Abstract
Hundreds of billions of cells undergo apoptosis in our body everyday and are removed by immunologically silent phagocytosis to maintain tissue homeostasis. Impairments in phagocytosis result in autoimmune and/or degenerative diseases. Eat-me signals are the key to the recognition of extracellular cargos and the initiation of the phagocytosis process by activating phagocytic receptors and signaling cascades, and are convenient targets for therapeutic modulation. Despite their importance, eat-me signals and other phagocytosis players are mostly identified on case-by-case basis with daunting challenges. This Commentary focuses on our latest knowledge of the extracellular players, highlights our approaches to systematically map unknown pathways by functional genetic and proteomic technologies, and discusses future direction to unravel the mystery of molecular phagocyte biology.
Keywords: Eat-me signal, Phagocytic receptor, Phagocytosis, Engulfment, Phagoligandomics, Phagocytomics
Apoptosis or programmed cell death is essential for morphogenesis, cell homeostasis, injury repair, immune tolerance and resolution of inflammation in multicellular organisms (Erwig and Henson, 2007; Ravichandran, 2010). Morphogenesis requires tissue remodeling by deleting unnecessary cells through apoptosis, as in neurogenesis and retinogenesis with up to 50% of superfluous neurons removed by apoptosis (Erwig and Henson, 2007). Approximately 2.4 million of red blood cells are generated per second and are expected to be eliminated at the same staggering rate by Kupffer cells, the resident macrophages in the liver (Sackmann, 1995). The turnover of circulating neutrophils is ~1 × 1011 cells per day (Erwig and Henson, 2007). Approximately 95% of developing T cells are deleted in thymus by negative selection as part of the central immune tolerance. Up to 2 tons of apoptotic cells are generated by bone marrow, lymph nodes and intestine through our 80-year lifespan (Hotchkiss et al., 2009).
The prompt and efficient clearance of apoptotic cells by phagocytosis is essential to maintain tissue homeostasis and prevent the secondary necrosis. Otherwise, the release of intracellular contents from necrotic cells might trigger inflammation and autoimmune diseases (Nagata et al., 2010). Phagocytosis can also be an integral part of physiological processes. Phagocytic clearance of shed distal tips of photoreceptor outer segments (POSs) by retinal pigment epithelial (RPE) cells is an essential process for maintenance of retinal homeostasis and regeneration of POSs (Strauss, 2005). The entire POSs are renewed in ~10 days. Based on the length of POSs, I calculate that each RPE cell phagocytoses ~75-fold of its own volume of shed POSs each year. Defects in RPE phagocytosis disrupt the visual cycle and retina homeostasis, leading to retinal degeneration (Kevany and Palczewski, 2010). Emerging evidence suggests that phagocytic clearance of apoptotic cells and cellular debris occurs in all tissues at all times to maintain immune balance and tissue homeostasis (Erwig and Henson, 2007).
Phagocytosis of foreign pathogens and autologous apoptotic cells has similar outcomes of ingesting extracellular cargos, but distinct underlying molecular mechanisms and immunological consequences. Phagocytosis of opsonized pathogens through complement receptors and Fc receptors triggers pro-inflammatory response, whereas ingestion of apoptotic cells results in anti-inflammatory response (Erwig and Henson, 2007). To distinguish the difference in these two processes, the latter is termed as efferocytosis (from effero—to carry to the grave to bury) or engulfment (Reddien and Horvitz, 2004; Vandivier et al., 2006).
Efferocytosis consists of several sequential events, including finding apoptotic cells by phagocytes, recognition of the cargos by phagocytic receptors, activation of intracellular signaling cascades with cytoskeletal rearrangements for cargo engulfment, and post-engulfment process for phagosome maturation and cargo degradation (Ravichandran and Lorenz, 2007). Molecular phagocyte biology deals with all players throughout the entire process. Given the importance of extracellular events in cargo selection and initiation of engulfment process, this Commentary focuses mainly on extracellular players, including find-me signals, eat-me signals and phagocytic receptors, with an emphasis on their systematic mapping by functional genetic and proteomic technologies. The intracellular events and players are covered by other excellent reviews (Ravichandran, 2010; Ravichandran and Lorenz, 2007).
Find-me signals
One of the fundamental questions in phagocytosis is whether phagocytes require chemotactic guidance to find apoptotic cells. The answer depends on the type of phagocytes. In general, professional phagocytes, such as macrophages and microglia, are mobile and are constantly surveying the body or tissue environment for apoptotic cells. They can be recruited through chemotaxis to apoptotic cells or lesion site as in spinal cord injury to perform the clearance (Napoli and Neumann, 2009). What are the chemotactic factors for phagocytes to find apoptotic cells? Nucleotides, ATP and UDP, are well-characterized find-me signals that are released from apoptotic cells through pannexin 1 channels (Chekeni et al., 2010; Koizumi et al., 2007). Released ATP and UDP generate concentration gradient for microglial chemotaxis by binding to P2Y12 and P2Y6 purinergic receptors (i.e. purinoceptors), respectively. The find-me signal of lysophosphatidylcholine (LPC) is released from phosphatidylcholine on apoptotic cells by a calcium-independent isoform of phospholipase A2 (iPLA2) and is capable of stimulating macrophage chemotaxis through G2A receptor (Lauber et al., 2003; Peter et al., 2008). Other identified find-me signals include sphingosine 1-phosphate (S1P) and CX3CL1/fractalkine (Gude et al., 2008; Truman et al., 2008). Our understanding of find-me signals is relatively primitive. It is well known that the integrity of the plasma membrane is compromised during apoptosis, leading to the leakage of intracellular proteins. Whether any leaked proteins can serve as find-me signals remains unclear.
In contrast, some non-professional phagocytes, such as RPE cells, are stationary without requiring any migration toward the shed POSs. The tips of POSs are embedded in the microvilli of RPE cells, and the shed POSs are conveniently captured and phagocytosed by RPE cells (Strauss, 2005). Other non-mobile phagocytes include endothelial and epithelial cells. Even some professional phagocytes, such as Kupffer cells in the liver, could be relatively stationary, waiting for aged red blood cells to pass by rather than chasing them in the circulation.
