Abstract
Background
Recent genome-wide association studies have identified multiple genetic loci that increase the risk of chronic kidney disease (CKD) in the general population. We hypothesized that knowledge of these loci might permit improved CKD risk prediction beyond that provided by traditional phenotypic risk factors.
Study design
Observational cohort study
Setting and participants
Participants who attended the 15th (1977–1979) and 24th (1995–1998) examination cycles of the Original cohort or the 6th (1995–1998) and 8th cycles (2005–2008) of the Offspring cohort of the Framingham Heart Study (n=2,489).
Predictors
Single-nucleotide polymorphisms (SNPs) at 16 stage 3 CKD loci were genotyped and used to construct a genetic risk score. Standard clinical predictors of incident stage 3 CKD were also used.
Outcomes and Measurements
Incident stage 3 CKD was defined as eGFR <60 mL/min/1.73m2 at follow-up. Participants with baseline stage 3 CKD were excluded. Logistic regression was used to generate C statistics, which measured the power of the genetic risk score to discriminate risk of incident CKD stage 3 with and without traditional risk factors.
Results
There were 270 new stage 3 CKD cases during an average of 10.8 years follow-up. The mean (±SD) genetic risk score was 17.5±2.8 among those who developed stage 3 CKD and 17.3±2.6 among those who did not (P-value for genotype score difference=0.2). The odds ratio for stage 3 CKD was 1.06 (95% CI, 1.01–1.11; p=0.03) per additional risk allele, adjusting for age and sex. In the age and sex-adjusted model, the C statistic was 0.748 without the genotype score and 0.751 with the score (P-value for difference=0.3). The risk score was not statistically significant in a multivariable model adjusted for standard stage 3 CKD risk factors (p=0.07).
Limitations
Participants all of European ancestry; genotype score may not be valid in different ancestral groups.
Conclusions
A genetic score generated from 16 known CKD risk alleles did not predict new cases of stage 3 CKD in the community beyond knowledge of common, clinical risk factors alone.
Chronic kidney disease (CKD) is a global public health epidemic, affecting populations of European,1–3 Asian4–6 and African descent.7 It is a cause of considerable morbidity,8–10 excess mortality from both cardiovascular 11 and non-cardiovascular causes,12 as well as predisposing to progressive kidney failure and end-stage renal disease.13 However, if detected early, loss of kidney function may be slowed 14,15 or even reversed,16 and these secondary complications may be averted. Fortunately, it may be possible to identify some at-risk individuals from the measurement of common, clinical risk factors.17–20
CKD also aggregates in families, with a positive family history of kidney disease increasing risk by a factor of 1.3 to 10.4.21 Recent genome-wide association studies have identified several novel loci associated with risk of stage 3 CKD in the community,22–24 each conferring a 4 to 25% increase in the relative odds of stage 3 CKD per risk allele. Although these loci account for only a small fraction of the total familial basis of stage 3 CKD, it is possible that they may be combined into a genetic risk score for enhanced detection of persons at risk for stage 3 CKD.
The promise of personalized medicine and individualized risk prediction based on genetic information has been proposed as a potential benefit from this phase of genetic discovery.25 However, the extent to which a comprehensive panel of genotypes will help in predicting incident stage 3 CKD beyond what is already achievable using known clinical risk factors is not known. To answer this question, we tested the ability of a genetic risk score, constructed from a panel of 16 single-nucleotide polymorphisms (SNPs) associated with stage 3 CKD risk,26 to predict new cases beyond information provided by known CKD risk factors in a large, prospectively examined, community-based cohort.
Methods
Study participants
Study participants were derived from the Framingham Heart Study (FHS) Original and Offspring cohorts. FHS is a population-based cohort study that began in 1948 in Framingham, Massachusetts. The original cohort comprised 5,209 individuals aged between 28 and 62 years at the first examination, and these participants have been examined every 2 years since study inception. In 1971, 5124 children of the original cohort and their spouses were invited to participate in the Framingham Offspring Study. Participants in the offspring cohort were re-examined 8 years after the first examination and every 4 years thereafter. Each such visit incorporates a detailed medical history, physical examination including blood pressure (BP) measurements, anthropometry, and laboratory assessment of risk factors. Detailed descriptions of the FHS Original and Offspring cohorts have been published previously.27,28
Participants who attended the 15th (1977–1979) and 24th (1995–1998) examination cycles of the Original cohort or the 6th (1995–1998) and 8th cycles (2005–2008) of the Offspring cohort were included in the present analysis. Of a total of 4,598 participants that attended their respective baseline and follow-up examinations, 2,934 had CKD status assessed on both occasions and were included in the CKD genomewide associationi study (GWAS). Following this, 236 were excluded due to baseline stage 3 CKD, resulting in a study sample of 2,698, of whom 2528 were brought forward for genotyping. Of these, 39 had more than 2 SNPs missing and were excluded, resulting in a final study sample of 2,489 (354 from the original cohort and 2,135 from the offspring cohort). All participants gave written, informed consent and the institutional review board of the Boston Medical Center, Boston, Massachusetts approved the study.
