The Tup1–Cyc8 complex has been thought to function as a corepressor for many genes in Saccharomyces cerevisiae. In this Perspective, the novel mechanism by which Tup1–Cyc8 acts is discussed, as shown by Wong and Struhl in this issue. The authors reveal that the complex acts by blocking the activation domain of DNA-binding proteins and preventing recruitment of transcriptional coactivators.
Keywords: Cyc8–Tup1, transcriptional corepressor, gene regulation, stress responses, transcriptional activation, transcriptional repression
Abstract
The Tup1–Cyc8 complex is responsible for repression of a large and diverse collection of genes in Saccharomyces cerevisiae. The predominant view has been that Tup1–Cyc8 functions as a corepressor, actively associating with regulatory proteins and organizing chromatin to block transcription. A new study by Wong and Struhl in this issue of Genes & Development (pp. 2525–2539) challenges nearly 20 years of models by demonstrating that Tup1–Cyc8 functions primarily as a shield to block DNA-binding proteins from recruiting transcriptional coactivators.
The Tup1–Cyc8 (also known as Tup1–Ssn6) complex is implicated in the repression of >300 genes in Saccharomyces cerevisiae in standard growth conditions (DeRisi et al. 1997; Green and Johnson 2004). A variety of mechanisms have been proposed to explain how Tup1–Cyc8 confers repression on its target genes, and, at individual genes, different mechanisms appear to influence expression to varying degrees. A study by Wong and Struhl (2011) in this issue of Genes & Development sheds light on the mechanism of Tup1–Cyc8 action by providing a new explanation for how repression is achieved. While Tup1–Cyc8 has long been considered a corepressor complex, Wong and Struhl (2011) suggest that these proteins do not usually function by actively repressing transcription. Rather, Tup1–Cyc8 interacts with a DNA-binding protein to mask or shield its activation domain, thereby preventing recruitment of the coactivators necessary for activated transcription.
The Tup1–Cyc8 complex: a prototypical corepressor
Tup1–Cyc8 was one of the first complexes to be defined as a transcriptional corepressor (Keleher et al. 1992; Tzamarias and Struhl 1994). As such, neither Tup1 nor Cyc8 binds directly to DNA. Instead, Tup1–Cyc8 associates with a variety of DNA-binding repressor proteins that direct its recruitment to particular subsets of genes. In this way, Tup1–Cyc8 is able to affect transcription of many genes regulated by glucose and oxygen availability, DNA damage, and cell type (Keleher et al. 1992; Balasubramanian et al. 1993; Komachi et al. 1994; Treitel and Carlson 1995; Huang et al. 1998; Park et al. 1999; Hanlon et al. 2011).
The importance of Tup1–Cyc8 is highlighted by the presence of homologs in Caenorhabditis elegans and Schizosaccharomyces pombe and the evolutionary relatives in metazoans, the Groucho/TLE family of corepressors (Malave and Dent 2006). The well-studied Drosophila melanogaster Groucho protein participates in many diverse processes throughout development, such as segmentation, dorsal ventral pattern formation, and sex determination. Likewise, the mammalian TLE proteins that function in embryonic development have been implicated in human disease.
The Tup1–Cyc8 complex is composed of four Tup1 subunits and one Cyc8 subunit (Varanasi et al. 1996). Multimerization of Tup1 is mediated by its N terminus, which is also required for interaction with Cyc8 (Tzamarias and Struhl 1994, 1995). The Tup1 C terminus contains seven 40-amino-acid WD40 repeat motifs that fold into a β propeller-like structure to mediate protein–protein interactions, such as with the repressor protein α2 (Sprague et al. 2000). Cyc8 has 10 tandem copies of a tetratricopeptide (TPR) motif, and different combinations of these motifs are necessary for interaction with Tup1 and repression of different genes (Tzamarias and Struhl 1995).
LexA fusion experiments have demonstrated that Tup1 is the primary repressor protein, as it can repress transcription of a reporter in the absence of Cyc8 (Tzamarias and Struhl 1994). In contrast, Cyc8 requires Tup1 for repression (Keleher et al. 1992; Tzamarias and Struhl 1994). Both proteins are necessary for repression at native genes. Studies have suggested that in many cases, Tup1 is the protein that carries out the repression, and Cyc8 mediates the interaction with DNA-binding repressor proteins.
