Abstract
Quality control processes regulate the proteome by determining whether a protein is to be folded or degraded. Hsp90 is a hub in the network of molecular chaperones that maintain this process because it promotes both folding and degradation, in addition to regulating expression of other quality control components. The significance of Hsp90’s role in quality control is enhanced by the function of its clients, which include protein kinases and transcription factors, in cellular signaling. Inhibition of Hsp90 with small molecules results in rapid degradation of such clients via the ubiquitin/proteasome pathway, and also in induction of the Hsp70 molecular chaperone. These two events result in markedly different outcomes depending on cell type. For tumor cells there is a profound loss of signaling in growth promoting pathways. By contrast, increased amounts of Hsp70 in neuronal cells ameliorate the toxicity that is associated with formation of aggregates observed in neurodegenerative conditions. In this review we discuss the mechanisms underlying these differential effects of Hsp90 inhibition on the quality control of distinct client proteins.
Introduction
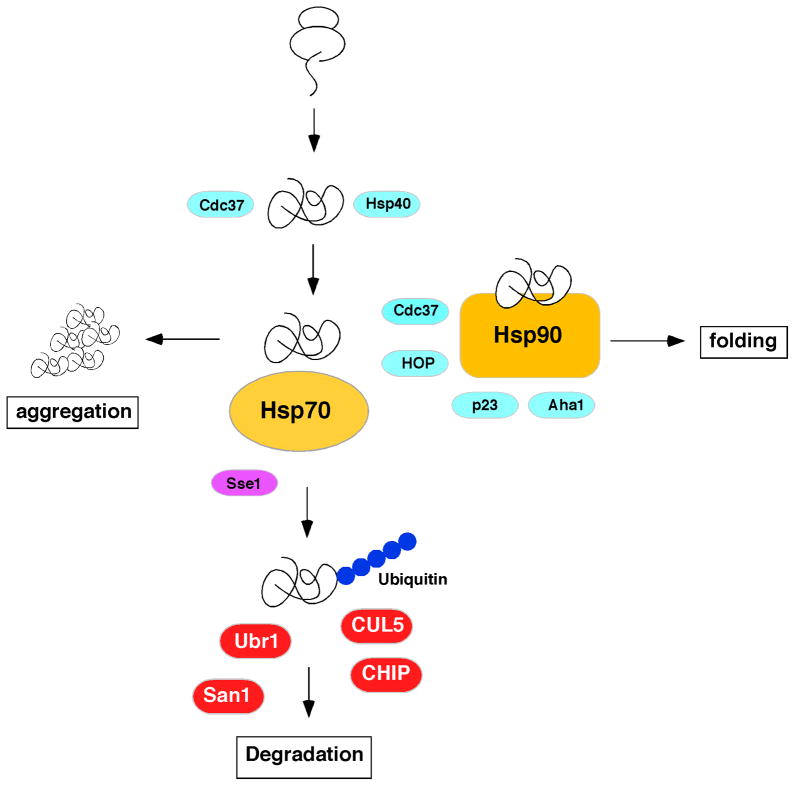
Quality control processes maintain proteome integrity by ensuring that newly made proteins fold rather than aggregate, and by promoting the degradation of misfolded proteins. Three cellular machineries control these processes: molecular chaperones interacting with nascent and unfolded/misfolded proteins to determine their fate, the ubiquitin/proteasome system for degradation of misfolded proteins, and the autophagic system for removal of aggregates and other misfolded proteins [1, 2]. Figure 1 shows in outline the relationship between molecular chaperones and components of the ubiquitin proteasome pathway in relation to the quality control process and the three fates for a protein: folding, degradation or aggregation.
FIGURE 1. Outline of the Cytosolic Quality Control Process.
Three major outcomes for a newly synthesized protein are shown: Folding, aggregation and degradation via the ubiquitin/proteasome system. Proteins shown in blue are co-chaperones that can promote client protein folding. Sse1, shown in purple, promotes degradation of Hsp90 clients. Select ubiquitin ligases known to promote degradation of Hsp90 clients are shown in red. Individual proteins are discussed throughout the review. The process of autophagy, which clears protein aggregates is not depicted.
Quality control pathways exist because proteins fold in a unfavorable cellular environment for this process due to molecular crowding and relatively high temperatures [3]. These two conditions, when combined, favor aggregation over folding. Molecular chaperones evolved to shift the equilibrium towards folding by preventing aggregation. This is achieved via weak interactions between the molecular chaperones and the exposed hydrophobic groups of unfolded or misfolded proteins. Under normal cellular conditions the expression level of molecular chaperones is matched to the overall level of protein synthesis so that folding is the expected fate for newly made proteins. Under stressful conditions, mature proteins unfold and exceed the capacity of chaperone networks to prevent aggregation. This type of proteotoxic stress induces feedback regulation which results in increased expression of genes encoding molecular chaperones, due to de-repression of the heat shock transcription factor, Hsf1. Importantly, Hsp90 plays a role in Hsf1 repression via direct interaction. Under stress conditions, or inhibition with small molecules, Hsp90 dissociates resulting in Hsf1 activation [4]. In addition to acutely stressful events, ageing is associated with the accumulation of aggregates containing oxidatively damaged proteins. It seems clear, however, that aggregation represents an end state that the cell attempts to avoid by directing damaged or misfolded proteins to the ubiquitin/proteasome system. It is only when this system becomes overwhelmed that aggregation ensues.
The role of the Hsp90 molecular chaperone in quality control processes is essential and its deregulation can affect several diseases from cystic fibrosis and tumor progression to neurodegenerative conditions [5–7]. All of these facets of Hsp90 function are linked directly to the clients it is helping to fold, which include many proteins important for cellular signaling, including transcription factors and protein kinases. In this review we describe the mechanisms by which Hsp90 controls the fate of its clients in the context of the quality control systems of the cell.