Outcome definition
The definition of stage 3 CKD from the KDOQI clinical practice guidelines of the National Kidney Foundation was used (estimated glomerular filtration rate (eGFR) <60 ml/min/1.73m2).29 Incident stage 3 CKD was defined as cases of stage 3 CKD present at exam cycle 24 (original cohort) and exam cycle 8 (offspring cohort) in individuals with eGFR > 60 ml/min/1.73m2 at the earlier respective baseline exam (i.e. exam 15 for the original cohort and exam 6 for the offspring cohort). GFR was estimated using the 4-variable Modification of Diet in Renal Disease (MDRD) Study equation.30 Serum creatinine in FHS was measured using a modified Jaffé reaction. A 2-step calibration process for serum creatinine was used to reduce potential inter-laboratory variability.19 Determinations of serum creatinine were made in a central laboratory, although the laboratories used for Original cohort and Offspring participants were not the same.
Covariate assessment
Covariates obtained at the baseline examination were used in the analyses. Two blood pressure measurements were taken using a mercury sphygmomanometer after 5 minutes rest, and the average was recorded. Hypertension was defined as a systolic BP ≥ 140 mm Hg or a diastolic BP ≥ 90 mm Hg, and/or the use of medications. Diabetes mellitus was defined as a fasting plasma glucose level of 126 mg/dL (7 mmol/L) or higher or the use of hypoglycemic medications in the Offspring and by a random glucose of at least 200 mg/dl or treatment in the Original Cohort. Proteinuria was assessed both by urine dipstick assay (Ames Labstix in Elkhardt, Indiana) and considered present when “trace or higher” was recorded.
Selection of Risk Alleles, Genotyping, and Genotype-Score Construction
We used the recent CKDGen GWAS26 to select 16 SNPs confirmed as being associated with eGFR and stage 3 CKD in populations of European ancestry. Details of these SNPs are presented in Table S1 (available as online supplementary material). Direct genotyping of these SNPs was performed at KBiosciences (www.kbioscience.com) using a TaqMan (Applied Biosystems, www.appliedbiosystems.com) real-time PCR assay. We constructed a single-point genotype score using these 16 loci, ranging from 0 to 32 on the basis of the number of risk alleles. Construction of the risk score is dependent on a full set of SNP data on all individuals in order to maintain the integrity of the distribution. Thus, individuals with 1 or 2 SNPs of the 16 SNPs missing (n=193) had the missing SNPs imputed in order to preserve the components of the risk score. Missing genotypes for each SNP were replaced by the population mean genotype value estimated using genotyped individuals.31 For example, if a SNP genotype frequency was 0.16, 0.48, 0.36 for genotypes 0, 1, 2, then all missing genotypes for that SNP were imputed with the value 1.2 (i.e. 0*0.16+1*0.48+2*0.36).
Statistical analyses
The cumulative incidence of stage 3 CKD was estimated by dividing the number of new stage 3 CKD cases as of the end of follow-up by the total number at risk. We analyzed two examination periods (Original cohort examinations 15 to 24, and Offspring examinations 6 to 8) to determine risk of stage 3 CKD over an average of 10.8 years of follow-up. Logistic-regression models, with generalized estimating equations to account for relatedness in the sample,32 were used to examine the association between the genotype score and the risk of incident stage 3 CKD during the follow-up period. Participants with stage 3 CKD at the baseline examination of each period were excluded, and new cases of stage 3 CKD were enumerated at the end of the last examination in the respective period. Two regression models were applied to test for association, the first adjusted for age and sex and the second adjusted for CKD risk factors identified in previous reports:33 age, sex, cohort status, baseline eGFR, hypertension, diabetes and proteinuria. Adjustments for hypertension, diabetes and proteinuria were done at baseline. Model discrimination was estimated by C statistic, which is the probability that a scoring system successfully ranks 2 randomly selected observations in terms of an outcome of interest.34 We tested the relative discriminatory capability of the models with the genotype score compared with models without the genotype score by comparing the respective C statistics using a nonparametric approach.35. Statistical analyses were performed using R and SAS version 9.1 (SAS Institute). A two-tailed P-value of 0.05 was the chosen threshold for statistical significance.