Eat-me signals
Eat-me signals, by definition, should be specifically displayed on the surface of apoptotic cells, but not on healthy cells, for selective clearance of dying cells by phagocytosis. Moreover, eat-me signals should be able to bind to their cognate receptors on phagocyte surface to trigger intracellular signaling cascades, cytoskeletal rearrangement and engulfment. Eat-me signals have attracted abundant attention because they are the keys to cargo selection and control the initiation of phagocytosis processes. Consequently, it would be more convenient to modulate efferocytosis activity for disease therapy with eat-me signals than with other intracellular signaling molecules.
Types of eat-me signals
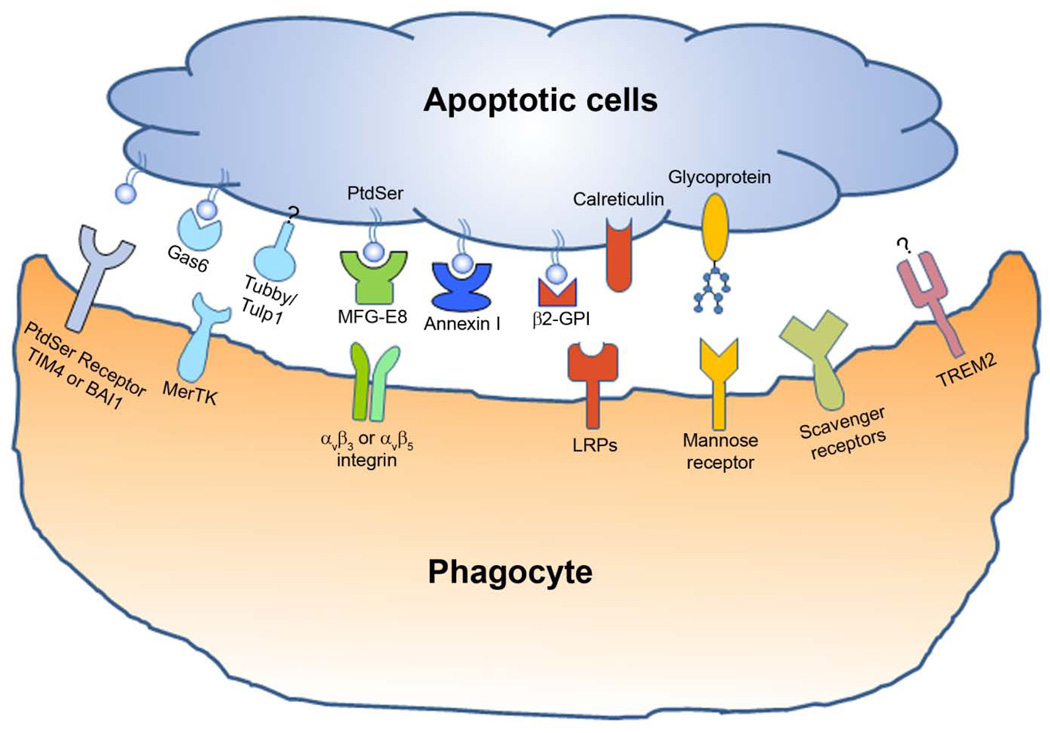
Eat-me signals or phagocytosis ligands can be classified into two major categories, membrane-anchored eat-me signals and soluble bridging molecules (Fig. 1). Phosphatidylserine (PtdSer) is one of the best characterized membrane-anchored eat-me signals. PtdSer normally resides on the inner leaflet of the plasma membrane bilayer but flips across the bilayer to be displayed on apoptotic cells during apoptosis (Ravichandran and Lorenz, 2007). The externalized PtdSer binds to its receptors on phagocytes to facilitate efferocytosis. Cleavage of sialic acids on the surface of apoptotic cells by neuraminidase enhances their engulfment, suggesting that newly exposed fucose residues in the apoptotic membrane blebs may serve as eat-me signals (Meesmann et al., 2010). In addition to non-protein molecules, membrane-anchored proteins can also function as eat-me signals. Calreticulin is upregulated on the surface of apoptotic cells to facilitate their engulfment by binding to and activating LDL-receptor-related protein 1 (LRP1, i.e. CD91) on phagocyte surface (Gardai et al., 2005).
Fig. 1.
Eat-me signals and phagocytic receptors for the engulfment of apoptotic cells. The apoptotic cells display various eat-me signals, which bind to their receptors on phagocyte surface either directly or indirectly through bridging molecules. Multiple eat-me signals and their receptors can form a cluster within the phagocytic cup as the engulfment synapse to facilitate the clearance of apoptotic cells. Eat-me signals and their cognate receptors are shown in the same color.
In contrast to membrane-anchored eat-me signals with only one receptor-binding domain, soluble bridging molecules are molecular adaptor with at least two binding domains, the receptor-binding domain and phagocytosis prey-binding domain (PPBD) (Caberoy et al., 2010c). Soluble bridging molecules, when bound to and displayed on apoptotic cells but not healthy cells through PPBD, serve as eat-me signals to facilitate discriminative clearance of apoptotic cells. Gas6 and protein S are two well-characterized MerTK ligands with sequence homology (Lemke and Rothlin, 2008). They bind to PtdSer through their N-terminal 11 γ-carboxyglutamic acid residues (Gla) and interact with phagocytic receptor MerTK through the C-terminal two globular laminin G-like domains (2 × LG). Tubby and tubby-like protein 1 (Tulp1) were recently identified as new ligands for MerTK phagocytic receptor with their N-terminal K/R(X)1–2KKK motifs for MerTK recognition and C-terminal PPBD for binding to apoptotic cells (Caberoy et al., 2010a; Caberoy et al., 2010c). Interestingly, deletional mutation of the C-terminal PPBD in either tubby or Tulp1 not only abolished their capacity to stimulate RPE engulfment but also caused retinal degeneration by undefined mechanisms (Carroll et al., 2004). MFG-E8 is another well-characterized soluble eat-me signal that recognizes PtdSer through its C-terminal discoidin-like domain and binds to αvβ3 or αvβ5 integrin on phagocytes through the N-terminal RGD motif (Hanayama et al., 2002). Two additional PtdSer-specific bridging molecules are β2-glycoprotein-I (β2-GPI) and annexin I (Arur et al., 2003; Balasubramanian et al., 1997). The former interacts with LRP family members on phagocytes (Maiti et al., 2008), whereas the receptor for annexin I remains elusive.
