Abstract
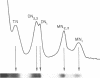
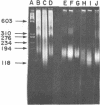
The chromatin structure of morphologically-similar, but increasingly-malignant erythroleukemia cells was investigated using milk micrococcal nuclease digestion of isolated nuclei. The maximum solubilization of chromatin was unique for each of the three cell types: the least malignant (our Stage II) released 61% of its chromatin DNA, the most malignant (Stage IV), 46%, and the intermediate (Stage III) released 36%. An analysis of the nucleosome oligomers liberated by digestion also demonstrated differences. After 15 minutes of digestion when release was reaching its maximum, a greater proportion of large nucleosomal oligomers (sizes > trinucleosome) was released from Stage II nuclei than from Stage III or IV nuclei. The cell types also differed in the relative amount of H1-depleted mononucleosomes released. Analysis of the size of the double-stranded DNA associated with mononucleosomal particles showed that Stage III mononucleosomes were smaller (148 bp) than Stage IV (167 bp) or Stage II (190 bp). In addition, while the DNA of mononucleosomes depleted in H1 was smaller than that in the H1-containing species, relative size differences among the different cell types were retained. These data suggested that the difference in the mononuocleosome particle size resistant to nuclease digestion was independent of histone H1. Differences in nucleosome repeat length were also noted among the cell types. These studies have demonstrated dramatic differences in chromatin structure associated with malignant potential of an otherwise morphologically identical cell type. These findings may reflect changes in the relative amounts of H2a variants which we have previously described among the different malignant cell types.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AXELRAD A. A., STEEVES R. A. ASSAY FOR FRIEND LEUKEMIA VIRUS: RAPID QUANTITATIVE METHOD BASED ON ENUMERATION OF MACROSCOPIC SPLEEN FOCI IN MICE. Virology. 1964 Nov;24:513–518. doi: 10.1016/0042-6822(64)90199-0. [DOI] [PubMed] [Google Scholar]
- Allan J., Staynov D. Z., Gould H. Reversible dissociation of linker histone from chromatin with preservation of internucleosomal repeat. Proc Natl Acad Sci U S A. 1980 Feb;77(2):885–889. doi: 10.1073/pnas.77.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafus N. L., Albright S. C., Todd R. D., Garrard W. T. A method for mapping the distributions of modified and variant histones among mono- and polynucleosomes. J Biol Chem. 1978 Apr 25;253(8):2568–2574. [PubMed] [Google Scholar]
- Bakayev V. V., Bakayeva T. G., Varshavsky A. J. Nucleosomes and subnucleosomes: heterogeneity and composition. Cell. 1977 Jul;11(3):619–629. doi: 10.1016/0092-8674(77)90079-4. [DOI] [PubMed] [Google Scholar]
- Bellard M., Gannon F., Chambon P. Nucleosome structure III: the structure and transcriptional activity of the chromatin containing the ovalbumin and globin genes in chick oviduct nuclei. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):779–791. doi: 10.1101/sqb.1978.042.01.078. [DOI] [PubMed] [Google Scholar]
- Blankstein L. A., Levy S. B. Changes in histone f2a2 associated with proliferation of Friend leukaemic cells. Nature. 1976 Apr 15;260(5552):638–640. doi: 10.1038/260638a0. [DOI] [PubMed] [Google Scholar]
- Blankstein L. A., Stollar B. D., Franklin S. G., Zweidler A., Levy S. B. Biochemical and immunological characterization of two distinct variants of histone H2A in Friend leukemia. Biochemistry. 1977 Oct 18;16(21):4557–4562. doi: 10.1021/bi00640a003. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., Stedman J. D. Histone 1 is proximal to histone 2A and to A24. Proc Natl Acad Sci U S A. 1979 May;76(5):2190–2194. doi: 10.1073/pnas.76.5.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown I. R. Postnatal appearance of short DNA repeat length in neurons of the cerebral cortex. Biochem Biophys Res Commun. 1978 Sep 29;84(2):285–292. doi: 10.1016/0006-291x(78)90168-7. [DOI] [PubMed] [Google Scholar]