Models of Tup1–Cyc8 repression
A variety of mechanisms have been proposed to explain how Tup1–Cyc8 represses genes. The three most predominant models include (1) recruitment of histone deacetylases (HDACs) to modify chromatin structure at the promoter, (2) directing the positioning of nucleosomes at the promoter, and (3) interference with the Mediator complex to more actively prevent transcription by RNA polymerase II. These models are not mutually exclusive, and it has been suggested that they serve as redundant mechanisms to ensure repression of target genes.
Several observations have suggested a mechanistic link between Tup1–Cyc8 repression and the deacetylation of chromatin. Lysine residues in the N-terminal tails of all four core histone proteins can be acetylated by histone acetyltransferases (HATs) and deacetylated by HDACs. In general, increased acetylation of histone tails is correlated with higher transcription, while decreased acetylation reduces the level of transcription. Tup1 can associate directly with histones, and, in vitro, Tup1 binds preferentially to the hypoacetylated tails of histones H3 and H4 (Edmondson et al. 1996). Histone tail mutations that disrupt the interaction with Tup1 cause derepression of reporter genes with binding sites for Tup1-recruiting repressors (Edmondson et al. 1996; Huang et al. 1997). Genetic and physical associations between Tup1–Cyc8 and several HDACs have been also been reported, including in vivo interactions with Rpd3 and Hos2 and an in vitro interaction with Hda1 (Watson et al. 2000; Wu et al. 2001; Robyr et al. 2002; Davie et al. 2003; Green and Johnson 2004). A general model to explain these observations is that a DNA-binding repressor recruits Tup1–Cyc8, which in turn recruits HDACs, leading to hypoacetylated chromatin at the promoter. The hypoacetylation not only diminishes transcription capacity, but also reinforces Tup1 association in a positive feed-forward mechanism (Davie et al. 2002). By this model, the particular HDACs involved and the pattern of histone deacetylation contributing to repression is gene-specific (Watson et al. 2000; Deckert and Struhl 2001; Wu et al. 2001).
A distinct model has Tup1–Cyc8 directing nucleosome positioning, which limits the accessibility of trans-acting factors to critical regions of the promoter, such as the TATA box. According to this model, hypoacetylation of histone tails contributes to Tup1–Cyc8 binding. This model is supported by the finding that positioned nucleosomes in the promoter region of a number of Tup1-regulated genes disappear in the absence of Tup1 (Cooper et al. 1994; Kastaniotis et al. 2000; Fleming and Pennings 2001, 2007; Rizzo et al. 2011). At some genes, Isw2 may also play a role in positioning the nucleosomes, although how Tup1 influences this process is unclear (Zhang and Reese 2004; Morohashi et al. 2006; Rizzo et al. 2011). It is important, however, to note that hypoacetylation is also required for repression at some promoters that do not have clearly positioned nucleosomes, suggesting that hypoacetylation can contribute to repression by means that do not involve nucleosome positioning.
Studies have also suggested that Tup1–Cyc8 may also function, at least in part, by interacting with the Mediator complex, possibly interfering with Mediator's association with activators. The S. cerevisiae Mediator complex associates with the C-terminal domain of the largest subunit of RNA polymerase II and contributes to both transcriptional activation and repression (Björklund and Gustafsson 2005; Kornberg 2005). A number of genetic screens that identified Tup1–Cyc8 as repressing gene transcription also identified Mediator subunits, suggesting that Tup1–Cyc8 and Mediator both contribute to repression (Wahi and Johnson 1995; Friesen et al. 1998; Mizuno et al. 1998; Kunoh et al. 2000). Additionally, mutations in unc-37/Groucho, the C. elegans Tup1 homolog, show genetic interactions with Mediator mutations (Zhang and Emmons 2002). Finally, biochemical studies show interactions between Tup1–Cyc8 and Mediator (Gromoller and Lehming 2000; Papamichos-Chronakis et al. 2000; Han et al. 2001). A model of Tup1–Cyc8 function involving Mediator components proposes that repression is achieved by competition between the repressor and activator for interaction with Mediator. The suggestion that Tup1–Cyc8 may interact with multiple subunits of Mediator provides versatility, allowing a basic mechanism to be adapted to different genes.
A new model: Tup1–Cyc8 masks activation domains
The models for Tup1–Cyc8 function presented above are not mutually exclusive. Rather, they have been proposed to provide complementary approaches to reinforce the repressive state. The repeated observation that mutation of any one chromatin or Mediator component has only a modest effect on repression, while combinations of these mutations have a synergistic effect, supports this idea of redundancy in the mechanism. Likewise, Tup1–Cyc8 repression in vitro is usually twofold to fourfold, much less than the repression levels observed at target genes in vivo (typically 15-fold to 50-fold) (Lee et al. 2000). Gene-specific differences are likely to occur as well, with one or more mechanisms predominating at particular loci.