Secondary analyses
We performed 5 secondary analyses. First, as the risk of stage 3 CKD may not be identical for all genetic markers, we constructed an allele-weighted genetic risk score, which weights each of the 16 renal loci by the magnitude of its respective beta-coefficient, and examined the ability of this model to predict incident stage 3 CKD as per the primary analysis. Second, to investigate the possibility of improved performance of the genetic score in younger participants, we performed an analysis stratified by age above and less than or equal to 60 years. Third, as the CKD-EPI equation may perform better than the MDRD Study equation at higher levels of GFR,36 we examined the performance of the risk score in predicting eGFR < 60 ml/min/1.73m2 by CKD-EPI. Fourth, we examined the performance of the score in participants free of diabetes. Finally, to assess whether a using a higher definition of proteinuria would improve specificity, we performed our primary analysis using proteinuria ≥1+ as the cut-off.
Results
Genetic Risk Score and Incident Stage 3 CKD
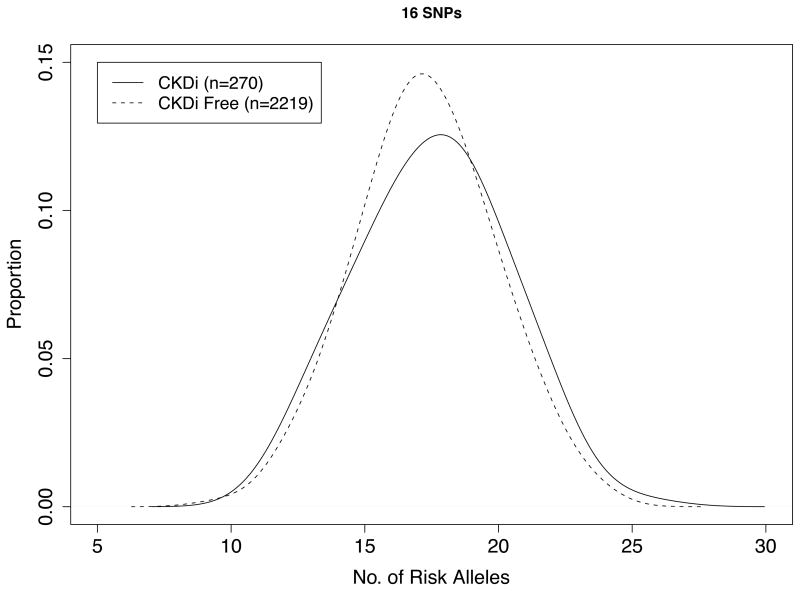
Baseline characteristics of the 2,489 participants are shown in Table 1. There were 270 cases (10.8%) of incident stage 3 CKD that developed over a mean of 10.8 years of follow-up. The mean (±SD) genetic risk score was 17.5±2.8 among those who developed stage 3 CKD and 17.3±2.6 among those who did not (P-value for genotype score difference=0.2; Figure 1).
Table 1.
Characteristics of Participants in the Framingham Original Cohort and Offspring Study Samples.
| Baseline* | Follow-up* | |
|---|---|---|
| Duration of follow-up, years | 10.8 +/− 3.3 | - |
| Number of participants | 2,489 | 2,489 |
| Age, years | 58 +/− 9 | 68 +/− 10 |
| Female sex, % | 54.3 (1351) | 54.3 (1351) |
| Hypertension,% | 34.8 (865) | 59.3 (1476) |
| Diabetes,% | 6.6 (165) | 26.4 (656) |
| CKD stage 3, %** | 0 | 10.8 (270) |
| Baseline eGFR, ml/min/1.73m2 | 92 +/− 24 | 81 +/− 18 |
| Dipstick proteinuria, %*** | 14.8 (365/2474) | - |
Data presented as mean +/− standard deviation for continuous variables and % (n) for categorical variables. Abbreviations: CKD, chronic kidney disease; eGFR estimated glomerular filtration rate.