- Carter C. W., Jr Histone packing in the nucleosome core particle of chromatin. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3649–3653. doi: 10.1073/pnas.75.8.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon P. Summary: the molecular biology of the eukaryotic genome is coming of age. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1209–1234. doi: 10.1101/sqb.1978.042.01.122. [DOI] [PubMed] [Google Scholar]
- Chapman G. E., Hartman P. G., Bradbury E. M. Studies on the role and mode of operation of the very-lysine-rich histone H1 in eukaryote chromatin. The isolation of the globular and non-globular regions of the histone H1 molecule. Eur J Biochem. 1976 Jan 2;61(1):69–75. doi: 10.1111/j.1432-1033.1976.tb09998.x. [DOI] [PubMed] [Google Scholar]
- Compton J. L., Bellard M., Chambon P. Biochemical evidence of variability in the DNA repeat length in the chromatin of higher eukaryotes. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4382–4386. doi: 10.1073/pnas.73.12.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornudella L., Rocha E. Nucleosome organization during germ cell development in the sea cucumber Holothuria tubulosa. Biochemistry. 1979 Aug 21;18(17):3724–3732. doi: 10.1021/bi00584a013. [DOI] [PubMed] [Google Scholar]
- Ermini M., Kuenzle C. C. The chromatin repeat length of cortical neurons shortens during early posnatal development. FEBS Lett. 1978 Jun 1;90(1):167–172. doi: 10.1016/0014-5793(78)80322-6. [DOI] [PubMed] [Google Scholar]
- Franklin S. G., Zweidler A. Non-allelic variants of histones 2a, 2b and 3 in mammals. Nature. 1977 Mar 17;266(5599):273–275. doi: 10.1038/266273a0. [DOI] [PubMed] [Google Scholar]
- Friend C., Scher W., Holland J. G., Sato T. Hemoglobin synthesis in murine virus-induced leukemic cells in vitro: stimulation of erythroid differentiation by dimethyl sulfoxide. Proc Natl Acad Sci U S A. 1971 Feb;68(2):378–382. doi: 10.1073/pnas.68.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotov B. O., Nikolaev L. G., Severin E. S. Histone H1--DNA interaction. On the mechanism of DNA strands crosslinking by histone H1. Nucleic Acids Res. 1978 Jul;5(7):2587–2605. doi: 10.1093/nar/5.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J. B., Pollock J. M., Jr, Rill R. L. Chromatin fractionation procedure that yields nucleosomes containing near-stoichiometric amounts of high mobility group nonhistone chromosomal proteins. Biochemistry. 1979 Aug 21;18(17):3739–3748. doi: 10.1021/bi00584a015. [DOI] [PubMed] [Google Scholar]
- Johnson E. M., Littau V. C., Allfrey V. G., Bradbury E. M., Matthews H. R. The subunit structure of chromatin from Physarum polycephalum. Nucleic Acids Res. 1976 Dec;3(12):3313–3329. doi: 10.1093/nar/3.12.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Levy A., Jakob K. M. Nascent DNA in nucleosome like structures from chromatin. Cell. 1978 Jun;14(2):259–267. doi: 10.1016/0092-8674(78)90112-5. [DOI] [PubMed] [Google Scholar]
- Loening U. E. The fractionation of high-molecular-weight ribonucleic acid by polyacrylamide-gel electrophoresis. Biochem J. 1967 Jan;102(1):251–257. doi: 10.1042/bj1020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Corden J., Tatchell K., Kovacic R. T., Van Holde K. E. Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1977 Jan;74(1):79–83. doi: 10.1073/pnas.74.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Kovacic R. T., Van Holde K. E. Quantitative analysis of the digestion of yeast chromatin by staphylococcal nuclease. Biochemistry. 1977 Feb 8;16(3):463–471. doi: 10.1021/bi00622a020. [DOI] [PubMed] [Google Scholar]
- Martin D. Z., Todd R. D., Lang D., Pei P. N., Garrard W. T. Heterogeneity in nucleosome spacing. J Biol Chem. 1977 Nov 25;252(22):8269–8277. [PubMed] [Google Scholar]
- Miki B. L., Neelin J. M. DNA repeat lengths of erythrocyte chromatins differing in content of histones H1 and H5. Nucleic Acids Res. 1980 Feb 11;8(3):529–542. doi: 10.1093/nar/8.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirand E. A., Steeves R. A., Avila L., Grace J. T., Jr Spleen focus formation by polycythemic strains of Friend leukemia virus. Proc Soc Exp Biol Med. 1968 Mar;127(3):900–904. doi: 10.3181/00379727-127-32831. [DOI] [PubMed] [Google Scholar]