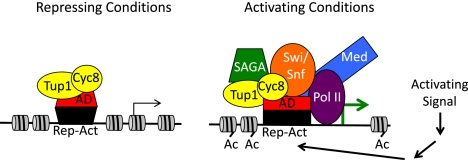
The study by Wong and Struhl (2011) suggests a new model (Fig. 1) that represents a paradigm shift in understanding the properties of the DNA-binding repressor protein and the Tup1–Cyc8 complex and the manner by which repression is achieved. Wong and Struhl (2011) propose that the DNA-binding protein is actually a “repressor–activator” capable of specifying both repression and activation and that Tup1–Cyc8 plays a more passive role in repression than previously appreciated. Rather than recruiting HDACs, positioning nucleosomes, or interfering with Mediator recruitment, Wong and Struhl (2011) provide evidence that Tup1–Cyc8 simply masks the activation domain of the DNA-bound protein, preventing recruitment of coactivators (including Mediator). An activating signal would then alter the repressor–activator or Tup1–Cyc8, allowing the activation domain to be unmasked and coactivators to be recruited.
Figure 1.
Model for Tup1–Cyc8 masking of activation domains. In repressing conditions, the complex of a Tup1 tetramer with a single Cyc8 protein associates with the activation domain (AD) of the repressor–activator (Rep–Act), preventing association of SAGA, Swi/Snf, and Mediator coactivators. The absence of recruited chromatin remodelers and HATs results in a “default” state of relatively deacetylated histones and compact nucleosome structure. An environmental signal (activating condition) initiates a cascade of events leading to modification of the repressor–activator and possibly a conformational change that alters its interaction with Tup1–Cyc8 to allow recruitment of SAGA, Swi/Snf, and Mediator and subsequent acetylation of histones and disruption of nucleosomes. Tup1–Cyc8 remains at the promoter and may participate in coactivator recruitment.
Anchor-away: an effective strategy to remove Tup1 and Cyc8 from cells
Wong and Struhl (2011) used the anchor-away method developed by Haruki et al. (2008) to assess the effects of depleting the nucleus of Tup1 or Cyc8. This system takes advantage of the tightly associated ternary complex of rapamycin with FKBP12 and FRB (components of the human TOR1 complex). FKBP12 is attached to the anchor protein (in this case, the Rpl13A ribosomal protein), and the FRB domain is attached to the target protein (in this case, either Tup1 or Cyc8). After translation in the cytoplasm, Rpl13A enters the nucleus to associate with rRNA, then returns to the cytoplasm to be assembled into the complete ribosome. In the presence of rapamycin, the Rpl13A-FKBP12 fusion anchor protein associates with Tup1-FRB in the nucleus. Upon leaving the nucleus, Rpl13A then effectively tethers Tup1 in the cytoplasm, where it cannot function.
The anchor-away method allows for rapid removal of Tup1 or Cyc8, such that direct effects can be monitored. This is especially important for proteins such as Tup1–Cyc8, where a gene disruption causes extreme sickness that can have many secondary effects unrelated to the direct action of Tup1 and Cyc8. As such, the experiments of Wong and Struhl (2011) more directly assess the role of these proteins than the artificial reporter systems used in many past studies. As a proof of principle, depletion of Tup1 or Cyc8 from the nucleus using the anchor-away method recapitulates key aspects of the tup1 phenotype previously observed; namely, cellular flocculation, reduced density of histone H3, increased H3 acetylation, and derepression of target genes. Importantly, the anchor-away method allows kinetic analysis of events following depletion of Tup1 or Cyc8, clearly identifying the direct effects of factor loss, as opposed to long-term or indirect effects.
Tup1–Cyc8 prevents recruitment of coactivators to target genes
Tup1–Cyc8 is thought to recruit HDACs, resulting in hypoacetylation and repression of the target gene. Wong and Struhl (2011) suggest an alternative strategy for enforcing repression. They propose that it is actually the loss of HATs that contributes to the repressive effects of Tup1–Cyc8, rather than the gain of HDACs. While chromatin immunoprecipitation (ChIP) with whole-genome sequencing (ChIP-seq) failed to reveal association of Hda1 with Tup1 target promoters, Wong and Struhl (2011) noticed that binding of Gcn5, the catalytic subunit of the SAGA HAT complex, increased at some target genes in the absence of Tup1. Moreover, the increased histone acetylation at these genes was GCN5-dependent. This observation suggests that, in a tup1 mutant, it could be recruitment of Gcn5 or other HATs to a Tup1-regulated promoter that causes the increased acetylation observed. The result is the same as if Tup1 recruited Hda1 or other HDACs, but the mechanism is different. As such, this model represents a change in the method by which Tup1–Cyc8 affects acetylation at target genes.
