Cotranscriptional splicing, in which mRNA is spliced as it is being transcribed, is thought to be necessary for proper gene regulation of many genes in eukaryotic cells. While studies have shown that splicing takes place cotranscriptionally in yeast, in higher eukaryotes, where genes contain multiple introns with widespread alternative splicing, the question of whether cotranscriptional splicing is a general phenomenon remains. Khodor et al. investigated what fractions of genes are cotranscriptionally spliced and what fraction of splicing occurs cotranscriptionally in Drosophila. The results demonstrate that the majority of introns show cotranscriptional splicing, although this process is intron-specific.
Keywords: nascent sequencing, cotranscriptional, pre-mRNA splicing, transcriptional coupling
Abstract
To determine the prevalence of cotranscriptional splicing in Drosophila, we sequenced nascent RNA transcripts from Drosophila S2 cells as well as from Drosophila heads. Eighty-seven percent of the introns assayed manifest >50% cotranscriptional splicing. The remaining 13% are cotranscriptionally spliced poorly or slowly, with ∼3% being almost completely retained in nascent pre-mRNA. Although individual introns showed slight but statistically significant differences in splicing efficiency, similar global levels of splicing were seen from both sources. Importantly, introns with low cotranscriptional splicing efficiencies are present in the same primary transcript with efficiently spliced introns, indicating that splicing is intron-specific. The analysis also indicates that cotranscriptional splicing is less efficient for first introns, longer introns, and introns annotated as alternative. Finally, S2 cells expressing the slow RpII215C4 mutant show substantially less intron retention than wild-type S2 cells.
In eukaryotes, nuclear mRNA processing includes at least three covalent events: 5′ capping, intron removal via splicing, and 3′ cleavage of the RNA and the addition of a poly(A) (pA) tail. Although initially studied as independent processes, these reactions have been shown to be coupled with each other as well as with transcription. For example, there is significant evidence that the CTD, the large C-terminal domain of RNA polymerase II (Pol II), provides a landing platform for capping as well as 3′ end formation factors and greatly enhances the rate of both of these reactions (Perales and Bentley 2009; Oesterreich et al. 2011).
The evidence for coupling between splicing and transcription is also strong. Electron microscopy spreads of Drosophila embryos have demonstrated cotranscriptional splicing (Beyer and Osheim 1988), and there are equally compelling visualization experiments from Daneholt and colleagues (Bauren and Wieslander 1994; Kiseleva et al. 1994) in Chironomous. Classical biochemical experiments indicate cotranscriptional splicing in Drosophila salivary glands (LeMaire and Thummel 1990), and multiple experiments have shown that the CTD enhances splicing (McCracken et al. 1997; Morris and Greenleaf 2000; Fong and Bentley 2001; Proudfoot et al. 2002; Bird et al. 2004). Moreover, there is good evidence for kinetic coupling between transcription and splicing (Das et al. 2006; de la Mata et al. 2010; Ip et al. 2011).
Cotranscriptional spliceosome assembly and/or splicing appear crucial for proper gene regulation in eukaryotes. In vitro studies showed that the spliceosome is rapidly associated with nascent transcripts, that its recruitment levels increase proportionally to transcript levels, and that recruitment is dependent on transcription by Pol II (Das et al. 2006). Further work showed that U1 snRNP proteins as well as proteins from the SR family of splicing regulators specifically associate with Pol II and that the presence of SR proteins during ongoing Pol II transcription is necessary for efficient cotranscriptional splicing (Das et al. 2007). In vivo experiments on select genes indicate that slowing the rate of transcription by Pol II enhances retention of alternative exons (de la Mata et al. 2010). Coupling can also be observed in competition between alternative 3′ splice sites (SSs), which may depend on the time interval between their synthesis, an idea that has been tested in both yeast and mammals by using different promoters, Pol II mutants, and inhibitors that slow elongation rates (Kadener et al. 2001; de la Mata et al. 2003). It is notable that a reverse relationship has been demonstrated in human cell lines; namely, a 5′SS can stimulate transcription even without active splicing (Damgaard et al. 2008).
Recent chromatin immunoprecipitation (ChIP) experiments demonstrate that spliceosome assembly as well as splicing also occur cotranscriptionally in yeast (Abruzzi et al. 2004; Gornemann et al. 2005; Lacadie and Rosbash 2005; Lacadie et al. 2006; Tardiff et al. 2006). However, ChIP approaches are indirect and only semiquantitative at best, precluding an accurate measurement of the fraction of global yeast splicing that takes place cotranscriptionally. Nevertheless, recent work using high-density tiling arrays for nascent RNA analysis indicates that most yeast splicing occurs cotranscriptionally (Carrillo Oesterreich et al. 2010). Furthermore, efficient cotranscriptional splicing may require a Pol II elongation pause around the 3′SS (Alexander et al. 2010; Carrillo Oesterreich et al. 2010).