Externalization of eat-me signals
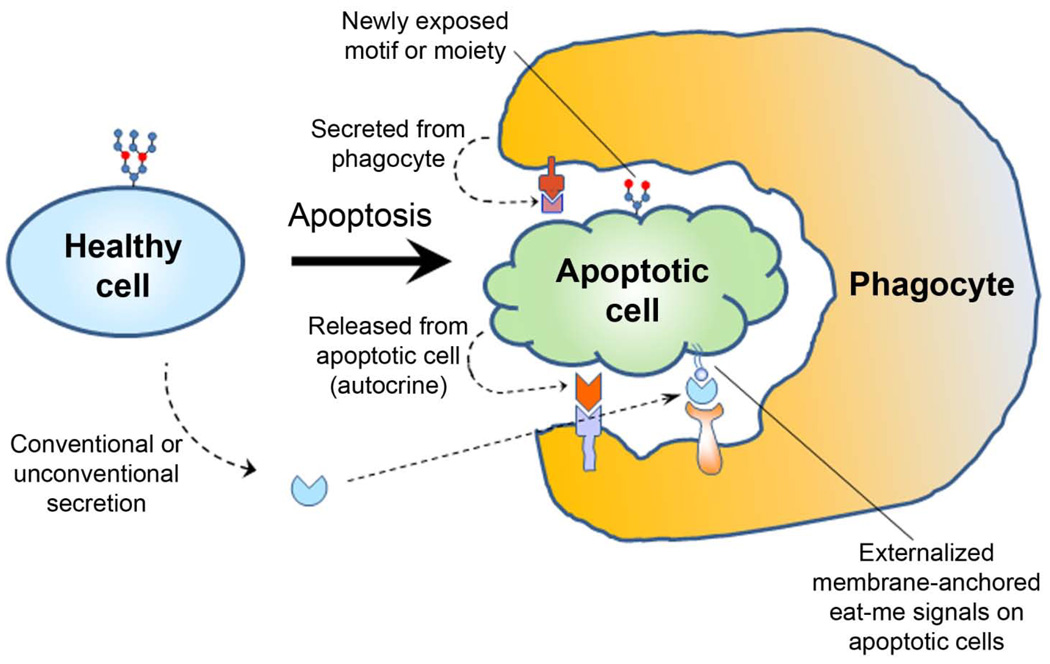
The extracellular trafficking of eat-me signals is necessary for them to be displayed on apoptotic cells. Four mechanisms are proposed for this trafficking: a) conventional secretion; b) unconventional secretion; c) passive release from apoptotic cells; and d) exposure by enzymatic modifications of the apoptotic cell surface (Fig. 2). Gas6, protein S and MEG-E8 are secreted with classic signal peptides, whereas tubby and Tulp1 lack the signal peptides and are secreted through unconventional pathways (Caberoy and Li, 2009). The proteins in these two categories are soluble bridging molecules.
Fig. 2.
Extracellular trafficking of eat-me signals through four different mechanisms. Soluble eat-me signals with or without signal peptide are secreted through conventional or unconventional pathways from phagocytes or non-phagocytes, and selectively bind to apoptotic cells. Non-secreted intracellular proteins or molecules, including membrane-anchored eat-me signals, are released during apoptosis through damaged plasma membrane. Finally, enzymes released or activated during apoptosis may modify the surface of apoptotic cells to create new motifs or moieties as eat-me signals.
Non-secreted cytoplasmic proteins can be released through damaged plasma membrane during apoptosis to function as eat-me signals. Annexin I is a non-secreted cytosolic protein, which leaks out during apoptosis to bind to externalized PtdSer on the surface of apoptotic cells and facilitates engulfment (Arur et al., 2003). Similarly, PtdSer and calreticulin are the membrane-anchored eat-me signals that are externalized during apoptosis. Many intracellular proteins are known to leak out from apoptotic cells. Whether any other leaked non-secreted proteins may function as eat-me signals remains to be determined. Finally, the cleavage of sialic acid on apoptotic cells creates newly exposed carbohydrate moieties, such as fucose resides on the apoptotic membrane blebs, to enhance engulfment (Meesmann et al., 2010). This represents a new mode to display eat-me signals on apoptotic cells. Further studies are needed to determine whether other enzymes released or activated during apoptosis, including proteases, can modify the cell surface to “create” new eat-me signals.
Don’t-eat-me signals
Emerging evidence suggests that healthy cells display “don’t-eat-me” signals on their surface to prevent them from unintentional ingestion by phagocytes. An example of a don’t-eat- me signal is CD47, which is widely distributed on cells and binds to a number of proteins, including SIRPα on phagocytes (Oldenborg et al., 2000). CD47-SIRPα interaction blocks the engulfment of apoptotic cells (Gardai et al., 2005). Another example of a don’t-eat-me signal is CD31 (Brown et al., 2002). Overall, our knowledge of don’t-eat-me signals is far less than that for eat-me signals.
Phagocytic receptors
Activation of phagocytic receptors on phagocytes is necessary to trigger intracellular signaling cascades and engulfment. PtdSer receptors have been extensively investigated owing to the controversial role of the original PtdSer receptor (PSR) in the clearance of apoptotic cells (Fadok et al., 2000; Ravichandran, 2010). BAI1 and TIM4 were subsequently identified and characterized as PtdSer receptors (Miyanishi et al., 2007; Park et al., 2007). Besides the direct interaction with its own receptors, PtdSer also facilitates engulfment through well characterized bridging molecules, e.g., Gas6, protein S, MFG-E8, β2-GPI and annexin I (Fig. 1) (Lauber et al., 2004). Given the importance of PtdSer in cellular signaling, it would not be surprising if more PtdSer-dependent bridging molecules are discovered in the near future.