Tup1–Cyc8 has been proposed to use nucleosome positioning to establish inhibitory chromatin structure at its target genes, in some cases in collaboration with the Isw2-repressive chromatin remodeler. Wong and Struhl (2011) considered the alternative model that Tup1–Cyc8 instead blocks association of activating nucleosome remodelers, resulting in nucleosomes positioned differently than in a tup1 mutant. They examined the kinetics following rapamycin addition and found that loss of Tup1 from target promoters is concurrent with, or slightly precedes, Swi/Snf recruitment and histone H3 depletion. They also used a “double anchor-away” method, allowing the sort of epistasis analysis used by geneticists to compare the phenotypes in single and double mutants. Depleting both Tup1 and Snf2 (the catalytic subunit of the Swi/Snf chromatin remodeler) or both Tup1 and Sth1 (the catalytic subunit of the RSC chromatin remodeler) caused diminished histone eviction relative to that observed simply in the absence of Tup1. This important finding shows that nucleosome eviction is not simply due to loss of Tup1, but that the remodeler is required for histone loss and that Tup1 blocks recruitment of the remodeler. The effects of depletion of Snf2 or Sth1 were gene-dependent, with some promoters affected more with one or the other of the two remodelers. While Tup1 loss resulted in gene derepression, the simultaneous removal of Tup1 in combination with Sth1 or Snf2 eliminated derepression at some target genes. This result supports the idea that Tup1 defines the repressed state at least in part by inhibiting recruitment of chromatin remodelers.
In support of the earlier data suggesting a connection between Tup1–Cyc8 and Mediator, depletion of Tup1 resulted in significantly increased Mediator recruitment. Wong and Struhl (2011) suggested that Tup1–Cyc8 inhibition of Mediator recruitment could contribute to the repressed state in a manner analogous to that described above for inhibiting recruitment of HATs and activating chromatin remodelers. In support of this idea, the transcriptional derepression observed in the Tup1 anchor-away strain was abrogated in a Tup1/Gal11 double anchor-away strain. This epistasis experiment shows that Mediator is required for gene activation in the tup1 mutant.
A ChIP-seq experiment provided additional compelling support for their model. Following Tup1 depletion, the coactivators Snf2 and Gal11 were recruited to genome-wide sites previously occupied by Tup1. Remarkable overlap was observed between sites of Tup1 localization and Gal11 and Snf2 localization. Tup1 was recruited to a highly localized site in promoters, and the Tup1 peaks were within 20 base pairs of the Gal11 and Snf2 peaks observed when Tup1 was depleted from the nucleus. These results suggest that, in most cases, the DNA-binding protein at the Tup1-regulated gene is capable of recruiting both the coactivators and Tup1. Thus, Wong and Struhl (2011) propose that the proteins previously suggested to be repressors are more aptly renamed “repressor–activators.”
An important feature of these experiments using anchor-away strains was the ability to track the kinetics of the derepression of Tup1–Cyc8 target genes. Introduction of a galactose-inducible Tup1 into the Tup1 anchor-away strain also allowed for kinetic monitoring of the re-establishment of repression. All of the coactivators were rapidly recruited upon Tup1 depletion, concurrent with histone acetylation and nucleosome eviction. The add-back experiment demonstrated that association of Tup1, loss of transcription, and dissociation of SAGA, Mediator, and Swi/Snf all occurred well before the redeposition of nucleosomes. This result suggests that repression of transcription occurs before the chromatin structure is re-established, arguing against the idea that Tup1 represses genes by imposing specific nucleosome positioning that is inhibitory to transcription.
Tup1–Cyc8 shields against association of activation domains, thereby converting activators to repressors
Wong and Struhl (2011) also investigated the mechanism by which Tup1 prevents the association of coactivators. Reporter genes were constructed with promoters containing an activator-binding site placed either upstream of or downstream from binding sites for factors that recruit Tup1. This experiment demonstrated that Tup1 does not sterically inhibit activator binding to the promoter—a mechanism that had been in the literature (Lee et al. 2000). Instead, Tup1 appears to interact with activation domains in vivo. LexA-Gcn4 and LexA-VP16 were shown to recruit Tup1 to LexA sites at a detectable level, although much less than the recruitment observed with known Tup1-recruiting proteins such as Nrg1, α2, and Rox1.