Nonetheless, yeast gene architecture is dramatically simpler than that of higher eukaryotes: Only 4%–5% of yeast genes have introns, the vast majority of these intron-containing genes have only a single intron, and there is almost no alternative splicing. In higher eukaryotes, the majority of genes contain multiple introns with widespread alternative splicing. Furthermore, many metazoan splicing regulators have no yeast orthologs, and differences also exist between the transcriptional machinery of yeast and higher eukaryotes. For example, the CTD of Pol II is required for splicing in human cells but not in yeast (McCracken et al. 1997; Licatalosi et al. 2002).
Investigation of metazoan cotranscriptional splicing has been restricted to a few specific genes (Beyer and Osheim 1988; LeMaire and Thummel 1990; Listerman et al. 2006; Pandya-Jones and Black 2009), making general cotranscriptional splicing uncertain. This issue has two quantitative aspects: (1) What fraction of genes are cotranscriptionally spliced? (2) For cotranscriptionally spliced genes, what fraction of splicing occurs cotranscriptionally?
To determine the prevalence of cotranscriptional splicing in Drosophila, we used a traditional fractionation protocol (Wuarin and Schibler 1994) to isolate nascent transcripts from Drosophila S2 cells as well as from Drosophila heads and sequenced the RNA on an Illumina Genome Analyzer. The vast majority, ∼87% of the introns assayed, are spliced cotranscriptionally >50% of the time. The remaining 13% of introns are spliced poorly or slowly, with ∼3% being almost completely retained cotranscriptionally. Because introns with low cotranscriptional splicing efficiencies are present in the same primary transcript with efficiently spliced introns, the distinction is controlled at the level of the individual introns. The genome-wide analysis indicates that splicing is less efficient for first introns, longer introns, and alternatively annotated introns. The Pol II elongation rate is also an important factor, as a Pol II mutant with a lower elongation rate caused a genome-wide increase in cotranscriptional splicing efficiency.
Results
Generation of Drosophila S2 cell nascent RNA for high-throughput sequencing
To obtain nascent RNA from Drosophila S2 cells, nuclei were isolated and lysed with NUN buffer, which contains a high concentration of NaCl, urea, and NP-40 detergent (Wuarin and Schibler 1994; Nechaev et al. 2010). This wash was previously shown to result in a tight pellet consisting of DNA, core histones, and elongating RNA polymerases containing nascent RNA (Supplemental Fig. S1A,B; see also Wuarin and Schibler 1994). Indeed, RNA isolated from this pellet (hereafter called NUN RNA) contained very little pA RNA (Supplemental Fig. S1C), and characterization on an Agilent Bioanalyzer indicated that it contained much more heterogeneous RNA than mature rRNA compared with S2 cell total RNA (Supplemental Fig. S1D).
However, real-time PCR indicated that a significant fraction of NUN RNA was still rRNA, presumably nascent rRNA. We therefore developed an rRNA subtraction scheme to reduce this fraction and thereby increase the fraction of sequencing reads containing nascent pre-mRNA (Supplemental Fig. S2A). We hybridized 32 biotinylated, antisense oligos to the primary transcript of 18S, 28S, 2S, and 5.3S rRNA as well as to 5S rRNA. The oligos were elongated via reverse transcription, and the resulting DNA:RNA hybrids were collected with streptavidin magnetic beads. The RNA remaining in solution showed an 85% decrease in rRNA signal (Supplemental Fig. S2B) and a substantial shift in the Bioanalyzer profile (Supplemental Fig. S2C). The residual rRNA peaks disappeared completely, and the remaining RNA was slightly smaller than untreated NUN RNA.
We also removed any contaminating pA RNA with oligo(dT) magnetic beads (Supplemental Fig. S2D) and used the supernatant in our high-throughput sequencing library preparation. Two biological replicate libraries were sequenced and analyzed separately, using TopHat (Trapnell et al. 2009) with Bowtie (Langmead et al. 2009) to map unique reads. As a control, two biological replicates of S2 cell pA RNA were purified from total RNA and also sequenced. The entire procedure is summarized in Supplemental Figure S2E.