While early studies focused on client protein folding, Hsp90’s integration into a larger quality control system was not appreciated until the benzoquinoid ansamycin, geldanamycin, was shown to be a specific inhibitor of the chaperone [8]. Subsequent studies revealed that client protein kinases and transcription factors were rapidly destroyed via the ubiquitin/proteasome pathway. This set the stage for a rapid expansion of the field that has led to the development of more than 13 small molecules currently in clinical trials as chemotherapeutics [9]. The consequence of Hsp90 inhibition to the emergence of a rare phenotype was later established with the demonstration that geldanamycin treatment led to emergence of hidden morphological characteristics in Drosophila melanogaster and Arabidopsis, as well as differential responses to stress in yeast [10–13]. These findings led to the hypothesis that Hsp90 acts as a capacitor by helping to fold polymorphic variants of the same protein even though their folding efficiencies vary. This was observed with v-Src, an unstable protein kinase, which has a greater requirement for Hsp90 than its cellular counterpart c-Src [14]. Similarly, mutant forms of p53 and B-Raf protein kinase have stronger interactions with molecular chaperones than do their wild type counterparts [15][16, 17]. Hsp90, therefore, sits at the nexus of folding and degradation pathways, with small molecule inhibitors pushing the equilibrium strongly towards the destructive fate.
Small Molecule Hsp90 Inhibitors and Client Protein Degradation: Role of molecular chaperones in fate determination
It is well established that Hsp90 sits at the distal end of the pathway of chaperone interactions for newly made transcription factors and protein kinases [18]. For example, newly made nuclear receptors such as the glucocorticoid receptor and the progesterone receptor interact with Hsp70 and Hsp40 chaperones prior to recruitment to Hsp90 by the actions of Hop/Sti1 – the Hsp organizing protein [19]. Likewise, protein kinases are recruited to Hsp90 via the actions of Sti1 and the kinase-specific chaperone Cdc37 [20–22]. The entry of Hsp90 into this system is associated with the folding of the client itself. Although the mechanism by which client protein folding is coordinated with Hsp90’s reaction cycle is largely unclear, there appears to be a closing in of the chaperone around its clients when they interact [23]. What is clear, however, is that many small molecule inhibitors act by means of competitive inhibition of Hsp90’s ATPase, which occurs in the N-domain of the chaperone. The specificity of such inhibitors is related to the unusual bent conformation of ATP as it resides in the N-domain [24].
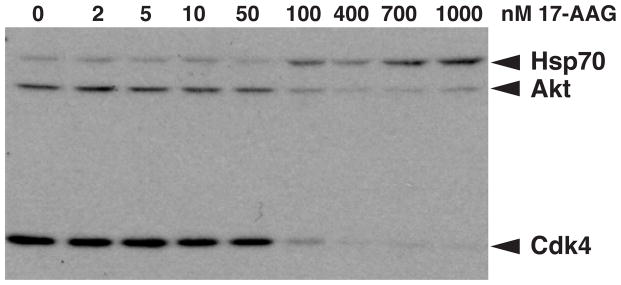
The inhibition of Hsp90 with N-terminal inhibitors results in rapid degradation via the ubiquitin/proteasome pathway for many, but not all, clients. This outcome is observed for many protein kinase clients including Akt and Cdk4 as shown in Figure 2. Note also how Hsp70 is induced in a reciprocal manner with protein kinase degradation (Fig. 2) as a result of Hsf1 de-repression. Protein kinases susceptible to loss of Hsp90 function include newly made protein kinases as well as mature clients that enter into multiple rounds of interaction [25, 26]. The mechanisms by which inhibitors promote degradation are not entirely clear. It seems likely that N-terminal inhibitors do not reprogram Hsp90 to actively promote degradation, but rather, that degradation is a consequence of blocking the folding pathway. N-terminal Hsp90 inhibitors appear to promote the reorganization of chaperone complexes. For protein kinases, there is an increase in Hsp70 binding to the client coupled with a reduction in Hsp90 binding. Such complexes are likely to contain Sti1/Hop, but not Cdc37 [27]. These findings correlate with the role of Hsp70 in degradation processes [28]. As described in more detail below, this may have a lot to do with Hsp70’s ability to coordinate with different ubiquitin ligases, including CHIP and Ubr1 [29–31].
FIGURE 2. Effect of Hsp90 Inhibition on Protein Kinase Clients and Induction of Hsp70.
A Western blot analysis is shown based on a dose-dependent study of 17-AAG in mouse Ba/F3 cells over a 24 hour period. Proteins indicated include the Hsp70 molecular chaperone as well as Akt and Cdk4 protein kinase clients of Hsp90.
Several molecular chaperones and co-chaperones other than Hsp70 function in both folding and degradation processes. Sse1, the Hsp110 chaperone that acts as a nucleotide exchange factor for Hsp70 [32] is required for degradation of client protein kinases in cells treated with geldanamycin [33]. This action requires direct interaction between Sse1 and Hsp70 and occurs before the protein is ubiquitinylated. By contrast, loss of function of the Hsp40 co-chaperone, Ydj1, leads to rapid degradation of protein kinases, suggesting it has a protective role for newly made Hsp90 clients [34]. However, several other studies showed that Hsp40 co-chaperones are required for degradation of unstable or mutant proteins, suggesting that this class of co-chaperone may have diverse functions in the quality control process [35–39]. Ydj1 also functions as an Hsp70 co-chaperone, and stimulates Hsp70’s ATPase, thereby stabilizing Hsp70:client complexes [40]. The distinct roles of Ydj1 and Sse1 as Hsp70 co-chaperones suggests a model in which the stable association of a client with Hsp70 is protective against degradation for newly made proteins. By contrast, complex dissociation is required for degradation when folding pathways are inhibited such as with geldanamycin.