The combined results presented by Wong and Struhl (2011) suggest a model in which the main function of Tup1–Cyc8 is to shield the activation domain of a repressor–activator DNA-binding protein, thereby preventing association of coactivators. This model provides an explanation as to why mutations of a variety of chromatin factors individually display only modest effects relative to deletion of the TUP1 or CYC8 genes. For example, if an HDAC is mutated, increased acetylation may occur at the gene, partially relieving repression, but since the Tup1–Cyc8 shield is still in place, chromatin remodelers and Mediator cannot associate and the gene cannot be fully activated. The only method of obtaining full activation is by eliminating the shield. Thus, rather than each previously described mechanism representing alternative or redundant methods of repression, Wong and Struhl (2011) suggest that there is a single unifying mechanism that prevents all coactivator components from associating with the promoter and activating transcription.
The Tup1–Cyc8 masking model explains several previous observations
The new model described by Wong and Struhl (2011) suggests an alternative interpretation of previous experimental results relative to models presented in earlier studies. For example, instead of viewing connections with HDACs as evidence that Tup1–Cyc8 recruits these complexes, a masking model suggests that it is the loss of HATs that links Tup1–Cyc8 to the deacetylated state. Previous models have suggested that the derepression and increased acetylation of Tup1–Cyc8 target genes in the absence of HDACs result from loss of HDACs targeted via recruitment by Tup1–Cyc8. However, the masking model suggests that these results are better explained by the HDACs acting in a global (i.e., nontargeted) fashion. Blocking HAT recruitment by Tup1–Cyc8 would allow the reciprocal HDAC with appropriate histone tail specificity to predominate at Tup1 target genes. Connections between the negative chromatin remodeler Isw2 and various components of the Mediator complex can also be reinterpreted in light of the new masking model by the loss of activating functions, rather than the gain of repressing functions. Similarly, nucleosome positioning at Tup1–Cyc8-regulated genes can be explained by the absence of activating chromatin remodelers that would alter chromatin structure at the gene. This would explain the fact that while there has been much suggestion of a role for Tup1–Cyc8 in positioning nucleosomes, there is, to date, no actual evidence that Tup1–Cyc8 itself actually performs this function.
The Tup1 masking model is also consistent with observations that have been difficult to explain by other models of Tup1–Cyc8 repression. Several studies have shown that Tup1 can remain associated with the formerly repressed gene upon activation. For example, ChIP–chip has demonstrated that Tup1 is bound to many glucose-repressible target genes even after glucose repression is relieved (Buck and Lieb 2006). In some cases, such as at the GAL1, GRE2, and AHP1 genes, Tup1–Cyc8 participates in the recruitment of chromatin-modifying and/or remodeling coactivators during activation (Papamichos-Chronakis et al. 2002; Proft and Struhl 2002). At these genes, the coactivators appear to be required only for overcoming the Tup1–Cyc8 repression, but the observations provide precedent for the idea that Tup1–Cyc8 may, in some cases, be capable of recruiting chromatin-modifying and remodeling complexes. However, at other genes, such as ANB1, there is no evidence to suggest that Tup1–Cyc8 plays a role in the activation process, even though Tup1 remains bound when repression is relieved (Mennella et al. 2003).
The suggestion that proteins that recruit Tup1 are capable of both activation and repression is supported by other experiments. The Sko1 DNA-binding protein, which activates genes induced in hyperosmotic stress, is converted from a repressor to an activator by phosphorylation (Proft and Struhl 2002). Crt1 also appears to function as a repressor–activator; its N-terminal Tup1–Cyc8 recruitment domain overlaps a TFIID and Swi/Snf association domain (Zhang and Reese 2005). Also in agreement with the new model by Wong and Struhl (2011), the Cin5, Phd1, Yap6, Skn7 DNA-binding proteins recently described as recruiting Tup1–Cyc8 (Hanlon et al. 2011) have been previously characterized as transcriptional activators.