Baseline examination was either at the 15th cycle of the Original cohort (1977–1979) or the 6th cycle of the Offspring cohort (1995–1998); follow-up was at the 24th cycle of the Original cohort (1995–1998) and the 8th cycle of the Offspring cohort (2005–2008) respectively.
defined as eGFR < 60 ml/min/1.73m2
Trace or above
Figure 1. Genotype Score Distribution among FHS Participants with and without stage 3 CKD.
Figure shows distribution of participants according to genotype score, stratified according to persons in whom stage 3 CKD developed over the follow-up period (n = 270) and those in whom stage 3 CKD did not develop (n = 2219).
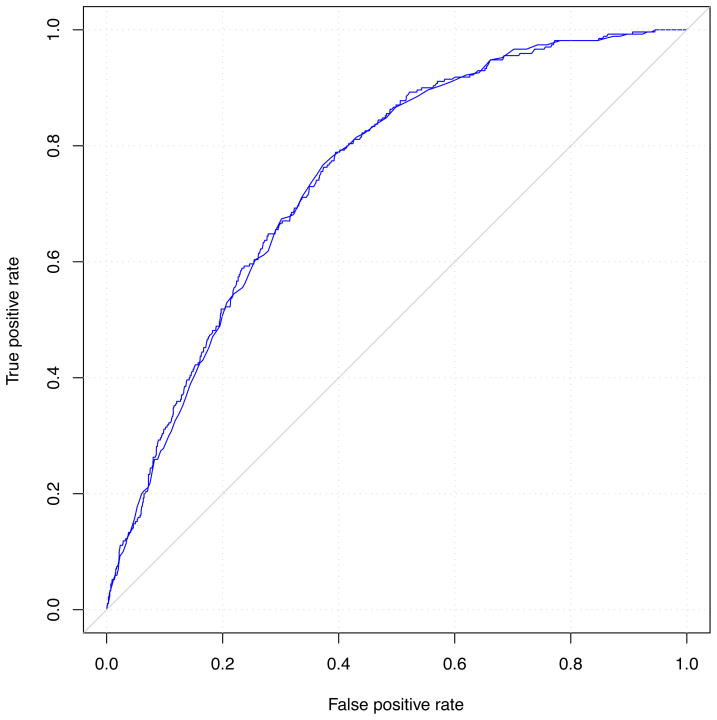
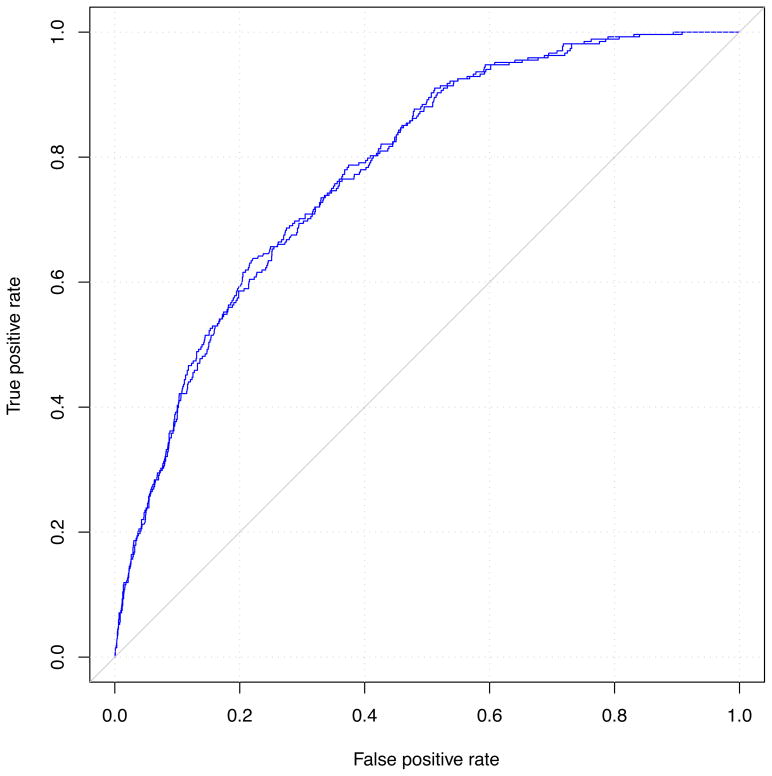
Logistic regression models for predicting incident stage 3 CKD are presented in Table 2. The age-sex adjusted odds ratio for stage 3 CKD was 1.06 per risk allele (95% CI, 1.01–1.11; p=0.03). The C statistic for this model was 0. 751, which offered no discriminatory improvement over the age-sex model without the genetic risk score (C statistic, 0.748; p-value for comparison of C statistics, 0.3). The genetic risk score was not a significant predictor of incident stage 3 CKD (p=0.07) in a multivariable model adjusted for known CKD risk factors (age, sex, baseline eGFR, hypertension, diabetes and proteinuria).