The protective role of Ydj1 for newly made Hsp90 client protein kinases is similar to the actions of Cdc37, the kinase specific chaperone. Cdc37 is well characterized for its ability to interact directly with protein kinases and also with Hsp90 [41]. This action serves to both protect the newly made protein kinase and also promote stable interaction with Hsp90 [22, 42, 43]. Indeed, Cdc37 has a wide- ranging role in the biogenesis of the protein kinome and serves to protect an estimated 75% of all protein kinases in yeast against rapid degradation either during or shortly after translation via the ubiquitin/proteasome pathway [42]. Importantly, the actions of Cdc37 and Ydj1 do not overlap since they cannot substitute for each other in the protection of newly made protein kinases [34]. This specificity suggests that they each interact with a unique region of the newly made kinase client prior to transfer to Hsp70 (via Ydj1) and Hsp90 (via Cdc37). These transfer reactions lead to further stabilization and folding unless the system is challenged by an Hsp90 inhibitor. In this case, the chaperone apparatus is reprogrammed away from its protective function and the pro-degradative roles of Hsp70 and Hsp110 are enabled.
Hsp90 can promote degradation
The sense that Hsp90 inhibitors promote degradation by loss of function can be misleading. There are several reports, for example, showing that Hsp90 is required for degradation of membrane proteins in the endoplasmic reticulum. These include cytochrome p450 2E1, mutant CFTRΔF508 and Apolipoprotein B. In each of these cases, geldanamycin was found to inhibit degradation in cell free systems when the substrate was added to cytosolic extracts in the form of microsomes. [44–46]. Degradation of a mutant form of the insulin receptor was also reduced upon injection of Hsp90 antibodies into cells [47]. By contrast, studies in yeast showed that a cytosolic protein, albeit one that was heterologously expressed can be degraded in an Hsp90-dependent manner. In this case, using VHL as a model protein, Hsp90 was shown to be required for degradation but not for folding in vivo [39].
The example of CFTRΔF508 is intriguing since this mutant form is stabilized upon Hsp90 inhibition in vitro, while the wild type counterpart is destabilized in cells treated with geldanamycin [48]. This could reflect differences in experimental approach, but a more compelling possibility is that a distinction in folding potential of the client may be important for determining the relative contribution of Hsp90 to the quality control process. Evidence for this comes from mass spectrometric analysis of wild type and ΔF508 versions of CFTR. Strikingly, the latter is complexed with Hsp90 co-chaperones not observed to associate with wild type CFTR, including Hop/Sti1 [7], which was also required for degradation of VHL [39]. Furthermore, modulation of Aha1, which catalyzes Hsp90’s ATPase, results in profound changes to the fate of CFTRΔF508. Decreasing Aha1 levels results in stabilization of CFTRΔF508 and a shift towards the folded form that translocates to the plasma membrane, while Aha1 overexpression leads to increased CFTRΔF508 turnover [7]. These findings suggest that the chaperone apparatus can be reprogrammed depending on the folding status of the client, and that clients whose folding potential is low eventually interact with a chaperone cohort that promotes degradation. However, a certain amount of caution is warranted in extrapolating results from the study of one client. While downregulation of Aha1 promotes stabilization of CFTRΔF508, it also sensitizes cells to Hsp90 inhibitors, which could reflect a relative destabilization of other clients [49].
In addition, recent studies suggest that some membrane proteins including CFTR can be removed from the plasma membrane in a chaperone-dependent manner for degradation via the lysosome rather than the proteasome [50–52].
Role of Ubiquitin ligases in the degradation of Hsp90 client proteins
Ubiquitinylation is important for the targeting of misfolded protein substrates to the proteasome. Ubiquitin is first activated via a thioester linkage catalyzed by an E1 enzyme prior to transfer to the E2 ubiquitin conjugating enzymes. Final transfer of ubiquitin to the substrate requires the action of the E3 or ubiquitin ligase, which acts in association with the E2s. Ubiquitin chains, that grow via lysine 48 linkage conventionally, become targets for receptors on the 19S regulatory particle of the proteasome [53, 54]. Ubiquitin is removed prior to substrate transfer to the 20S catalytic core of the protease. The family of ubiquitin ligases is large and diverse and several are known to specifically target misfolded proteins to the proteasome and to work with molecular chaperones in this process [55, 56].
CHIP
The best-characterized ubiquitin ligase that functions in cytosolic quality control is CHIP (Carboxyl terminus of Hsc70-Interacting Protein). CHIP is a 35 kDa protein containing three tandem tetratricopeptide (TPR) motifs in its NH2-terminal region that interact directly with Hsp70 and Hsp90 [57–59]. At its C-terminus is the U-box [60] that is a modified version of the RING finger domain that is responsible for the intrinsic E3 ubiquitin ligase activity [61]. The E2s that function with CHIP belong to the mammalian UBCH5 family that is closely related to yeast Ubc4 and Ubc5 [60, 61]. CHIP has been characterized to help in the disposal of a great many Hsp70 client proteins, and our discussion here will focus on those that also require Hsp90.
One of the best-studied clients of Hsp90 is the glucocorticoid receptor (GR;[62]). The assembly of the GR in vitro needs a minimum of 5 components; Hsp90, Hsp70, Hsp40, HOP and p23. In the absence of a ligand, Hsp90 binds to GR and maintains the receptor in a hormone binding competent conformation. Treatment of cells with geldanamycin results in loss of p23 binding to GR, disruption of the hormone-binding activity, and increased proteasomal degradation [63]. The addition of CHIP leads to a similar remodeling of the Hsp90 complex, with reduced association of HOP and release of p23 [29]. Most importantly, incubation of CHIP with in vitro translated GR led to formation of a high molecular mass, ubiquitinylated species of the receptor. The Hsp70 co-chaperone BAG-1M, which is also known to inhibit GR assembly and hormone binding [64] had no effect on GR ubiquitinylation, suggesting that the ubiquitinylation of GR is not simply a result of the remodeling of the Hsp90 complex.