Tup1–Cyc8 as a shield: future questions
The physical associations between Tup1–Cyc8 and Mediator components, HDACs, and histone tails must be re-examined in light of the new masking model of Wong and Struhl (2011). Experimentally observed interactions between Tup1–Cyc8 and transcriptionally positive Mediator subunits were formerly interpreted as a mechanism by which Tup1–Cyc8 could interfere directly with Mediator function and thus repress transcription. In the new model, these interactions are instead explained by an alternative mechanism in which Tup1–Cyc8 remains at some promoters during activation and helps to recruit Mediator in a manner analogous to the recruitment of chromatin modifiers and remodelers that has previously been observed. The interactions between Tup1–Cyc8 and histone tails or HDACs are not required for coherence of the masking model. Perhaps an association between Tup1 and histones helps to reinforce the repressed state. Alternatively, Wong and Struhl (2011) suggest that Tup1–Cyc8 may function in part as a corepressor at some genes by interacting with histone tails or HDACs, but that this role is secondary to its major role in masking activation domains. Gene-specific variations may also contribute to the complexity; while Tup1–Cyc8 may generally function as a mask for activation domains, it is possible that different mechanisms are used at particular genes. The model of Tup1–Cyc8 acting as an anti-coactivator shield should inform future experiments to examine the role of Tup1–Cyc8 at specific genes and should clarify any role of interactions with Mediator, histone tails, or HDACs.
How is transcriptional activation achieved at genes repressed by Tup1–Cyc8?
There appear to be at least two distinct modes for activation of these target genes. It is believed that, at some genes, transcriptional activation is accompanied by loss of Tup1–Cyc8 binding. Future studies will need to focus on the mechanism by which this is accomplished. Is the DNA-binding protein post-translationally modified to terminate the interaction with Tup1–Cyc8? A distinct possibility is that nuclear entry by a different DNA-binding activator that does not interact with Tup1–Cyc8 effectively replaces the repressing DNA-binding protein at the promoter.
At other genes, there is evidence that Tup1–Cyc8 remains bound to the promoter following transcriptional activation (Fig. 1). This raises important mechanistic questions about how these genes are activated. Is the repressor–activator modified to alter the interaction with Tup1–Cyc8 such that coactivators can now be recruited? Or is Tup1–Cyc8 the target of modification, either changing its interaction with the repressor–activator to unmask the activation domain or allowing it to actually recruit the coactivators? Modification of the repressor–activator seems most likely, as this would allow activation of a subset of Tup1–Cyc8 targets in a specific condition. In support of this idea, the Sko1 DNA-binding protein is phosphorylated upon hyperosmotic stress, converting it from a repressor to an activator, and Tup1–Cyc8 remains associated and participates in recruitment of SAGA and Swi/Snf coactivators (Proft and Struhl 2002). A major mechanistic question then becomes how the transcriptional activation signal modifies the interaction of the DNA-binding protein with Tup1–Cyc8 so that Tup1–Cyc8 no longer acts as a shield to inhibit coactivator recruitment.
The DNA-binding proteins that inhibit transcription by recruiting Tup1–Cyc8 to promoters have been called repressors, but Wong and Struhl (2011) suggest that we refer to these proteins as repressor–activators, based on their apparent dual function. Building on this idea, genetic experiments need to be performed to determine the effect of deleting the binding site for the repressor–activator at Tup1-regulated promoters. Deleting the binding site for a classic repressor should cause derepression, while a gene should be off when a repressor–activator site is deleted. This model also predicts that the activation and repression domains of repressor–activators can never be separated. Of course, some promoters may have nearby binding sites for other activators, and then the gene might be active upon deletion of the repressor–activator site. This latter situation raises the important question of how Tup1–Cyc8, recruited by a repressor–activator, can inhibit coactivator recruitment by an activator bound nearby. Finally, the major mechanistic question for future study is how Tup1–Cyc8, with their multiple WD40 and TPR motifs, function as a shield, repelling the recruitment of coactivators.
In conclusion, the new model for Tup1–Cyc8 repression presented by Wong and Struhl (2011) represents a significant advance in the field, unifying multiple seemingly diverse observations concerning Tup1–Cyc8 into a single parsimonious explanation. At specific genes, additional levels of complexity may exist, accounting for observations that are not necessarily consistent with the masking model. Nevertheless, this model provides a new framework for understanding how Tup1–Cyc8 repression is achieved and will certainly spark both the re-examination and potential reinterpretation of previous findings and the design of further experiments to determine the precise mechanisms used at individual target genes.
Acknowledgments
We thank Dean Tantin for helpful comments on the manuscript.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.181768.111.