MerTK is an important phagocytic receptor whose mutations cause impairment in engulfment, leading to autoimmunity and retinal degeneration (Erwig and Henson, 2007; Strick and Vollrath, 2010). MerTK belongs to the TAM receptor tyrosine kinase (RTK) subfamily with three members, including Tyro3, Axl and MerTK (Rothlin and Lemke, 2010). Activation of TAM RTKs results in signaling for anti-apoptosis, survival, proliferation, phagocytosis, migration and adhesion (Hafizi and Dahlback, 2006). Elimination of MerTK nearly abolishes efferocytosis by macrophages and RPE cells, whereas ablation of Axl, Tyro3, or both, reduces macrophage efferocytosis by approximately half but appears to have no effect on the RPE (Seitz et al., 2007). The TAM receptors provide pivotal inhibitory signaling to prevent innate inflammatory response, and TAM-deficient mice develop broad-spectrum autoimmune disease (Rothlin and Lemke, 2010). Gas6 and protein S are well-known MerTK ligands identified more than 15 years ago (Lemke and Rothlin, 2008). Tubby and Tulp1 were recently characterized as new MerTK ligands (Caberoy et al., 2010c).
Other engulfment receptors include αvβ3 and αvβ5 integrins for MFG-E8 (Hanayama et al., 2002), LRP (CD91) for calreticulin (Gardai et al., 2005), scavenger receptors, mannose receptors and triggering receptors expressed by myeloid cells-2 (TREM2). β5-deficient mice, which specifically lack αvβ5 integrin, develop retinal degeneration and age-related blindness owing to the loss of synchronized RPE phagocytosis (Nandrot et al., 2004). Scavenger receptors can also participate in phagocytic clearance of cellular debris. CD36 is a scavenger receptor that binds to thrombospondin-1 and oxidized lipids (Lauber et al., 2004). Similar to scavenger receptors, mannose receptor is another pattern recognition receptor, whose natural ligands include self or microbial glycoproteins and glycolipids.
TREM2 belongs to DAP12-associated receptor protein family with an important role in innate immunity and is mainly expressed on microglia in the central nervous system (CNS) (Hsieh et al., 2009). Loss-of-function mutations of either TREM2 or DAP12 cause Nasu-Hakola disease, a rare and fatal neurodegenerative disease. The mechanisms of the neurodegeneration are unknown, but one hypothesis is that lack of either TREM2 or DAP12 impairs microglial engulfment of apoptotic cells (Thrash et al., 2009). An absence of TREM2 expression on microglia impairs their capacity to engulf cell membrane debris and increases their gene transcription of pro-inflammatory cytokines (Neumann and Takahashi, 2007). The binding activity for TREM2 is upregulated on the surface of apoptotic cells to facilitate their TREM2-mediated engulfment (Hsieh et al., 2009). However, the identity of endogenous TREM2 ligands remains elusive.
How to systematically unravel the mystery of molecular phagocyte biology
Although a number of molecular players in efferocytosis signaling pathways have been identified on case-by-case basis with challenges, we really do not how many eat-me signals and receptors remain to be identified. Lack of global understanding of engulfment signaling pathways hinders our capacity to assess the relative importance of the known players. The obstacle is how to systematically identify unknown signaling pathways in the absence of any molecular probes. Some unbiased approaches of functional genetics and proteomics in the literature are summarized here.
Genetic approach
A live screen of accumulated cell corpses in Caenorhabditis elegans by optical microscopy was described more than 28 years ago to identify chemically-induced random genetic mutations critical for engulfment of cell corpses (Hedgecock et al., 1983). Several crucial genes were identified, including ced-1, 6, 7 and dyn-1 in one signaling pathway, and ced-2, 5, 10, and 12 in the other pathway (Mangahas and Zhou, 2005). CED-7 protein, a homologue of mouse ABCA1, is essential for the recognition of cell corpses by CED-1 on engulfing cells. In theory, this approach is capable of identifying all signaling proteins in efferocytosis pathways, including eat-me signals, phagocytic receptors and intracellular signaling molecules. However, this approach is not applicable to organisms that are not transparent for live screening by optical microscopy. The absence of professional phagocytes in C. elegans has compromised this approach to dissect complicated signaling pathways in mammalian phagocytes (Reddien and Horvitz, 2004).
Mass spectrometry and yeast two-hybrid system
Emergence of various functional proteomic technologies provides new means to explore the molecular mechanisms of efferocytosis. Arur et al. used mass spectrometry technology to analyze proteins associated with the membranes of apoptotic cells but not healthy cells, and identified annexin I as an eat-me signal (Arur et al., 2003). The significance of this study is the unbiased identification of a novel eat-me signal in the absence of any information for its binding partner at the protein level. This approach was to globally analyze and compare the protein expression profiles in the apoptotic and healthy membranes by expressional proteomics, rather than by functional proteomics. The drawback is that identified proteins are not always functionally relevant to phagocytosis pathways. The broad applicability and efficiency of this approach to identify other eat-me signals remains to be demonstrated.
Many other groups used various technologies of functional proteomics, such as the popular yeast two-hybrid system, to map signaling cascades of efferocytosis (Park et al., 2007). However, most of these approaches require initial knowledge of some molecular probes.