Similar results on the effect of CHIP on GR have been reported in studies of the estrogen receptor (ERα). The ligand free receptor is short lived, but in the presence of a ligand the turnover of ERα can be increased or decreased. Increased turnover is promoted by the binding of the cognate ligand 17β-estradiol, ATP depletion, or by Hsp90 inhibition by geldanamycin or radicicol. Overexpression of CHIP was also found to increase the ubiquitinylation and subsequent degradation of ERα through the proteasomal pathway, while CHIP depletion by small interfering RNA resulted in increased levels of ERα and increased reporter gene transactivation. Upon GA treatment the interaction between CHIP and ERα was enhanced and the ERα degradation was further increased [65]. In the case of the androgen receptor (AR), overexpression of CHIP reduces the levels of AR in steady state and induces ubiquitylation. CHIP functions during translation or shortly after translation with its effects being prior to ligand binding and Hsp90 binding [66].
CHIP has also been shown to function in the degradation of Hsp90 client protein kinases. ErbB2/Her2 is a member of the ErbB receptor tyrosine kinase family, which includes the epidermal growth factor receptor EGFR/ErbB1 and is frequently found over-expressed in different types of cancer. Her2 homodimerizes or heterodimerizes with other members of the ErbB family and promotes survival signals. EGFR homodimers are degraded via the actions of the c-Cbl ubiquitin ligase, however activated ErbB2 has reduced association with c-Cbl. The stability of both newly synthesized and mature ErbB2 depends on Hsp90 [67, 68]. Upon geldanamycin treatment the Hsp90/ErbB2 complex destabilizes and ErbB2 becomes ubiquitinylated and degraded via the proteasome [69, 70]. The sensitivity of ErbB2 to CHIP depends on the kinase domain of the protein but not on the activity of the kinase. The effects of CHIP are enhanced with the addition of GA and are independent of c-Cbl E3 ligase activity. Several lines of evidence converge to demonstrate that CHIP acts via molecular chaperones in the degradation of ErbB2. For example, mutations in the TPR domain abrogated CHIP activity on ErbB2 and co-immunoprecipitation experiments demonstrated that CHIP binding with ErbB2 is also dependent on the TPR-domain [69].
Although CHIP can bind to both Hsp70 and Hsp90 via its TPR motifs, it is most likely to function in association with Hsp70 to promote client ubiquitinylation. CHIP competes with HOP for Hsp90 binding, suggesting that a complex that could promote folding and degradation at the same time does not exist. Based on the in vivo concentrations of Hsp70, Hsp90, HOP and CHIP, and the dissociation constants of the possible complexes, Kundrat et al. [71] determined that there should be 10 times more CHIP-Hsp70 complex compared with free CHIP under non-stress conditions, suggesting that decisions regarding degradation are taken while the client is bound to Hsp70. Upon inhibition of Hsp90, the concentration of Hsp70 in the cell increases 4-fold, much more so than for other chaperones. Therefore, upon Hsp90 inhibition, folding complexes containing Hsp70-HOP-Hsp90 complex become unproductive, and since CHIP and HOP are competitive for both chaperones, there must be a transition of the client from Hsp90:HOP:Hsp70 to Hsp70:CHIP to promote degradation [71].
While CHIP is clearly important for degradation of the client types described above, it is not unique in having this function. For example, there is reduced degradation of ErbB2 in the CHIP −/− MEFs treated with geldanamycin, but it is not abolished. This finding indicated the presence of other ubiquitin ligases that could substitute for CHIP. Also, treatment of CHIP−/− MEFs with the Hsp90 inhibitor geldanamycin promotes the degradation of endogenous GR at the same rate as in CHIP+/+ cells [72]. Furthermore, the transcription factor HIF-1α is an Hsp90 client, but its degradation upon GA treatment is independent of CHIP [73, 74]. As described below, there are now several examples of ubiquitin ligases that function in addition to CHIP in the quality control of Hsp90 clients.
Ubr1
Ubr1 was shown to act on Hsp90 clients by virtue of its ability to promote degradation of protein kinases in geldanamycin treated yeast cells [31]. The finding was surprising given that Ubr1 was previously found to function via N-end rule degradation, when the identity of the N-terminal amino acid determines a protein’s half life [75]. However, Ubr1’s action in cytosolic quality control has been observed by other investigators using mislocalized and unstable proteins [76, 77]. For Hsp90 clients, UBR1 deletion resulted in reduced degradation protein kinase clients. Similarly, Ubr1 was also required for degradation of protein kinases in cdc37 mutant cells, confirming that the quality control pathways dependent on Cdc37 and Hsp90 are similar [31]. Biochemical studies revealed that Ubr1 acts via direct interaction with its misfolded protein substrates, because it only bound with ubiquitinylated denatured luciferase and not its native counterpart. This action, however, must be viewed in the context of its relative abundance, which is thought to be very low, especially in comparison with molecular chaperones such as Hsp90, or even Cdc37 [78, 79]. Accordingly, Hsp70 itself stimulated ubiquitinylation by Ubr1 in a purified system. Together these findings suggest that Ubr1 acts in an analogous manner to CHIP.