Most S2 cell introns are efficiently spliced cotranscriptionally
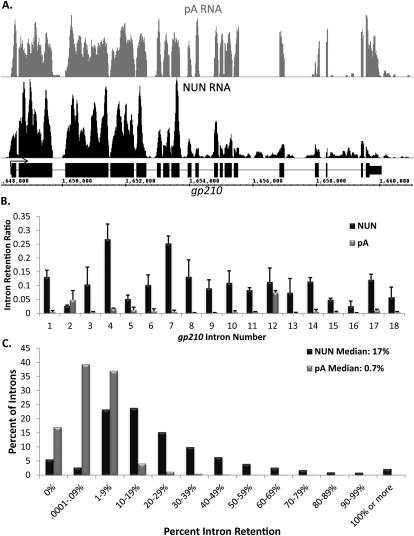
Nascent RNA shows two expected characteristics compared with pA RNA (Fig. 1A,C). First, more nascent RNA reads align to 5′ exons than to 3′ exons, as most elongating RNA polymerases should contain 5′ exon RNA and less 3′ exon RNA. This gradient is always absent or much less apparent in pA RNA. Second, NUN RNA should have more intron reads compared with pA RNA. The reverse was never observed. For example, the gp210 gene shows this 5′-to-3′ gradient in the NUN sample (Fig. 1A, black) but a constant signal across all exons in the pA RNA sample (Fig. 1A, gray; for more examples, see Supplemental Fig. S3). Furthermore, intron signal is apparent in the NUN sample but absent from the pA sample.
Figure 1.
Most introns in Drosophila S2 cells are efficiently spliced cotranscriptionally. (A) An image of the sequencing reads for a typical gene, gp210, in the Affymetrix Integrated Genome Browser. pA RNA is in gray, NUN RNA is in black, and gene structure is in black. Note breaks in sequencing reads coinciding with intron position. (B) Quantitation of intron retention for the introns of gp210. Intron retention = reads per base pair in introns/reads per base pair in all exons. (C) A histogram of the percent of all introns of abundantly transcribed genes, grouped by intron retention. pA RNA is in gray, and NUN RNA is in black.
To quantify the extent of cotranscriptional splicing, we developed an intron retention ratio metric: the ratio of an individual intron signal to the signal of all exons in that gene, normalized for length. This metric was used to filter out any variation due to a possible imbalance among different exons and also mitigates against data difficulties due to small exons. The bar graph of the gene illustrates the greater intron retention as well as the big difference in the retention of different introns in the NUN sample compared with the pA sample (Fig. 1B).
To examine global splicing patterns, we restricted our analyses to genes where adequate sequencing coverage provides unambiguous data. To this end, we only examined introns from genes containing an average of at least three reads per base pair in all exons in both replicates of the NUN samples, which is ∼43% of all Drosophila introns (∼35% of all genes; ∼65% of genes that meet the same criteria in the pA sequencing [pA-seq] data). With this requirement, intron retention is well-correlated (Spearman's ρ = 0.715, P < 0.01) between replicates (Supplemental Fig. S4A). Increasing the stringency to 10 reads per base pair improved the correlation (Supplemental Fig. S4B) but made little difference to the other analyses. We therefore chose the more permissive threshold because it includes a larger number of introns.
There are roughly 20-fold more unspliced introns in the NUN fraction than in the control pA sample; e.g., 17% median retention for NUN versus 0.7% for pA. The data also indicate that only ∼13% of introns analyzed from the S2 NUN samples show at least 50% cotranscriptional retention (Fig. 1C).
To verify that our metric was not biasing the sample toward greater cotranscriptional splicing by double counting reads in overlapping intronic regions, we performed this analysis in several different ways and obtained similar results (see the Materials and Methods; Supplemental Fig. S5A,B). Due to the possibility that some of the intron signal results from elongating Pol II molecules that have not yet reached a 3′SS, cotranscriptional splicing efficiency was also calculated in another way: the ratio of reads just prior to the 3′SS compared with just after the 3′SS. The conclusions are very similar to the intron retention metric, with a median 3′SS ratio of 0.24 (Supplemental Fig. S5C). However, the correlation between the biological replicates is lower (Supplemental Fig. S6), most likely due to greater variability in reads over such short lengths surrounding the 3′SS. When the 3′SS ratio analysis is limited to only those ∼10,000 introns demonstrating good reproducibility between sequencing runs (standard deviation of <0.1), the median 3′SS ratio decreases to 0.14. This shift suggests that the difference between the intron retention and 3′SS ratio medians is due to the increased variability of the skewed distribution.