Open reading frame (ORF) phage display
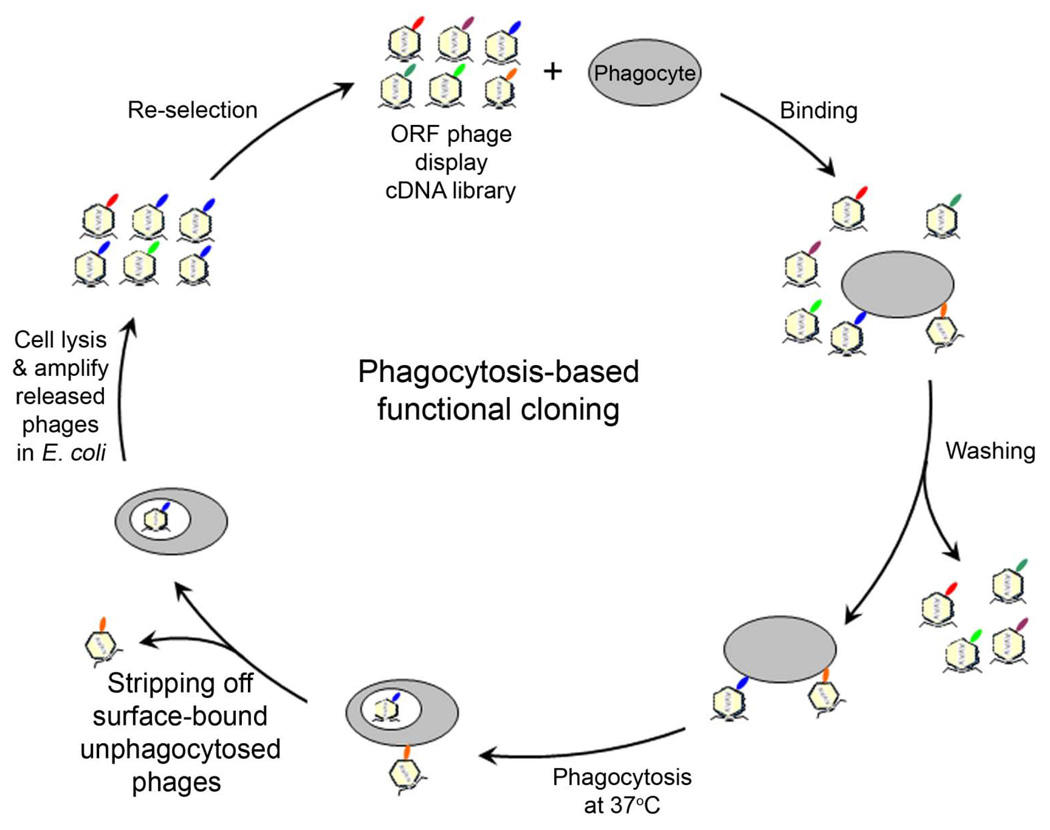
In an attempt to develop a broadly applicable approach for unbiased identification of eat-me signals in the absence of any molecular probe, we developed a new strategy of phagocytosis-based functional cloning by ORF phage display (Fig. 3) (Caberoy et al., 2010a). Owing to uncontrollable protein reading frames, phage display with conventional cDNA libraries identifies high percentage (>90%) of non-open reading frames (non-ORFs) (Li and Caberoy, 2010), which encode unnatural short peptides with minimal implication in cellular protein interaction networks. ORF phage display with minimal reading frame problem is a new technology of functional proteomics with versatile applications (Caberoy et al., 2010b; Li and Caberoy, 2010). The feasibility of phagocytosis-based functional selection to enrich phage clones displaying eat-me signals was demonstrated in macrophages, microglia and RPE cells, suggesting its broad applicability (Caberoy et al., 2009). The validity and biological relevance of this approach were demonstrated by unbiased identification of Tulp1 as a new eat-me signal and subsequent verification of Tulp1 as a new ligand for MerTK (Caberoy et al., 2010a; Caberoy et al., 2010c). Unlike most conventional molecular cloning techniques, this approach mixes a heterogeneous ORF phage display cDNA library with heterogeneous phagocytic receptors on phagocyte surface without any molecular probe. Intriguingly, multi-round phagocytosis-based selection to exploit the only common functional characteristic shared by all phagocytes is able to identify unknown eat-me signals in the absence of receptor information.
Fig. 3.
Phagocytosis-based functional cloning by ORF phage display to identify eat-me signals in the absence of receptor information. Phagocytes are incubated with an ORF phage display cDNA library at 4°C in culture without phagocytosis. After washing, phage-cell complexes are incubated at 37°C to allow the bound phages to be phagocytosed. After stripping off unphagocytosed surface-bound phages with a low pH isotonic buffer, the internalized phages are released by cell lysis with a hypotonic buffer, amplified in E. coli and used as input for the next round of functional selection (Caberoy et al., 2010a). Multiple rounds of phagocytosis-based selection enrich phage clones encoding proteins capable of stimulating phagocytosis. Individual clones can be analyzed for their internalization activity, identified by DNA sequencing and independently validated.
The advantage of this approach is its efficiency and sensitivity owing to the clone enrichment by multi-round phage selection and amplification. Large-scale screening of enriched clones will enable thorough mapping of eat-me signals. A drawback of this approach is that non-protein eat-me signals, such as PtdSer and carbohydrate moieties, will not be identified. Fortunately, protein eat-me signals should far outnumber non-protein eat-me signals based on our current knowledge (Lauber et al., 2004; Ravichandran and Lorenz, 2007). The incapacity to identify carbohydrate moieties could be addressed by combining ORF strategy with a mammalian display systems, such as retroviral display (Buchholz et al., 2008) and mammalian cell display (Ho and Pastan, 2009), which are less efficient and technically more complicated than ORF phage display.
It is worth noting that not all proteins identified by ORF phage display are genuine eat-me signals. Endogenous proteins internalized through other receptors, such as scavenger receptors, complement receptors or Fc receptors, may be identified by ORF phage display as well. Although all these endogenous ligands will provide a comprehensive understanding of phagocytic clearance of various extracellular debris through different mechanisms, identified proteins should be independently validated as genuine eat-me signals by the following four criteria. First, identified bridging molecules should selectively bind to apoptotic cells but not to healthy cells. Identified membrane-anchored eat-me signals should be displayed only on apoptotic cells. Second, eat-me signals should have access to their receptors on phagocyte surface, namely, identified proteins should be externalized or secreted through one of the four modes discussed above. Third, identified proteins should bind to their receptors on phagocytes. Even though their receptors may be unknown, the phagocyte-binding activity of identified eat-me signals, including soluble bridging molecules or externalized domain of membrane proteins, can be conveniently analyzed by flow cytometry. Finally, genuine eat-me signals should be able to facilitate engulfment of apoptotic cell or cellular debris. Their physiological or pathological relevance should be further verified in vivo.