References
- Balasubramanian B, Lowry CV, Zitomer RS 1993. The Rox1 repressor of the Saccharomyces cerevisiae hypoxic genes is a specific DNA-binding protein with a high-mobility-group motif. Mol Cell Biol 13: 6071–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björklund S, Gustafsson CM 2005. The yeast Mediator complex and its regulation. Trends Biochem Sci 30: 240–244 [DOI] [PubMed] [Google Scholar]
- Buck MJ, Lieb JD 2006. A chromatin-mediated mechanism for specification of conditional transcription factor targets. Nat Genet 38: 1446–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JP, Roth SY, Simpson RT 1994. The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev 8: 1400–1410 [DOI] [PubMed] [Google Scholar]
- Davie JK, Trumbly RJ, Dent SY 2002. Histone-dependent association of Tup1–Ssn6 with repressed genes in vivo. Mol Cell Biol 22: 693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie JK, Edmondson DG, Coco CB, Dent SY 2003. Tup1–Ssn6 interacts with multiple class I histone deacetylases in vivo. J Biol Chem 278: 50158–50162 [DOI] [PubMed] [Google Scholar]
- Deckert J, Struhl K 2001. Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol Cell Biol 21: 2726–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680–686 [DOI] [PubMed] [Google Scholar]
- Edmondson DG, Smith MM, Roth SY 1996. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev 10: 1247–1259 [DOI] [PubMed] [Google Scholar]
- Fleming AB, Pennings S 2001. Antagonistic remodelling by Swi–Snf and Tup1–Ssn6 of an extensive chromatin region forms the background for FLO1 gene regulation. EMBO J 20: 5219–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AB, Pennings S 2007. Tup1–Ssn6 and Swi–Snf remodelling activities influence long-range chromatin organization upstream of the yeast SUC2 gene. Nucleic Acids Res 35: 5520–5531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen H, Tanny JC, Segall J 1998. Spe3, which encodes spermidine synthase, is required for full repression through NRE(DIT) in Saccharomyces cerevisiae. Genetics 150: 59–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SR, Johnson AD 2004. Promoter-dependent roles for the Srb10 cyclin-dependent kinase and the Hda1 deacetylase in Tup1-mediated repression in Saccharomyces cerevisiae. Mol Biol Cell 15: 4191–4202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromoller A, Lehming N 2000. Srb7p is a physical and physiological target of Tup1p. EMBO J 19: 6845–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SJ, Lee JS, Kang JS, Kim YJ 2001. Med9/Cse2 and Gal11 modules are required for transcriptional repression of distinct group of genes. J Biol Chem 276: 37020–37026 [DOI] [PubMed] [Google Scholar]
- Hanlon SE, Rizzo JM, Tatomer DC, Lieb JD, Buck MJ 2011. The stress response factors Yap6, Cin5, Phd1, and Skn7 direct targeting of the conserved co-repressor Tup1–Ssn6 in S. cerevisiae. PLoS ONE 6: e19060 doi: 10.1371/journal.pone.0019060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK 2008. The anchor-away technique: Rapid, conditional establishment of yeast mutant phenotypes. Mol Cell 31: 925–932 [DOI] [PubMed] [Google Scholar]
- Huang L, Zhang W, Roth SY 1997. Amino termini of histones H3 and H4 are required for a1-α2 repression in yeast. Mol Cell Biol 17: 6555–6562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Zhou Z, Elledge SJ 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94: 595–605 [DOI] [PubMed] [Google Scholar]
- Kastaniotis AJ, Mennella TA, Konrad C, Torres AM, Zitomer RS 2000. Roles of transcription factor Mot3 and chromatin in repression of the hypoxic gene ANB1 in yeast. Mol Cell Biol 20: 7088–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher CA, Redd MJ, Schultz J, Carlson M, Johnson AD 1992. Ssn6–Tup1 is a general repressor of transcription in yeast. Cell 68: 709–719 [DOI] [PubMed] [Google Scholar]
- Komachi K, Redd MJ, Johnson AD 1994. The WD repeats of Tup1 interact with the homeo domain protein α 2. Genes Dev 8: 2857–2867 [DOI] [PubMed] [Google Scholar]
- Kornberg RD 2005. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci 30: 235–239 [DOI] [PubMed] [Google Scholar]
- Kunoh T, Sakuno T, Furukawa T, Kaneko Y, Harashima S 2000. Genetic characterization of rbt mutants that enhance basal transcription from core promoters in Saccharomyces cerevisiae. J Biochem 128: 575–584 [DOI] [PubMed] [Google Scholar]