Table 2.
Risk of Incident stage 3 CKD Associated with Genotype Score and Phenotypic CKD Risk Factors.
| Model | Prediction Model | P-value | |
|---|---|---|---|
| Without Genotype Score | With Genotype Score | ||
| Model 1: Age and sex adjusted | |||
| Genotype score OR (95% CI) | - | 1.06 (1.01–1.11) | 0.03 |
| C statistic | 0.748 | 0.751 | 0.3b |
| Model 2: Multivariable modela | |||
| Genotype score OR (95% CI) | - | 1.05 (1.00–1.11) | 0.07 |
| C statistic | 0.780 | 0.781 | 0.6b |
Results presented as odds of incident stage 3 CKD per 1-point increase in the genotype score using an additive model CKD, chronic kidney disease; OR: Odds ratio; CI: confidence intervals.
Multivariable model adjusted for age, sex, cohort status, baseline estimated glomerular filtration rate, hypertension, diabetes, and proteinuria.
P value for difference in C statistic in the model with and without the genotype score
Secondary analyses
We also considered whether the genotype score might perform better in younger participants by stratifying by age above or below 60 years. Among those aged ≤60 years, there were 1,530 participants with 77 cases of stage 3 CKD (5.0%). There were 959 participants aged over 60 years, with 193 cases of CKD (20.1%). We found no evidence of improved performance of the genotype score in younger participants, in whom the risk score was non-significant (age-sex adjusted odds ratio for stage 3 CKD in participants ≤60 years, 1.04 per risk allele; 95% CI, 0.95–1.14; p=0.4). The risk score was also not statistically significant in participants over 60 years of age (age-sex adjusted odds ratio in > 60 years, 1.06 per risk allele; 95% CI, 0.99–1.13; p-value=0.1).
There were 2,323 participants without diabetes at baseline and 232 (10.0%) developed stage 3 CKD during follow-up. Although not statistically significant, the age- and sex-adjusted odds ratio was of comparable to the primary analysis (OR, 1.05 per copy; 95% CI, 0.99–1.11; p=0.09). The risk score did not improve discrimination in the age- and sex-adjusted (C statistic without genetic score, 0.749; C statistic with genetic score, 0.751; p-value for comparison of C statistics, 0.4) and multivariable-adjusted models (C statistic without genetic score, 0.780; C statistic with genetic score, 0.780; p-value for comparison of C statistics = 0.2) in this diabetes-free subgroup.
We performed our primary analysis using eGFR < 60ml/min/1.73 m2 by the CKD-EPI equation to define stage 3 CKD.36 The mean (±SD) genetic risk score was 17.4±2.7 among those who developed CKD and 17.3±2.6 among those who did not (P-value for genotype score difference=0.4). The age- and sex-adjusted odds ratio for stage 3 CKD was 1.04 per additional risk allele (95% CI, 0.99–1.09). Although the β–coefficient was comparable to that derived from the MDRD Study equation, it was not significant for the CKD-EPI equation (p=0.09). Furthermore, the risk score was not statistically significant in a multivariable model adjusted for standard CKD risk factors (p=0.2).
We modeled an additional risk score with each SNP weighted in proportion to the magnitude of its respective β-coefficient from our primary meta-analysis.24 The mean (±SD) weighted genetic risk score was 18.2 among those who developed stage 3 CKD and 17.7 among those who did not (P-value for genotype score difference=0.01). The age- and sex-adjusted odds ratio for stage 3 CKD was 1.08 per 1-point increase in the score (95% CI, 1.03–1.13; p=0.002), comparable in magnitude to our primary analysis using a single-point SNP risk score. However, addition of the risk score did not improve discrimination in models including age and sex (p-value for difference in C statistics = 0.2) or standard CKD risk factors (p-value for difference in C statistics = 0.4).
To assess whether a using a higher cut-off for the proteinuria covariate would improve specificity, we performed our primary analysis using proteinuria ≥1+ as the cutoff. This captured 1.4% of our sample (n=36). The results were similar to the primary analysis, but did not reach statistical significance (OR, 1.05; 95% CI, 1.00–1.11; p=0.07).