Dual functional cloning
Given the difficulties to identify receptor-specific eat-me signals, many phagocytic receptors, such as TREM2 and signal-regulatory protein-β1 (SIRPβ1), still lack a known ligand (Hsieh et al., 2009; Neumann and Takahashi, 2007). To address the challenge, an advanced dual functional cloning strategy of ORF phage display, in which phagocytosis-based functional cloning was coupled to receptor-based affinity cloning, was developed to directly identify receptor-specific eat-me signals (Caberoy et al., unpublished data). Both Tulp1 and galectin-3 (Gal-3) were identified as MerTK-specific ligands by this strategy using MerTK-expressing D407 RPE cells and Mer-Fc (MerTK extracellular domain fused to Fc). Gal-3 was independently validated for its capacity to facilitate engulfment of apoptotic cells with MerTK activation. Excess Mer-Fc blocked Gal-3-mediated engulfment. This new strategy will improve our capacity to identify receptor-specific eat-me signals in an unbiased manner.
Perspectives
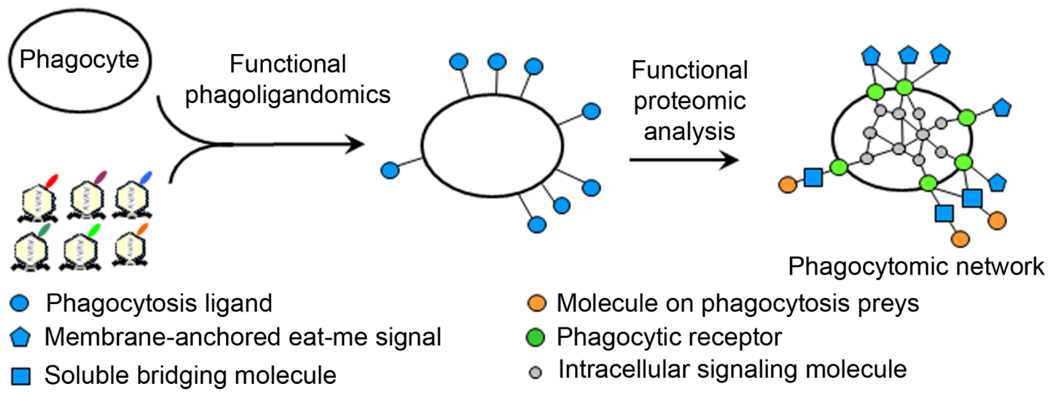
Phagocytosis-based functional cloning by ORF phage display provides a broadly applicable technology for unbiased identification of eat-me signals in the absence of receptor information. Coupled with the dual functional cloning, these new approaches will lead to systematic mapping of phagocytosis ligands based on their functional activity or “functional phagoligandomics” (Fig. 4). Identified proteins could be characterized as membrane-anchored eat-me signals or bridging molecules. Validated proteins can be used as molecular probes to further map their cognate receptors, their binding partners on apoptotic cells or intracellular signaling cascades by various technologies of functional proteomics, including yeast two-hybrid system, mass spectrometry or even ORF phage display with affinity selection (Caberoy et al., 2010b), as shown in a recent study (Caberoy et al., 2010c). This will lead to systematic mapping of the entire phagocytosis signaling pathways or “functional phagocytomics” to unravel the mystery of molecular phagocyte biology.
Fig. 4.
Systematic mapping of molecular phagocyte biology. Proteins identified by phagocytosis-based functional cloning of ORF phage display can be characterized as membrane-anchored eat-me signals or soluble bridging molecules and used as molecular probes to map their cognate receptors and intracellular signaling cascades by technologies of functional proteomics. This will lead to functional phagoligandomics for systematic mapping of phagocytosis ligands based on their functional activity and functional phagocytomics for systematic mapping of the signaling networks of phagocytes.
The engulfment field is relatively young, and we are only at the beginning of systematic mapping of molecular phagocyte biology for different phagocytes. Global understanding of their signaling pathways will improve our capacity to tackle some formidable questions. For example, can we stimulate immunologically silent efferocytosis of deleterious metabolic products, such as amyloid β peptide or amyloid plaque in Alzheimer’s disease or glaucoma, to minimize tissue aging or age-related degeneration? Can we switch pro-inflammatory macrophages/microglial phagocytosis to anti-inflammatory engulfment for the clearance of damaged cells or cellular debris in spinal cord injury or multiple sclerosis to prevent inflammation-induced secondary damage and promote tissue regeneration? Can we minimize microglial senescent activation by stimulating anti-inflammatory efferocytosis signaling pathways in aged brain and age-related neurodegeneration (Luo et al., 2010)? Can we boost RPE phagocytosis function in age-related macular degeneration (AMD) to delay the disease process (Kevany and Palczewski, 2010)? Identification of bridging molecules with different properties to control cargo recognition and receptor activation is the key to tackling these provocative questions and will improve our capacity to harness the therapeutic potentials of various phagocytes in the next phase of engulfment research.
Acknowledgement
This work was supported by NIH R01EY016211, R01EY016211-05S1 and an institutional grant from Research to Prevent Blindness. The author thanks Dr. Nora Caberoy for discussion.