Yeast Ubr1 has a paralog, Ubr2 that does not function via the N-end rule. Nevertheless, it does appear to have some function in protein kinase degradation but the phenotype resulting from its deletion was mild compared with a ubr1Δ strain. Similarly, deletion of the nuclear localized San1 also led to reduced degradation of a protein kinase in the presence of geldanamycin [31]. Like Ubr1, San1 acts via direct interaction with its misfolded substrate [80]. At this time it is unclear whether mammalian homologs of Ubr1 and Ubr2 have conserved roles in cytosolic quality control. Yeast San1 functions in nuclear quality control when expressed in mammalian cells, and proteins with similar function to San1 have been identified [81].
Cul5
Cul5 is one of seven members of the cullin family of E3 ubiquitin ligases. It interacts with both Hsp70 and Hsp90 and appears to play a direct role in client kinase degradation upon Hsp90 inhibition in mammalian cells [74]. Inhibition of Hsp90 resulted in impaired degradation of ErbB2, when Cul5 was knocked down or when a dominant negative form of the ubiquitin ligase was overexpressed. In addition, Cul5 was required for efficient degradation of Hif1α after Hsp90 inhibition. Although Cul5 usually functions in association with adaptor proteins ElonginB and ElonginC, these were not required for its function in the quality control of Hsp90 clients. This indicates that ubiquitin ligases engaged in quality control may function both with specific clients that depend on protein sequence, and with others that depend on protein conformation independent of sequence considerations.
Implications for Hsp90’s role in Cellular Quality Control in Neurodegeneration
Hsp90 inhibition has direct application as chemotherapy because it can induce apoptosis and other forms of cell death [82]. In addition, there is a great interest in Hsp90 inhibitors which cross the blood:brain barrier that could be used to ameliorate the symptoms of several neurodegenerative conditions [6]. The underlying principle of such an action is embodied in the dual role of Hsp90 inhibitors to promote toxic client elimination and in Hsp70 induction as a result of Hsf1 derepression.
Several recent studies have demonstrated how increasing Hsp70 levels led to a reduction in the toxicity associated with the expression of proteins that promote neurodegenerative conditions. These include mutant forms of Huntingtin, which aggregate as a consequence of expanded polyglutamine tracts [83]. Other examples include α-synuclein (Parkinson’s disease) and Aβ (Alzheimer’s disease); in each case, Hsp70 acted to reduce aggregation or fibril formation [84, 85]. What is intriguing is how Hsp90 inhibitors help to reduce toxicity of such proteins by stimulating derepression of Hsf and promoting Hsp70 induction (for review see ref. [6]). This suggests a possible pathway to a new class of therapeutics that cross the blood brain barrier.
In contrast to the effects of upregulating Hsp70, there are also examples where Hsp90 inhibition can lead to degradation of proteins that are toxic themselves or promote toxicity and neurodegeneration. Examples include the polyglutamine expanded form of the androgen receptor, which is dependent on Hsp90 for proper folding [86] and the protein kinase LRKK2 which is associated with Parkinson’s disease [87]. In Alzheimer’s disease, hyperphosphorylation of tau, which leads to its aggregation, can be inhibited by Hsp90 inhibitors that affect the stability of Cdk5 activating proteins [88].
Concluding Remarks
Hsp90 inhibition with small molecules alters quality control processes in ways that promote death of cancer cells and reduced toxicity in neurons. This seeming paradox shows how Hsp90 sits at the hub of the quality control triage process by controlling folding and degradation processes while also signaling to Hsf to maintain chaperone balance. In addition to affecting outcomes in disease models, Hsp90 also plays an important role in determining phenotype during development. The central role of Hsp90 in this process appears to be related to its capacity for folding proteins that are integral to the signaling capability of the cell, including protein kinases and transcription factors. As mentioned above, Cdc37, together with Hsp90, may be responsible for stabilizing and folding of over 75% of all protein kinases [42], at least in yeast, although the extent of its influence over transcription factor folding is still unclear. While the use of Hsp90 inhibitors in chemotherapy has received mixed results [9], there is great interest in their use in a broad spectrum of neurodegenerative conditions. Further exploration of the mechanisms by which Hsp90 influences proteome quality should lead to a more definitive understanding of the place that such inhibitors have in the treatment of a wide range of late onset diseases.
Highlights.
Small molecule Hsp90 inhibitors promote client protein degradation via the actions of molecular chaperones.
Hsp90 can promote degradation as well as protect against degradation of misfolded proteins.
Several ubiquitin ligases in the degradation of Hsp90 client proteins including CHIP, Ubr1 and Cul5.
Inhibition of Hsp90 can protect against neurodegeneration.