- Mirzabekov A. D., Shick V. V., Belyavsky A. V., Bavykin S. G. Primary organization of nucleosome core particle of chromatin: sequence of histone arrangement along DNA. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4184–4188. doi: 10.1073/pnas.75.9.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris N. R. Nucleosome structure in Aspergillus nidulans. Cell. 1976 Jul;8(3):357–363. doi: 10.1016/0092-8674(76)90147-1. [DOI] [PubMed] [Google Scholar]
- Murphy R. F., Wallace R. B., Bonner J. Altered nucleosome spacing in newly replicated chromatin from Friend leukemia cells. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5903–5907. doi: 10.1073/pnas.75.12.5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyldermans S., Lasters I., Wyns L. Histone H1 can be removed selectively from chicken erythrocyte chromatin at near physiological conditions. Nucleic Acids Res. 1980 Feb 25;8(4):731–739. [PMC free article] [PubMed] [Google Scholar]
- Noll M. Differences and similarities in chromatin structure of Neurospora crassa and higher eucaryotes. Cell. 1976 Jul;8(3):349–355. doi: 10.1016/0092-8674(76)90146-x. [DOI] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Ostertag W., Melderis H., Steinheider G., Kluge N., Dube S. Synthesis of mouse haemoglobin and globin mRNA in leukaemic cell cultures. Nat New Biol. 1972 Oct 25;239(95):231–234. doi: 10.1038/newbio239231a0. [DOI] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D. Relation of nucleosomes to DNA sequences. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):103–108. doi: 10.1101/sqb.1978.042.01.011. [DOI] [PubMed] [Google Scholar]
- Reuben R. C., Khanna P. L., Gazitt Y., Breslow R., Rifkind R. A., Marks P. A. Inducers of erythroleukemic differentiation. Relationship of structure to activity among planar-polar compounds. J Biol Chem. 1978 Jun 25;253(12):4214–4218. [PubMed] [Google Scholar]
- Rill R. L., Nelson D. A., Oosterhof D. K., Hozier J. C. Structural repeat units of Chinese hamster ovary chromatin. Evidence for variations in repeat unit DNA size in higher eukaryotes. Nucleic Acids Res. 1977 Apr;4(4):771–789. doi: 10.1093/nar/4.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel R. A., Haye K. R., Litwack A. H., Phelps B. M. Nucleosome repeat lengths in the definitive erythroid series of the adult chicken. Biochim Biophys Acta. 1980 Feb 29;606(2):316–330. doi: 10.1016/0005-2787(80)90041-6. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Spadafora C., Bellard M., Compton J. L., Chambon P. The DNA repeat lengths in chromatins from sea urchin sperm and gastrule cells are markedly different. FEBS Lett. 1976 Oct 15;69(1):281–285. doi: 10.1016/0014-5793(76)80704-1. [DOI] [PubMed] [Google Scholar]
- Tatchell K., Van Holde K. E. Compact oligomers and nucleosome phasing. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3583–3587. doi: 10.1073/pnas.75.8.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Thompson R. J. Variation in chromatin structure in two cell types from the same tissue: a short DNA repeat length in cerebral cortex neurons. Cell. 1977 Apr;10(4):633–640. doi: 10.1016/0092-8674(77)90096-4. [DOI] [PubMed] [Google Scholar]
- Vindelov L. L. Flow microfluorometric analysis of nuclear DNA in cells from solid tumors and cell suspensions. A new method for rapid isolation and straining of nuclei. Virchows Arch B Cell Pathol. 1977 Aug 10;24(3):227–242. [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- West M. H., Bonner W. M. Histone 2A, a heteromorphous family of eight protein species. Biochemistry. 1980 Jul 8;19(14):3238–3245. doi: 10.1021/bi00555a022. [DOI] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Simpson R. T. Removal of histone H1 exposes a fifty base pair DNA segment between nucleosomes. Biochemistry. 1976 Jul 27;15(15):3307–3314. doi: 10.1021/bi00660a022. [DOI] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Simpson R. T. Removal of histone H1 exposes a fifty base pair DNA segment between nucleosomes. Biochemistry. 1976 Jul 27;15(15):3307–3314. doi: 10.1021/bi00660a022. [DOI] [PubMed] [Google Scholar]
- Wilhelm M. L., Mazen A., Wilhelm F. X. Comparison of the DNA repeat length in H1- and H3-containing chromatin. FEBS Lett. 1977 Jul 15;79(2):404–408. doi: 10.1016/0014-5793(77)80831-4. [DOI] [PubMed] [Google Scholar]
- Yaneva M., Tasheva B., Dessev G. Nuclease digestion of reconstituted chromatin. FEBS Lett. 1976 Nov;70(1):67–70. doi: 10.1016/0014-5793(76)80727-2. [DOI] [PubMed] [Google Scholar]