Contract grant Sponsor: National Institute of Health; Contract grant number: R01EY016211
Contract grant Sponsor: National Institute of Health; Contract grant number: R01EY016211-05S1
Literature Cited
- Arur S, Uche UE, Rezaul K, Fong M, Scranton V, Cowan AE, Mohler W, Han DK. Annexin I is an endogenous ligand that mediates apoptotic cell engulfment. Dev Cell. 2003;4(4):587–598. doi: 10.1016/s1534-5807(03)00090-x. [DOI] [PubMed] [Google Scholar]
- Balasubramanian K, Chandra J, Schroit AJ. Immune clearance of phosphatidylserine-expressing cells by phagocytes. The role of beta2-glycoprotein I in macrophage recognition. J Biol Chem. 1997;272(49):31113–31117. doi: 10.1074/jbc.272.49.31113. [DOI] [PubMed] [Google Scholar]
- Brown S, Heinisch I, Ross E, Shaw K, Buckley CD, Savill J. Apoptosis disables CD31-mediated cell detachment from phagocytes promoting binding and engulfment. Nature. 2002;418(6894):200–203. doi: 10.1038/nature00811. [DOI] [PubMed] [Google Scholar]
- Buchholz CJ, Duerner LJ, Funke S, Schneider IC. Retroviral display and high throughput screening. Comb Chem High Throughput Screen. 2008;11(2):99–110. doi: 10.2174/138620708783744543. [DOI] [PubMed] [Google Scholar]
- Caberoy NB, Li W. Unconventional secretion of tubby and tubby-like protein 1. FEBS Lett. 2009;583:3057–3062. doi: 10.1016/j.febslet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caberoy NB, Maiguel D, Kim Y, Li W. Identification of tubby and tubby-like protein 1 as eat-me signals by phage display. Exp Cell Res. 2010a;316(2):245–257. doi: 10.1016/j.yexcr.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caberoy NB, Zhou Y, Jiang X, Alvarado G, Li W. Efficient identification of tubby-binding proteins by an improved system of T7 phage display. J Mol Recognit. 2010b;23(1):74–83. doi: 10.1002/jmr.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caberoy NB, Zhou Y, Li W. Can phage display be used as a tool to functionally identify endogenous eat-me signals in phagocytosis? J Biomol Screen. 2009;14(6):653–661. doi: 10.1177/1087057109335679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caberoy NB, Zhou Y, Li W. Tubby and tubby-like protein 1 are new MerTK ligands for phagocytosis. EMBO J. 2010c;29(23):3898–3910. doi: 10.1038/emboj.2010.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll K, Gomez C, Shapiro L. Tubby proteins: the plot thickens. Nat Rev Mol Cell Biol. 2004;5(1):55–63. doi: 10.1038/nrm1278. [DOI] [PubMed] [Google Scholar]
- Chekeni FB, Elliott MR, Sandilos JK, Walk SF, Kinchen JM, Lazarowski ER, Armstrong AJ, Penuela S, Laird DW, Salvesen GS, Isakson BE, Bayliss DA, Ravichandran KS. Pannexin 1 channels mediate 'find-me' signal release and membrane permeability during apoptosis. Nature. 2010;467(7317):863–867. doi: 10.1038/nature09413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwig LP, Henson PM. Immunological consequences of apoptotic cell phagocytosis. Am J Pathol. 2007;171(1):2–8. doi: 10.2353/ajpath.2007.070135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok VA, Bratton DL, Rose DM, Pearson A, Ezekewitz RA, Henson PM. A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature. 2000;405(6782):85–90. doi: 10.1038/35011084. [DOI] [PubMed] [Google Scholar]
- Gardai SJ, McPhillips KA, Frasch SC, Janssen WJ, Starefeldt A, Murphy-Ullrich JE, Bratton DL, Oldenborg PA, Michalak M, Henson PM. Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell. 2005;123(2):321–334. doi: 10.1016/j.cell.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Gude DR, Alvarez SE, Paugh SW, Mitra P, Yu J, Griffiths R, Barbour SE, Milstien S, Spiegel S. Apoptosis induces expression of sphingosine kinase 1 to release sphingosine-1-phosphate as a "come-and-get-me" signal. Faseb J. 2008;22(8):2629–2638. doi: 10.1096/fj.08-107169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafizi S, Dahlback B. Signalling and functional diversity within the Axl subfamily of receptor tyrosine kinases. Cytokine Growth Factor Rev. 2006;17(4):295–304. doi: 10.1016/j.cytogfr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Hanayama R, Tanaka M, Miwa K, Shinohara A, Iwamatsu A, Nagata S. Identification of a factor that links apoptotic cells to phagocytes. Nature. 2002;417(6885):182–187. doi: 10.1038/417182a. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Sulston JE, Thomson JN. Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans. Science. 1983;220(4603):1277–1279. doi: 10.1126/science.6857247. [DOI] [PubMed] [Google Scholar]
- Ho M, Pastan I. Mammalian cell display for antibody engineering. Methods Mol Biol. 2009;525:337–352. xiv. doi: 10.1007/978-1-59745-554-1_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss RS, Strasser A, McDunn JE, Swanson PE. Cell death. N Engl J Med. 2009;361(16):1570–1583. doi: 10.1056/NEJMra0901217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CL, Koike M, Spusta SC, Niemi EC, Yenari M, Nakamura MC, Seaman WE. A role for TREM2 ligands in the phagocytosis of apoptotic neuronal cells by microglia. J Neurochem. 2009;109(4):1144–1156. doi: 10.1111/j.1471-4159.2009.06042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevany BM, Palczewski K. Phagocytosis of retinal rod and cone photoreceptors. Physiology (Bethesda) 2010;25(1):8–15. doi: 10.1152/physiol.00038.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi S, Shigemoto-Mogami Y, Nasu-Tada K, Shinozaki Y, Ohsawa K, Tsuda M, Joshi BV, Jacobson KA, Kohsaka S, Inoue K. UDP acting at P2Y6 receptors is a mediator of microglial phagocytosis. Nature. 2007;446(7139):1091–1095. doi: 10.1038/nature05704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber K, Blumenthal SG, Waibel M, Wesselborg S. Clearance of apoptotic cells: getting rid of the corpses. Mol Cell. 2004;14(3):277–287. doi: 10.1016/s1097-2765(04)00237-0. [DOI] [PubMed] [Google Scholar]