Acknowledgments
The authors thank Dr. Effie Maclachlan for help with editing the manuscript. Work in the author’s lab is supported by grants from the National Institutes of Health: RO1GM70596 and U54CA132378.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 2.Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 3.Ellis RJ. Protein misassembly: macromolecular crowding and molecular chaperones. Advances in experimental medicine and biology. 2007;594:1–13. doi: 10.1007/978-0-387-39975-1_1. [DOI] [PubMed] [Google Scholar]
- 4.Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- 5.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 6.Luo W, Sun W, Taldone T, Rodina A, Chiosis G. Heat shock protein 90 in neurodegenerative diseases. Mol Neurodegener. 2010;5:24. doi: 10.1186/1750-1326-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Venable J, LaPointe P, Hutt DM, Koulov AV, Coppinger J, Gurkan C, Kellner W, Matteson J, Plutner H, Riordan JR, Kelly JW, Yates JR, 3rd, Balch WE. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 8.Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci U S A. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 11.Jarosz DF, Lindquist S. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science. 2010;330:1820–1824. doi: 10.1126/science.1195487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 13.Jarosz DF, Taipale M, Lindquist S. Protein homeostasis and the phenotypic manifestation of genetic diversity: principles and mechanisms. Annual review of genetics. 2010;44:189–216. doi: 10.1146/annurev.genet.40.110405.090412. [DOI] [PubMed] [Google Scholar]
- 14.Xu Y, Lindquist S. Heat-shock protein hsp90 governs the activity of pp60v-src kinase. Proc Natl Acad Sci U S A. 1993;90:7074–7078. doi: 10.1073/pnas.90.15.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esser C, Scheffner M, Hohfeld J. The chaperone-associated ubiquitin ligase CHIP is able to target p53 for proteasomal degradation. J Biol Chem. 2005;280:27443–27448. doi: 10.1074/jbc.M501574200. [DOI] [PubMed] [Google Scholar]
- 16.Whitesell L, Sutphin PD, Pulcini EJ, Martinez JD, Cook PH. The physical association of multiple molecular chaperone proteins with mutant p53 is altered by geldanamycin, an hsp90-binding agent. Mol Cell Biol. 1998;18:1517–1524. doi: 10.1128/mcb.18.3.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grbovic OM, Basso AD, Sawai A, Ye Q, Friedlander P, Solit D, Rosen N. V600E B-Raf requires the Hsp90 chaperone for stability and is degraded in response to Hsp90 inhibitors. Proc Natl Acad Sci U S A. 2006;103:57–62. doi: 10.1073/pnas.0609973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nature reviews Molecular cell biology. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 19.Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood) 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 20.Lee P, Shabbir A, Cardozo C, Caplan AJ. Sti1 and Cdc37 can stabilize Hsp90 in chaperone complexes with a protein kinase. Molecular biology of the cell. 2004;15:1785–1792. doi: 10.1091/mbc.E03-07-0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee P, Rao J, Fliss A, Yang E, Garrett S, Caplan AJ. The Cdc37 protein kinase-binding domain is sufficient for protein kinase activity and cell viability. The Journal of cell biology. 2002;159:1051–1059. doi: 10.1083/jcb.200210121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grammatikakis N, Lin JH, Grammatikakis A, Tsichlis PN, Cochran BH. p50(cdc37) acting in concert with Hsp90 is required for Raf-1 function. Molecular and cellular biology. 1999;19:1661–1672. doi: 10.1128/mcb.19.3.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Street TO, Lavery LA, Agard DA. Substrate binding drives large-scale conformational changes in the Hsp90 molecular chaperone. Mol Cell. 2011;42:96–105. doi: 10.1016/j.molcel.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 25.Xu W, Yuan X, Xiang Z, Mimnaugh E, Marcu M, Neckers L. Surface charge and hydrophobicity determine ErbB2 binding to the Hsp90 chaperone complex. Nat Struct Mol Biol. 2005 doi: 10.1038/nsmb885. [DOI] [PubMed] [Google Scholar]
- 26.Caplan AJ, Mandal AK, Theodoraki MA. Molecular chaperones and protein kinase quality control. Trends Cell Biol. 2007;17:87–92. doi: 10.1016/j.tcb.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 27.An WG, Schulte TW, Neckers LM. The heat shock protein 90 antagonist geldanamycin alters chaperone association with p210bcr-abl and v-src proteins before their degradation by the proteasome. Cell Growth Differ. 2000;11:355–360. [PubMed] [Google Scholar]
- 28.Kettern N, Dreiseidler M, Tawo R, Hohfeld J. Chaperone-assisted degradation: multiple paths to destruction. Biological Chemistry. 2010;391:481–489. doi: 10.1515/BC.2010.058. [DOI] [PubMed] [Google Scholar]
- 29.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 30.Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 31.Nillegoda NB, Theodoraki MA, Mandal AK, Mayo KJ, Ren HY, Sultana R, Wu K, Johnson J, Cyr DM, Caplan AJ. Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Molecular biology of the cell. 2010;21:2102–2116. doi: 10.1091/mbc.E10-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaner L, Morano KA. All in the family: atypical Hsp70 chaperones are conserved modulators of Hsp70 activity. Cell Stress Chaperones. 2007;12:1–8. doi: 10.1379/CSC-245R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandal AK, Gibney PA, Nillegoda NB, Theodoraki MA, Caplan AJ, Morano KA. Hsp110 chaperones control client fate determination in the hsp70-Hsp90 chaperone system. Molecular biology of the cell. 2010;21:1439–1448. doi: 10.1091/mbc.E09-09-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandal AK, Nillegoda NB, Chen JA, Caplan AJ. Ydj1 protects nascent protein kinases from degradation and controls the rate of their maturation. Molecular and cellular biology. 2008;28:4434–4444. doi: 10.1128/MCB.00543-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee DH, Sherman MY, Goldberg AL. Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4773–4781. doi: 10.1128/mcb.16.9.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park SH, Bolender N, Eisele F, Kostova Z, Takeuchi J, Coffino P, Wolf DH. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol Biol Cell. 2007;18:153–165. doi: 10.1091/mbc.E06-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, Michaelis S. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem. 2004;279:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- 38.Han S, Liu Y, Chang A. Cytoplasmic Hsp70 promotes ubiquitination for endoplasmic reticulum-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, PMA1. J Biol Chem. 2007;282:26140–26149. doi: 10.1074/jbc.M701969200. [DOI] [PubMed] [Google Scholar]
