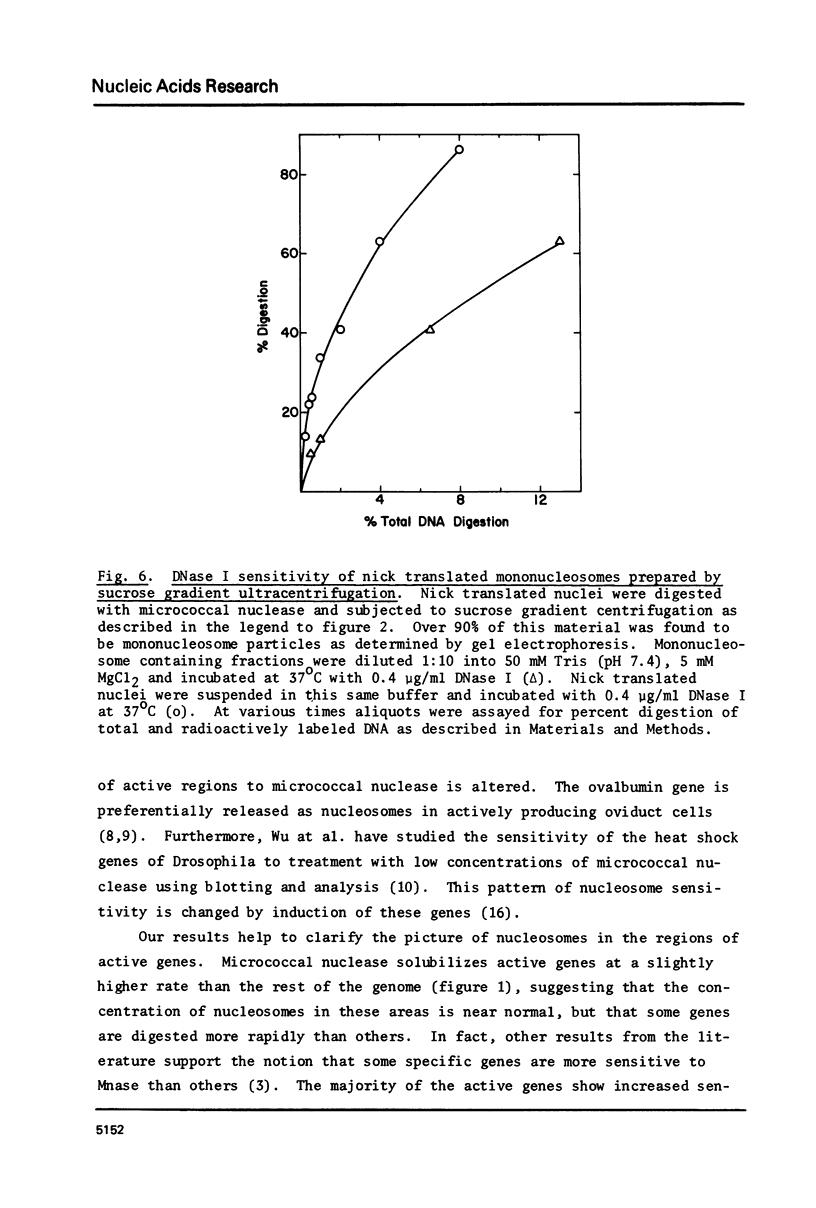
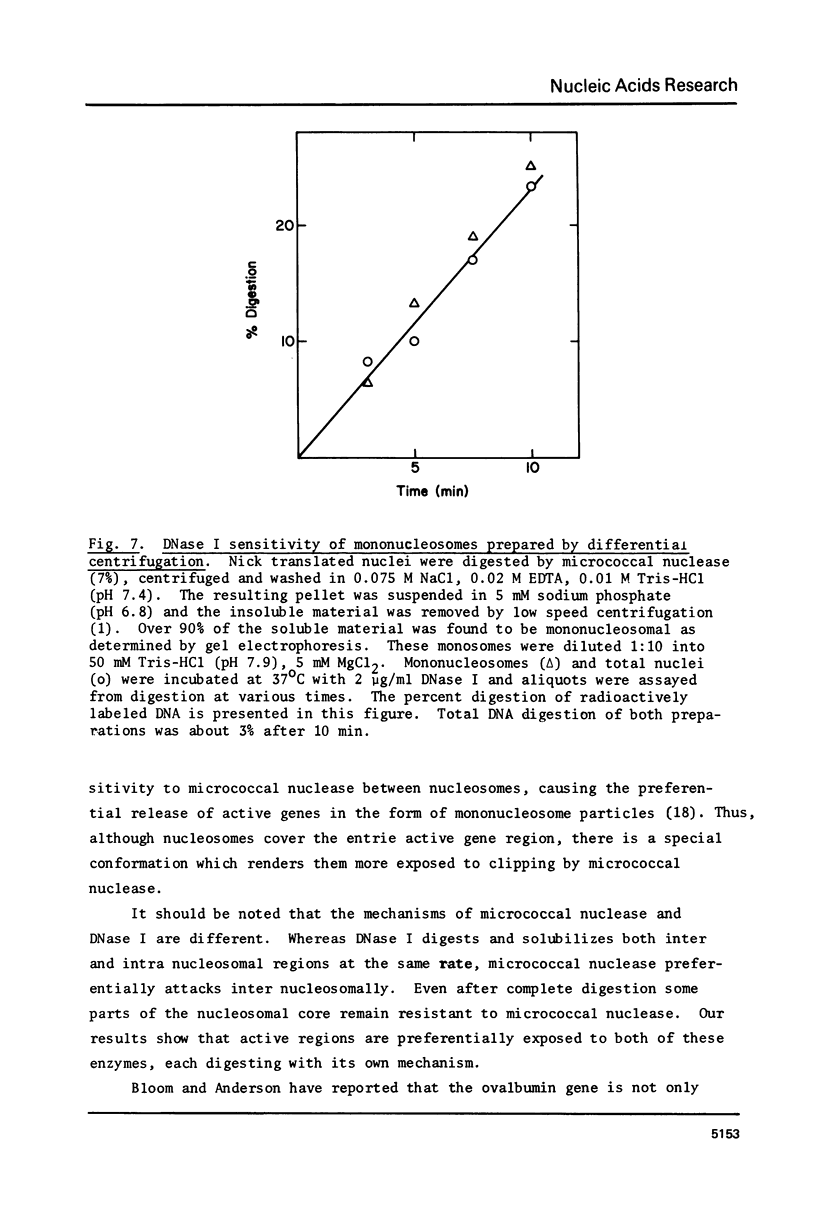


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bloom K. S., Anderson J. N. Conformation of ovalbumin and globin genes in chromatin during differential gene expression. J Biol Chem. 1979 Oct 25;254(20):10532–10539. [PubMed] [Google Scholar]
- Bloom K. S., Anderson J. N. Fractionation of hen oviduct chromatin into transcriptionally active and inactive regions after selective micrococcal nuclease digestion. Cell. 1978 Sep;15(1):141–150. doi: 10.1016/0092-8674(78)90090-9. [DOI] [PubMed] [Google Scholar]
- Chambon P. Summary: the molecular biology of the eukaryotic genome is coming of age. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1209–1234. doi: 10.1101/sqb.1978.042.01.122. [DOI] [PubMed] [Google Scholar]
- Garel A., Zolan M., Axel R. Genes transcribed at diverse rates have a similar conformation in chromatin. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4867–4871. doi: 10.1073/pnas.74.11.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit B., Panet A., Cedar H. Reconstitution of a deoxyribonuclease I-sensitive structure on active genes. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1787–1790. doi: 10.1073/pnas.77.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy E., Axel R. Analysis of DNA of isolated chromatin subunits. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3978–3982. doi: 10.1073/pnas.72.10.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt A., Axel R., Cedar H. Nick translation of active genes in intact nuclei. Dev Biol. 1979 Apr;69(2):496–505. doi: 10.1016/0012-1606(79)90307-5. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Miller O. L., Jr Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell. 1976 Jun;8(2):305–319. doi: 10.1016/0092-8674(76)90014-3. [DOI] [PubMed] [Google Scholar]
- Panet A., Cedar H. Selective degradation of integrated murine leukemia proviral DNA by deoxyribonucleases. Cell. 1977 Aug;11(4):933–940. doi: 10.1016/0092-8674(77)90304-x. [DOI] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D. Relation of nucleosomes to DNA sequences. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):103–108. doi: 10.1101/sqb.1978.042.01.011. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weisbrod S., Groudine M., Weintraub H. Interaction of HMG 14 and 17 with actively transcribed genes. Cell. 1980 Jan;19(1):289–301. doi: 10.1016/0092-8674(80)90410-9. [DOI] [PubMed] [Google Scholar]
- Weisbrod S., Weintraub H. Isolation of a subclass of nuclear proteins responsible for conferring a DNase I-sensitive structure on globin chromatin. Proc Natl Acad Sci U S A. 1979 Feb;76(2):630–634. doi: 10.1073/pnas.76.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Bingham P. M., Livak K. J., Holmgren R., Elgin S. C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979 Apr;16(4):797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- Wu C., Wong Y. C., Elgin S. C. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979 Apr;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- Zasloff M., Camerini-Otero R. D. Limited DNase I nicking as a probe of gene conformation. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1907–1911. doi: 10.1073/pnas.77.4.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]