Discussion
The findings of the present study are twofold. First, we found that a genetic risk score for incident stage 3 CKD, comprising 16 risk variants, was associated with a 6% increase in the relative risk of stage 3 CKD per risk allele, after adjusting for age and sex. Second, the genetic risk score did not offer improved discrimination over what can be achieved from knowledge of existing clinical CKD risk factors.
Although this is the first study to construct and assess a genotype score for kidney disease, our findings are consistent with similar studies that have been performed for other phenotypes. For example, several studies have attempted to predict incident type 2 diabetes using combinations of SNPs. In a prior FHS study, a risk score based on 18 type 2 diabetes-associated genotypes did predict new cases, but provided a negligible improvement in risk prediction over knowledge of common, clinical risk factors alone.37 Similarly, in a subsequent study from the United Kingdom, a panel of 20 risk alleles for type 2 diabetes was inferior to the Cambridge and Framingham offspring clinical type 2 diabetes risk scores in discriminating incident cases of type 2 diabetes, and inclusion of the genetic panel failed to improve discrimination, calibration, or reclassification when added to these models.38 Similar attempts to use genotype-based risk modeling for the prediction of incident myocardial infarction,39 coronary heart disease40 or coronary artery disease, myocardial and stroke in women41 have also failed to improve on risk predictions based on traditional risk factors.
There are several possible reasons for the lack of improvement in discriminatory power provided by our genotype score. For example, the 16 SNPs we included account for only a minority of eGFR heritability. Taken together, all of the genetic loci explain only 1.43% of population variability in eGFR.26 As more CKD risk variants are uncovered, including them in the genotype score may explain more of the genetic risk implied by heritability studies and could therefore improve discrimination.42 However, as yet-to-be identified common variants would be expected to have smaller effect sizes, the scope for dramatic improvements in predictive power is limited. As such, rare variants may explain more of the “missing heritability” and are currently the focus of ongoing discovery efforts. Alternatively, it is possible that acquired risk factors are substantially more powerful determinants of the near-term stage 3 CKD risk than are genetic influences.
These results argue against the idea of genetic fatalism. They suggest that personalized medicine, or individualized disease prediction based on genetic information, are unlikely to be a major translational application of this phase of genetic discovery. Instead, the utility of recently identified stage 3 CKD and GFR-associated genotypes is more likely to stem from the insight they provide into disease pathogenesis and potential novel therapeutic targets.
Our study has several strengths, including the large, community-based sample, the long duration of follow-up and the rigorous and detailed assessment of risk factors including measures of baseline kidney function and proteinuria. A few limitations also warrant mention. First, Framingham participants are of European ancestry, and our genotype score may not be valid in different ancestral groups due to allelic variation. Also, a potential reason for the failure of the genetic risk score may relate to the heterogeneity of underlying conditions that may result in stage 3 CKD.
A genetic risk score generated from 16 known CKD risk alleles did not predict new cases of stage 3 CKD in the community beyond knowledge of common, clinical risk factors alone. These findings suggest that a continued focus on the early identification of adverse clinical risk factors should remain the cornerstone of kidney disease risk prediction.
Supplementary Material
Table S1: Details of the 16 SNPs used to construct the genotype score
Figure 2. Comparison of receiver-operating characteristic curves for models with and without the genetic risk score and incident stage 3 CKD.
Panel A is adjusted for age and sex; panel B is adjusted for standard CKD risk factors.
Acknowledgments
Support: This research was conducted in part using data and resources from the Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine. This work was partially supported by the National Heart, Lung and Blood Institute’s Framingham Heart Study (Contract No. N01-HC-25195). The sponsors had no role in the conduct or interpretation of the study. The corresponding author had full access to all data in the study and had final responsibility for the decision to submit for publication.