- 39.McClellan AJ, Scott MD, Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121:739–748. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 40.Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearl LH. Hsp90 and Cdc37 -- a chaperone cancer conspiracy. Curr Opin Genet Dev. 2005;15:55–61. doi: 10.1016/j.gde.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 42.Mandal AK, Lee P, Chen JA, Nillegoda N, Heller A, DiStasio S, Oen H, Victor J, Nair DM, Brodsky JL, Caplan AJ. Cdc37 has distinct roles in protein kinase quality control that protect nascent chains from degradation and promote posttranslational maturation. The Journal of cell biology. 2007;176:319–328. doi: 10.1083/jcb.200604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaughan CK, Gohlke U, Sobott F, Good VM, Ali MM, Prodromou C, Robinson CV, Saibil HR, Pearl LH. Structure of an Hsp90-Cdc37-Cdk4 complex. Molecular cell. 2006;23:697–707. doi: 10.1016/j.molcel.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goasduff T, Cederbaum AI. CYP2E1 degradation by in vitro reconstituted systems: role of the molecular chaperone hsp90. Arch Biochem Biophys. 2000;379:321–330. doi: 10.1006/abbi.2000.1870. [DOI] [PubMed] [Google Scholar]
- 45.Fuller W, Cuthbert AW. Post-translational disruption of the delta F508 cystic fibrosis transmembrane conductance regulator (CFTR)-molecular chaperone complex with geldanamycin stabilizes delta F508 CFTR in the rabbit reticulocyte lysate. J Biol Chem. 2000;275:37462–37468. doi: 10.1074/jbc.M006278200. [DOI] [PubMed] [Google Scholar]
- 46.Gusarova V, Caplan AJ, Brodsky JL, Fisher EA. Apoprotein B degradation is promoted by the molecular chaperones hsp90 and hsp70. The Journal of biological chemistry. 2001;276:24891–24900. doi: 10.1074/jbc.M100633200. [DOI] [PubMed] [Google Scholar]
- 47.Imamura T, Haruta T, Takata Y, Usui I, Iwata M, Ishihara H, Ishiki M, Ishibashi O, Ueno E, Sasaoka T, Kobayashi M. Involvement of heat shock protein 90 in the degradation of mutant insulin receptors by the proteasome. J Biol Chem. 1998;273:11183–11188. doi: 10.1074/jbc.273.18.11183. [DOI] [PubMed] [Google Scholar]
- 48.Loo MA, Jensen TJ, Cui L, Hou Y, Chang XB, Riordan JR. Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the proteasome. Embo J. 1998;17:6879–6887. doi: 10.1093/emboj/17.23.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holmes JL, Sharp SY, Hobbs S, Workman P. Silencing of HSP90 cochaperone AHA1 expression decreases client protein activation and increases cellular sensitivity to the HSP90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2008;68:1188–1197. doi: 10.1158/0008-5472.CAN-07-3268. [DOI] [PubMed] [Google Scholar]
- 50.Okiyoneda T, Barriere H, Bagdany M, Rabeh WM, Du K, Hohfeld J, Young JC, Lukacs GL. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science. 2010;329:805–810. doi: 10.1126/science.1191542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Apaja PM, Xu H, Lukacs GL. Quality control for unfolded proteins at the plasma membrane. J Cell Biol. 2010;191:553–570. doi: 10.1083/jcb.201006012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okiyoneda T, Apaja PM, Lukacs GL. Protein quality control at the plasma membrane. Current opinion in cell biology. 2011 doi: 10.1016/j.ceb.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 54.Finley D, Sadis S, Monia BP, Boucher P, Ecker DJ, Crooke ST, Chau V. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Molecular and cellular biology. 1994;14:5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hochstrasser M. Ubiquitin-dependent protein degradation. Annual review of genetics. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 56.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 57.Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 59.Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 60.Murata S, Minami Y, Minami M, Chiba T, Tanaka K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2001;2:1133–1138. doi: 10.1093/embo-reports/kve246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 62.Sanchez ER, Toft DO, Schlesinger MJ, Pratt WB. Evidence that the 90-kDa phosphoprotein associated with the untransformed L-cell glucocorticoid receptor is a murine heat shock protein. J Biol Chem. 1985;260:12398–12401. [PubMed] [Google Scholar]
- 63.Whitesell L, Cook P. Stable and specific binding of heat shock protein 90 by geldanamycin disrupts glucocorticoid receptor function in intact cells. Mol Endocrinol. 1996;10:705–712. doi: 10.1210/mend.10.6.8776730. [DOI] [PubMed] [Google Scholar]
- 64.Kanelakis KC, Morishima Y, Dittmar KD, Galigniana MD, Takayama S, Reed JC, Pratt WB. Differential effects of the hsp70-binding protein BAG-1 on glucocorticoid receptor folding by the hsp90-based chaperone machinery. J Biol Chem. 1999;274:34134–34140. doi: 10.1074/jbc.274.48.34134. [DOI] [PubMed] [Google Scholar]
- 65.Fan M, Park A, Nephew KP. CHIP (carboxyl terminus of Hsc70-interacting protein) promotes basal and geldanamycin-induced degradation of estrogen receptor-alpha. Mol Endocrinol. 2005;19:2901–2914. doi: 10.1210/me.2005-0111. [DOI] [PubMed] [Google Scholar]