- Lee M, Chatterjee S, Struhl K 2000. Genetic analysis of the role of Pol II holoenzyme components in repression by the Cyc8–Tup1 corepressor in yeast. Genetics 155: 1535–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malave TM, Dent SY 2006. Transcriptional repression by Tup1–Ssn6. Biochem Cell Biol 84: 437–443 [DOI] [PubMed] [Google Scholar]
- Mennella TA, Klinkenberg LG, Zitomer RS 2003. Recruitment of Tup1–Ssn6 by yeast hypoxic genes and chromatin-independent exclusion of TATA binding protein. Eukaryot Cell 2: 1288–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Nakazawa N, Remgsamrarn P, Kunoh T, Oshima Y, Harashima S 1998. The Tup1–Ssn6 general repressor is involved in repression of IME1 encoding a transcriptional activator of meiosis in Saccharomyces cerevisiae. Curr Genet 33: 239–247 [DOI] [PubMed] [Google Scholar]
- Morohashi N, Yamamoto Y, Kuwana S, Morita W, Shindo H, Mitchell AP, Shimizu M 2006. Effect of sequence-directed nucleosome disruption on cell-type-specific repression by α2/Mcm1 in the yeast genome. Eukaryot Cell 5: 1925–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Conlan RS, Gounalaki N, Copf T, Tzamarias D 2000. Hrs1/Med3 is a Cyc8–Tup1 corepressor target in the RNA polymerase II holoenzyme. J Biol Chem 275: 8397–8403 [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Petrakis T, Ktistaki E, Topalidou I, Tzamarias D 2002. Cti6, a PHD domain protein, bridges the Cyc8–Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol Cell 9: 1297–1305 [DOI] [PubMed] [Google Scholar]
- Park SH, Koh SS, Chun JH, Hwang HJ, Kang HS 1999. Nrg1 is a transcriptional repressor for glucose repression of STA1 gene expression in Saccharomyces cerevisiae. Mol Cell Biol 19: 2044–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M, Struhl K 2002. Hog1 kinase converts the Sko1–Cyc8–Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell 9: 1307–1317 [DOI] [PubMed] [Google Scholar]
- Rizzo JM, Mieczkowski PA, Buck MJ 2011. Tup1 stabilizes promoter nucleosome positioning and occupancy at transcriptionally plastic genes. Nucleic Acids Res 39: 8803–8819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, Grunstein M 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109: 437–446 [DOI] [PubMed] [Google Scholar]
- Sprague ER, Redd MJ, Johnson AD, Wolberger C 2000. Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast. EMBO J 19: 3016–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treitel MA, Carlson M 1995. Repression by SSN6–TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci 92: 3132–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzamarias D, Struhl K 1994. Functional dissection of the yeast Cyc8–Tup1 transcriptional co-repressor complex. Nature 369: 758–761 [DOI] [PubMed] [Google Scholar]
- Tzamarias D, Struhl K 1995. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8–Tup1 corepressor complex to differentially regulated promoters. Genes Dev 9: 821–831 [DOI] [PubMed] [Google Scholar]
- Varanasi US, Klis M, Mikesell PB, Trumbly RJ 1996. The Cyc8 (Ssn6)–Tup1 corepressor complex is composed of one Cyc8 and four Tup1 subunits. Mol Cell Biol 16: 6707–6714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahi M, Johnson AD 1995. Identification of genes required for α 2 repression in Saccharomyces cerevisiae. Genetics 140: 79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson AD, Edmondson DG, Bone JR, Mukai Y, Yu Y, Du W, Stillman DJ, Roth SY 2000. Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev 14: 2737–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KH, Struhl K 2011. The Cyc8–Tup1 complex inhibits transcription primarily by masking the activation domain of the recruiting protein. Genes Dev (this issue) doi: 10.1101/gad.179275.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Suka N, Carlson M, Grunstein M 2001. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol Cell 7: 117–126 [DOI] [PubMed] [Google Scholar]
- Zhang H, Emmons SW 2002. Caenorhabditis elegans unc-37/groucho interacts genetically with components of the transcriptional mediator complex. Genetics 160: 799–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Reese JC 2004. Ssn6-Tup1 requires the ISW2 complex to position nucleosomes in Saccharomyces cerevisiae. EMBO J 23: 2246–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Reese JC 2005. Molecular genetic analysis of the yeast repressor Rfx1/Crt1 reveals a novel two-step regulatory mechanism. Mol Cell Biol 25: 7399–7411 [DOI] [PMC free article] [PubMed] [Google Scholar]