Footnotes
Financial Disclosure: The authors declare that they have no other relevant financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Coresh J, Selvin E, Stevens LA, et al. Prevalence of chronic kidney disease in the United States. JAMA. 2007 Nov 7;298(17):2038–2047. doi: 10.1001/jama.298.17.2038. [DOI] [PubMed] [Google Scholar]
- 2.Zhang QL, Rothenbacher D. Prevalence of chronic kidney disease in population-based studies: systematic review. BMC Public Health. 2008;8:117. doi: 10.1186/1471-2458-8-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chadban SJ, Briganti EM, Kerr PG, et al. Prevalence of kidney damage in Australian adults: The AusDiab kidney study. J Am Soc Nephrol. 2003 Jul;14(7 Suppl 2):S131–138. doi: 10.1097/01.asn.0000070152.11927.4a. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Wildman RP, Gu D, et al. Prevalence of decreased kidney function in Chinese adults aged 35 to 74 years. Kidney Int. 2005 Dec;68(6):2837–2845. doi: 10.1111/j.1523-1755.2005.00757.x. [DOI] [PubMed] [Google Scholar]
- 5.Perkovic V, Cass A, Patel AA, et al. High prevalence of chronic kidney disease in Thailand. Kidney Int. 2008 Feb;73(4):473–479. doi: 10.1038/sj.ki.5002701. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka H, Shiohira Y, Uezu Y, Higa A, Iseki K. Metabolic syndrome and chronic kidney disease in Okinawa, Japan. Kidney Int. 2006 Jan;69(2):369–374. doi: 10.1038/sj.ki.5000050. [DOI] [PubMed] [Google Scholar]
- 7.Tareen N, Zadshir A, Martins D, Pan D, Nicholas S, Norris K. Chronic kidney disease in African American and Mexican American populations. Kidney Int Suppl. 2005 Aug;97:S137–140. doi: 10.1111/j.1523-1755.2005.09723.x. [DOI] [PubMed] [Google Scholar]
- 8.Muntner P, Vupputuri S, Coresh J, Uribarri J, Fox CS. Metabolic abnormalities are present in adults with elevated serum cystatin C. Kidney Int. 2009 Jul;76(1):81–88. doi: 10.1038/ki.2009.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu CY, McCulloch CE, Curhan GC. Epidemiology of anemia associated with chronic renal insufficiency among adults in the United States: results from the Third National Health and Nutrition Examination Survey. J Am Soc Nephrol. 2002 Feb;13(2):504–510. doi: 10.1681/ASN.V132504. [DOI] [PubMed] [Google Scholar]
- 10.Ensrud KE, Lui LY, Taylor BC, et al. Renal function and risk of hip and vertebral fractures in older women. Arch Intern Med. 2007 Jan 22;167(2):133–139. doi: 10.1001/archinte.167.2.133. [DOI] [PubMed] [Google Scholar]
- 11.Fried LF, Shlipak MG, Crump C, et al. Renal insufficiency as a predictor of cardiovascular outcomes and mortality in elderly individuals. J Am Coll Cardiol. 2003 Apr 16;41(8):1364–1372. doi: 10.1016/s0735-1097(03)00163-3. [DOI] [PubMed] [Google Scholar]
- 12.Weiner DE, Tighiouart H, Amin MG, et al. Chronic kidney disease as a risk factor for cardiovascular disease and all-cause mortality: a pooled analysis of community-based studies. J Am Soc Nephrol. 2004 May;15(5):1307–1315. doi: 10.1097/01.asn.0000123691.46138.e2. [DOI] [PubMed] [Google Scholar]
- 13.McClellan WM, Flanders WD. Risk factors for progressive chronic kidney disease. J Am Soc Nephrol. 2003 Jul;14(7 Suppl 2):S65–70. doi: 10.1097/01.asn.0000070147.10399.9e. [DOI] [PubMed] [Google Scholar]
- 14.de Zeeuw D, Ramjit D, Zhang Z, et al. Renal risk and renoprotection among ethnic groups with type 2 diabetic nephropathy: a post hoc analysis of RENAAL. Kidney Int. 2006 May;69(9):1675–1682. doi: 10.1038/sj.ki.5000326. [DOI] [PubMed] [Google Scholar]
- 15.Ruggenenti P, Perna A, Gherardi G, et al. Renoprotective properties of ACE-inhibition in non-diabetic nephropathies with non-nephrotic proteinuria. Lancet. 1999 Jul 31;354(9176):359–364. doi: 10.1016/S0140-6736(98)10363-X. [DOI] [PubMed] [Google Scholar]
- 16.Makino H, Nakamura Y, Wada J. Remission and regression of diabetic nephropathy. Hypertens Res. 2003 Jul;26(7):515–519. doi: 10.1291/hypres.26.515. [DOI] [PubMed] [Google Scholar]
- 17.Kshirsagar AV, Bang H, Bomback AS, et al. A simple algorithm to predict incident kidney disease. Arch Intern Med. 2008 Dec 8;168(22):2466–2473. doi: 10.1001/archinte.168.22.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fox CS, Gona P, Larson MG, et al. A multi-marker approach to predict incident CKD and microalbuminuria. J Am Soc Nephrol. Dec;21(12):2143–2149. doi: 10.1681/ASN.2010010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fox CS, Larson MG, Leip EP, Culleton B, Wilson PW, Levy D. Predictors of new-onset kidney disease in a community-based population. JAMA. 2004 Feb 18;291(7):844–850. doi: 10.1001/jama.291.7.844. [DOI] [PubMed] [Google Scholar]