- Lauber K, Bohn E, Krober SM, Xiao YJ, Blumenthal SG, Lindemann RK, Marini P, Wiedig C, Zobywalski A, Baksh S, Xu Y, Autenrieth IB, Schulze-Osthoff K, Belka C, Stuhler G, Wesselborg S. Apoptotic cells induce migration of phagocytes via caspase-3-mediated release of a lipid attraction signal. Cell. 2003;113(6):717–730. doi: 10.1016/s0092-8674(03)00422-7. [DOI] [PubMed] [Google Scholar]
- Lemke G, Rothlin CV. Immunobiology of the TAM receptors. Nat Rev Immunol. 2008;8(5):327–336. doi: 10.1038/nri2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Caberoy NB. New perspective for phage display as an efficient and versatile technology of functional proteomics. Appl Microbiol Biotechnol. 2010;85(4):909–919. doi: 10.1007/s00253-009-2277-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo XG, Ding JQ, Chen SD. Microglia in the aging brain: relevance to neurodegeneration. Mol Neurodegener. 2010;5:12. doi: 10.1186/1750-1326-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti SN, Balasubramanian K, Ramoth JA, Schroit AJ. Beta-2-glycoprotein 1-dependent macrophage uptake of apoptotic cells. Binding to lipoprotein receptor-related protein receptor family members. J Biol Chem. 2008;283(7):3761–3766. doi: 10.1074/jbc.M704990200. [DOI] [PubMed] [Google Scholar]
- Mangahas PM, Zhou Z. Clearance of apoptotic cells in Caenorhabditis elegans. Semin Cell Dev Biol. 2005;16(2):295–306. doi: 10.1016/j.semcdb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Meesmann HM, Fehr EM, Kierschke S, Herrmann M, Bilyy R, Heyder P, Blank N, Krienke S, Lorenz HM, Schiller M. Decrease of sialic acid residues as an eat-me signal on the surface of apoptotic lymphocytes. J Cell Sci. 2010;123(Pt 19):3347–3356. doi: 10.1242/jcs.066696. [DOI] [PubMed] [Google Scholar]
- Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450(7168):435–439. doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- Nagata S, Hanayama R, Kawane K. Autoimmunity and the clearance of dead cells. Cell. 2010;140(5):619–630. doi: 10.1016/j.cell.2010.02.014. [DOI] [PubMed] [Google Scholar]
- Nandrot EF, Kim Y, Brodie SE, Huang X, Sheppard D, Finnemann SC. Loss of synchronized retinal phagocytosis and age-related blindness in mice lacking alphavbeta5 integrin. J Exp Med. 2004;200(12):1539–1545. doi: 10.1084/jem.20041447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I, Neumann H. Microglial clearance function in health and disease. Neuroscience. 2009;158(3):1030–1038. doi: 10.1016/j.neuroscience.2008.06.046. [DOI] [PubMed] [Google Scholar]
- Neumann H, Takahashi K. Essential role of the microglial triggering receptor expressed on myeloid cells-2 (TREM2) for central nervous tissue immune homeostasis. J Neuroimmunol. 2007;184(1–2):92–99. doi: 10.1016/j.jneuroim.2006.11.032. [DOI] [PubMed] [Google Scholar]
- Oldenborg PA, Zheleznyak A, Fang YF, Lagenaur CF, Gresham HD, Lindberg FP. Role of CD47 as a marker of self on red blood cells. Science. 2000;288(5473):2051–2054. doi: 10.1126/science.288.5473.2051. [DOI] [PubMed] [Google Scholar]
- Park D, Tosello-Trampont AC, Elliott MR, Lu M, Haney LB, Ma Z, Klibanov AL, Mandell JW, Ravichandran KS. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450(7168):430–434. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- Peter C, Waibel M, Radu CG, Yang LV, Witte ON, Schulze-Osthoff K, Wesselborg S, Lauber K. Migration to apoptotic "find-me" signals is mediated via the phagocyte receptor G2A. J Biol Chem. 2008;283(9):5296–5305. doi: 10.1074/jbc.M706586200. [DOI] [PubMed] [Google Scholar]
- Ravichandran KS. Find-me and eat-me signals in apoptotic cell clearance: progress and conundrums. J Exp Med. 2010;207(9):1807–1817. doi: 10.1084/jem.20101157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran KS, Lorenz U. Engulfment of apoptotic cells: signals for a good meal. Nat Rev Immunol. 2007;7(12):964–974. doi: 10.1038/nri2214. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Horvitz HR. The engulfment process of programmed cell death in caenorhabditis elegans. Annu Rev Cell Dev Biol. 2004;20:193–221. doi: 10.1146/annurev.cellbio.20.022003.114619. [DOI] [PubMed] [Google Scholar]
- Rothlin CV, Lemke G. TAM receptor signaling and autoimmune disease. Curr Opin Immunol. 2010;22(6):740–746. doi: 10.1016/j.coi.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackmann E. Biological membrane architecture and function. In: Lipowsky R, Sackmann E, editors. Handbook of Biological Physics. vol 1. Elservier; 1995. [Google Scholar]
- Seitz HM, Camenisch TD, Lemke G, Earp HS, Matsushima GK. Macrophages and dendritic cells use different Axl/Mertk/Tyro3 receptors in clearance of apoptotic cells. J Immunol. 2007;178(9):5635–5642. doi: 10.4049/jimmunol.178.9.5635. [DOI] [PubMed] [Google Scholar]
- Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85(3):845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- Strick DJ, Vollrath D. Focus on molecules: MERTK. Exp Eye Res. 2010;91(6):786–787. doi: 10.1016/j.exer.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash JC, Torbett BE, Carson MJ. Developmental regulation of TREM2 and DAP12 expression in the murine CNS: implications for Nasu-Hakola disease. Neurochem Res. 2009;34(1):38–45. doi: 10.1007/s11064-008-9657-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman LA, Ford CA, Pasikowska M, Pound JD, Wilkinson SJ, Dumitriu IE, Melville L, Melrose LA, Ogden CA, Nibbs R, Graham G, Combadiere C, Gregory CD. CX3CL1/fractalkine is released from apoptotic lymphocytes to stimulate macrophage chemotaxis. Blood. 2008;112(13):5026–5036. doi: 10.1182/blood-2008-06-162404. [DOI] [PubMed] [Google Scholar]
- Vandivier RW, Henson PM, Douglas IS. Burying the dead: the impact of failed apoptotic cell removal (efferocytosis) on chronic inflammatory lung disease. Chest. 2006;129(6):1673–1682. doi: 10.1378/chest.129.6.1673. [DOI] [PubMed] [Google Scholar]