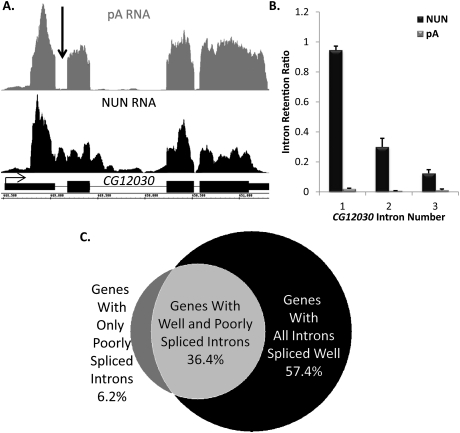
Despite the large fraction of introns with efficient cotranscriptional splicing, the minor fraction with inefficient cotranscriptional splicing was significant and reproducible: 1689 genes had 2793 introns with >50% cotranscriptional intron retention. Most genes with these inefficiently spliced introns also have efficient introns, with only a few genes containing one or two inefficient introns (Fig. 2A [visualization], B [quantitation]; for more examples, see Supplemental Fig. S7). This is based on an examination of individual genes as well as a genome-wide analysis (Fig. 2C).
Figure 2.
Select introns are dramatically retained in the nascent transcript, while others in the same gene are efficiently spliced. (A) A gene, CG12030, that has an intron with high retention (arrow) in the NUN RNA sample (black) and no matching intron signal in the pA (gray) control sample. Gene structure is in black. (B) Quantification of intron retention for CG12030. (C) A Venn diagram demonstrating that poorly spliced introns are found in ∼43% of analyzed genes and coexist with introns undergoing efficient cotranscriptional splicing in 36% of cases.
Most fly head introns are also efficiently spliced cotranscriptionally
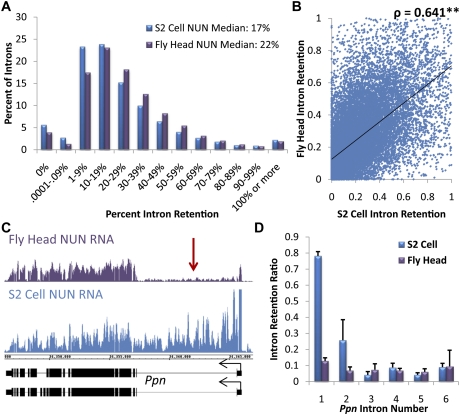
The terminally differentiated tissues of the adult fly are significantly different from rapidly dividing S2 cells, which are derived from embryonic tissue. To compare cotranscriptional splicing between these two sources, we sequenced NUN RNA from the fly head. There was a similar global level of cotranscriptional intron retention, as median intron retention scores differed by only 5% from S2 cells (Fig. 3A). However, a comparison of individual introns from genes sufficiently expressed in both tissues (as above; more than three reads per base pair in all exons) (Supplemental Fig. S8A) showed significantly different intron retention scores (P < 0.001, Wilcoxon signed-rank test) (Fig. 3B) relative to the comparisons between the individual replicates from a single source (Spearman's ρ = 0.641 vs. ρ = 0.715 for S2 cell replicates and ρ = 0.711 for fly head) (Supplemental Figs. S4A, S8B). A particularly striking example of a gene with an intron retention difference between S2 cells and the fly head is shown and quantified (Fig. 3C,D). More representative comparisons with pA tracts are also shown (Supplemental Fig. S9).
Figure 3.
A similar degree of efficient cotranscriptional splicing is present in heterogeneous fly head tissue as well as S2 cells. (A) A histogram of the percent of all introns of abundantly transcribed genes, grouped by intron retention. S2 cell NUN RNA is in blue, and fly head NUN RNA is in purple. Retention = reads per base pair in introns/reads per base pair in all exons. (B) A scatter plot of intron retention values for individual introns in both S2 cells and fly heads. Although there are small variances in splicing of individual introns between tissues, there is a high correlation (Spearman's ρ = 0.641, P < 0.01). (C) An example gene, Ppn, whose first intron (red arrow) has dramatic differences in cotranscriptional splicing between the fly head (purple) and S2 cell (blue) NUN RNA fractions. (D) Quantification of intron retention for the introns of Ppn.