- 20.O’Seaghdha CM, Hwang SJ, Upadhyay A, Meigs JB, Fox CS. Predictors of incident albuminuria in the Framingham Offspring cohort. Am J Kidney Dis. Nov;56(5):852–860. doi: 10.1053/j.ajkd.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lei HH, Perneger TV, Klag MJ, Whelton PK, Coresh J. Familial aggregation of renal disease in a population-based case-control study. J Am Soc Nephrol. 1998 Jul;9(7):1270–1276. doi: 10.1681/ASN.V971270. [DOI] [PubMed] [Google Scholar]
- 22.Chambers JC, Zhang W, Lord GM, et al. Genetic loci influencing kidney function and chronic kidney disease. Nature genetics. 2010 May;42(5):373–375. doi: 10.1038/ng.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kottgen A, Glazer NL, Dehghan A, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nature genetics. 2009 Jun;41(6):712–717. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kottgen A, Pattaro C, Boger CA, et al. New loci associated with kidney function and chronic kidney disease. Nature genetics. 2010 May;42(5):376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allison M. Is personalized medicine finally arriving? Nat Biotechnol. 2008 May;26(5):509–517. doi: 10.1038/nbt0508-509. [DOI] [PubMed] [Google Scholar]
- 26.Kottgen A, Pattaro C, Boger CA, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. Apr 11; doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dawber TR, Kannel WB. The Framingham study. An epidemiological approach to coronary heart disease. Circulation. 1966 Oct;34(4):553–555. doi: 10.1161/01.cir.34.4.553. [DOI] [PubMed] [Google Scholar]
- 28.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families. The Framingham offspring study. Am J Epidemiol. 1979 Sep;110(3):281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 29.National Kidney Foundation. K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am J Kidney Dis. 2002 Feb;39(2 Suppl 1):S1–266. [PubMed] [Google Scholar]
- 30.Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med. 1999 Mar 16;130(6):461–470. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 31.Little RJARD. Statistical Analysis with Missing Data. 2002. p. 61. [Google Scholar]
- 32.Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986 Mar;42(1):121–130. [PubMed] [Google Scholar]
- 33.O’Seaghdha CMLA, Massaro JM, Coresh J, Astor BC, Fox CS. J Am Soc Nephrol. Vol. 21. ASN World Congress of Nephrology; Denver CO: 2010. Development of a Risk Score for Chronic Kidney Disease in Population-Based Studies [abstract] [Google Scholar]
- 34.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982 Apr;143(1):29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 35.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988 Sep;44(3):837–845. [PubMed] [Google Scholar]
- 36.Levey AS, Stevens LA, Schmid CH, et al. A new equation to estimate glomerular filtration rate. Annals of internal medicine. 2009 May 5;150(9):604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meigs JB, Shrader P, Sullivan LM, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med. 2008 Nov 20;359(21):2208–2219. doi: 10.1056/NEJMoa0804742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talmud PJ, Hingorani AD, Cooper JA, et al. Utility of genetic and non-genetic risk factors in prediction of type 2 diabetes: Whitehall II prospective cohort study. BMJ. 340:b4838. doi: 10.1136/bmj.b4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi L, Ma J, Qi Q, Hartiala J, Allayee H, Campos H. Genetic Risk Score and Risk of Myocardial Infarction in Hispanics. Circulation. Jan 17; doi: 10.1161/CIRCULATIONAHA.110.976613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ripatti S, Tikkanen E, Orho-Melander M, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. Oct 23;376(9750):1393–1400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paynter NP, Chasman DI, Pare G, et al. Association between a literature-based genetic risk score and cardiovascular events in women. JAMA. Feb 17;303(7):631–637. doi: 10.1001/jama.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cook NR. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation. 2007 Feb 20;115(7):928–935. doi: 10.1161/CIRCULATIONAHA.106.672402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Details of the 16 SNPs used to construct the genotype score