- 66.Cardozo CP, Michaud C, Ost MC, Fliss AE, Yang E, Patterson C, Hall SJ, Caplan AJ. C-terminal Hsp-interacting protein slows androgen receptor synthesis and reduces its rate of degradation. Arch Biochem Biophys. 2003;410:134–140. doi: 10.1016/s0003-9861(02)00680-x. [DOI] [PubMed] [Google Scholar]
- 67.Chavany C, Mimnaugh E, Miller P, Bitton R, Nguyen P, Trepel J, Whitesell L, Schnur R, Moyer J, Neckers L. p185erbB2 binds to GRP94 in vivo. Dissociation of the p185erbB2/GRP94 heterocomplex by benzoquinone ansamycins precedes depletion of p185erbB2. J Biol Chem. 1996;271:4974–4977. doi: 10.1074/jbc.271.9.4974. [DOI] [PubMed] [Google Scholar]
- 68.Miller P, DiOrio C, Moyer M, Schnur RC, Bruskin A, Cullen W, Moyer JD. Depletion of the erbB-2 gene product p185 by benzoquinoid ansamycins. Cancer Res. 1994;54:2724–2730. [PubMed] [Google Scholar]
- 69.Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc Natl Acad Sci U S A. 2002;99:12847–12852. doi: 10.1073/pnas.202365899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou P, Fernandes N, Dodge IL, Reddi AL, Rao N, Safran H, DiPetrillo TA, Wazer DE, Band V, Band H. ErbB2 degradation mediated by the co-chaperone protein CHIP. J Biol Chem. 2003;278:13829–13837. doi: 10.1074/jbc.M209640200. [DOI] [PubMed] [Google Scholar]
- 71.Kundrat L, Regan L. Balance between folding and degradation for Hsp90-dependent client proteins: a key role for CHIP. Biochemistry. 2010;49:7428–7438. doi: 10.1021/bi100386w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morishima Y, Wang AM, Yu Z, Pratt WB, Osawa Y, Lieberman AP. CHIP deletion reveals functional redundancy of E3 ligases in promoting degradation of both signaling proteins and expanded glutamine proteins. Hum Mol Genet. 2008;17:3942–3952. doi: 10.1093/hmg/ddn296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Isaacs JS, Jung YJ, Mimnaugh EG, Martinez A, Cuttitta F, Neckers LM. Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1 alpha-degradative pathway. J Biol Chem. 2002;277:29936–29944. doi: 10.1074/jbc.M204733200. [DOI] [PubMed] [Google Scholar]
- 74.Ehrlich ES, Wang T, Luo K, Xiao Z, Niewiadomska AM, Martinez T, Xu W, Neckers L, Yu XF. Regulation of Hsp90 client proteins by a Cullin5-RING E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2009;106:20330–20335. doi: 10.1073/pnas.0810571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mogk A, Schmidt R, Bukau B. The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol. 2007;17:165–172. doi: 10.1016/j.tcb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 76.Eisele F, Wolf DH. Degradation of misfolded protein in the cytoplasm is mediated by the ubiquitin ligase Ubr1. FEBS Lett. 2008;582:4143–4146. doi: 10.1016/j.febslet.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 77.Heck JW, Cheung SK, Hampton RY. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci U S A. 2010;107:1106–1111. doi: 10.1073/pnas.0910591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xia Z, Webster A, Du F, Piatkov K, Ghislain M, Varshavsky A. Substrate-binding sites of UBR1, the ubiquitin ligase of the N-end rule pathway. J Biol Chem. 2008;283:24011–24028. doi: 10.1074/jbc.M802583200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 80.Fredrickson EK, Rosenbaum JC, Locke MN, Milac TI, Gardner RG. Exposed hydrophobicity is a key determinant of nuclear quality control degradation. Molecular Biology of the Cell. 2011;22:2384–2395. doi: 10.1091/mbc.E11-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Iwata A, Nagashima Y, Matsumoto L, Suzuki T, Yamanaka T, Date H, Deoka K, Nukina N, Tsuji S. Intranuclear degradation of polyglutamine aggregates by the ubiquitin-proteasome system. J Biol Chem. 2009;284:9796–9803. doi: 10.1074/jbc.M809739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rodina A, Vilenchik M, Moulick K, Aguirre J, Kim J, Chiang A, Litz J, Clement CC, Kang Y, She Y, Wu N, Felts S, Wipf P, Massague J, Jiang X, Brodsky JL, Krystal GW, Chiosis G. Selective compounds define Hsp90 as a major inhibitor of apoptosis in small-cell lung cancer. Nat Chem Biol. 2007;3:498–507. doi: 10.1038/nchembio.2007.10. [DOI] [PubMed] [Google Scholar]
- 83.Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl FU. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci U S A. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang C, Cheng H, Hao S, Zhou H, Zhang X, Gao J, Sun QH, Hu H, Wang CC. Heat shock protein 70 inhibits alpha-synuclein fibril formation via interactions with diverse intermediates. Journal of molecular biology. 2006;364:323–336. doi: 10.1016/j.jmb.2006.08.062. [DOI] [PubMed] [Google Scholar]
- 85.Evans CG, Wisen S, Gestwicki JE. Heat shock proteins 70 and 90 inhibit early stages of amyloid beta-(1-42) aggregation in vitro. J Biol Chem. 2006;281:33182–33191. doi: 10.1074/jbc.M606192200. [DOI] [PubMed] [Google Scholar]
- 86.Waza M, Adachi H, Katsuno M, Minamiyama M, Sang C, Tanaka F, Inukai A, Doyu M, Sobue G. 17-AAG, an Hsp90 inhibitor, ameliorates polyglutamine-mediated motor neuron degeneration. Nat Med. 2005;11:1088–1095. doi: 10.1038/nm1298. [DOI] [PubMed] [Google Scholar]
- 87.Wang L, Xie C, Greggio E, Parisiadou L, Shim H, Sun L, Chandran J, Lin X, Lai C, Yang WJ, Moore DJ, Dawson TM, Dawson VL, Chiosis G, Cookson MR, Cai H. The chaperone activity of heat shock protein 90 is critical for maintaining the stability of leucine-rich repeat kinase 2. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:3384–3391. doi: 10.1523/JNEUROSCI.0185-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Luo W, Dou F, Rodina A, Chip S, Kim J, Zhao Q, Moulick K, Aguirre J, Wu N, Greengard P, Chiosis G. Roles of heat-shock protein 90 in maintaining and facilitating the neurodegenerative phenotype in tauopathies. Proc Natl Acad Sci U S A. 2007;104:9511–9516. doi: 10.1073/pnas.0701055104. [DOI] [PMC free article] [PubMed] [Google Scholar]