Retention is dependent on intron characteristics
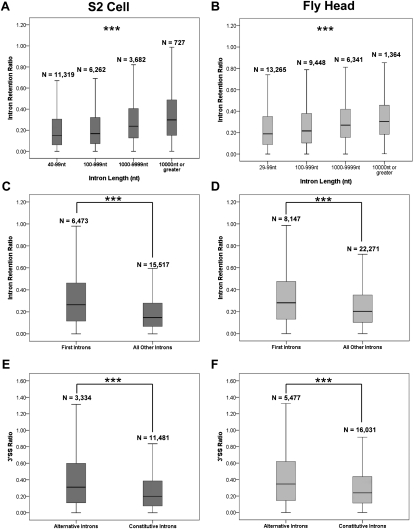
We then asked whether any “rules” govern global splicing in S2 cells or fly heads. For example, intron length can affect splicing efficiency as well as the alternative splicing of flanking exons, in part through coupling of elongation rate and splicing rate (for reviews, see Neugebauer 2002; Proudfoot 2003). Moreover, other work has shown that long Drosophila introns are recursively spliced (Burnette et al. 2005), which may affect their cotranscriptional retention. Our analysis indeed indicates that intron length significantly correlates with intron retention (P < 0.001, Kruskal-Wallis test) (Fig. 4A,B). Furthermore, the increased retention of longer introns persists regardless of position within a gene or its alternative or constitutive splicing status (P < 0.001, Kruskal-Wallis test) (Supplemental Fig. S10). Because longer introns may contain more Pol II that has not reached the 3′SS, the intron retention metric may be inherently biased toward greater retention of longer introns. The 3′SS ratio was therefore used to examine intron length, which is still a predictor of greater intron retention (Supplemental Fig. S11); only the few introns >10 kb are exceptions.
Figure 4.
Intron retention is dependent on intron length and position and whether the intron is annotated as alternative or constitutive. (A,B) Longer introns show greater retention in both S2 cells (A) and fly head (B) populations (P < 0.001, Kruskal-Wallis test, all pairwise comparisons). (C,D) First introns show greater retention than all other introns in both S2 cells (C) and fly heads (D) (P < 0.001, Mann-Whitney U-test). (E,F) Alternatively annotated introns show a higher 3′ SS ratio than constitutive introns in both S2 cell (E) and fly head (F) populations (P < 0.001, Mann-Whitney U-test).
Previous work in Chironomus suggested an overall 5′-to-3′ order of cotranscriptional intron excision—e.g., first introns are excised before second introns (Wetterberg et al. 1996)—and that only one complete spliceosome can assemble on a transcript at a time (Wetterberg et al. 2001). Recent work on select human genes also supports ordered and processive intron removal (Pandya-Jones and Black 2009). Other recent work shows a positive correlation of splicing machinery accumulation on nascent transcripts in vivo with an increase in intron number (Brody et al. 2011). These findings are in contrast to other data that support an ordered but not processive model of intron excision (Kessler et al. 1993; Schwarze et al. 1999). We therefore determined whether intron position has any effect on cotranscriptional splicing efficiency.
Our analysis indicates that first introns are more retained—i.e., less efficiently cotranscriptionally spliced—than other introns (P < 0.001, Mann-Whitney U-test) (Fig. 4C,D). Although first introns are, on average, longer than others in Drosophila, this is insufficient to explain the increased retention of first introns (P < 0.001, Mann-Whitney U-test across all categories) (Supplemental Fig. S12A,B). Alternative as well as constitutive introns manifest a greater retention of first introns (P < 0.001, Mann-Whitney U-test for all populations) (Supplemental Fig. S12C,D), which is also the case with the 3′SS ratio analysis (data not shown).
We next addressed whether alternative or constitutive splicing correlated with cotranscriptional splicing efficiency. Less efficient splicing of alternative introns might reflect the time it takes the splicing machinery to distinguish between competing splice sites, and recent work on nascent RNA from human cell lines showed that specific alternative introns are more retained than their constitutive neighbors (Pandya-Jones and Black 2009). Indeed, annotated alternative introns are more retained than constitutive introns in both S2 cells and fly heads (Fig. 4E,F). Annotated first introns are probably different because most of them arise from alternative initiation sites, rather than alternative splice sites (data not shown). Nonetheless, fly head alternative first introns also showed an increased 3′SS ratio relative to constitutive first introns (Supplemental Fig. S13B), but there were no significant differences between alternative and constitutive first introns in the S2 cell data (P = 0.188, Mann-Whitney U-test) (Supplemental Fig. S13A).
Slower Pol II elongation rate results in greater cotranscriptional splicing
Previous work has linked Pol II elongation to the magnitude of cotranscriptional splicing (Kadener et al. 2001; de la Mata et al. 2003, 2010), but it has not been determined whether global splicing efficiency is sensitive to elongation rate. To address this issue, we stably integrated a slow Pol II elongation mutant (RpII215C4) into wild-type S2 cells (Supplemental Fig. S14A). In vitro and in vivo work demonstrated that the RpII215C4 mutant is resistant to α-amanitin and has an elongation rate less than half of wild-type Pol II (Coulter and Greenleaf 1985; Boireau et al. 2007). After treating the stably integrated cells with α-amanitin for 24 h, we isolated and sequenced nascent RNA as described above (Supplemental Fig. S14B,C).
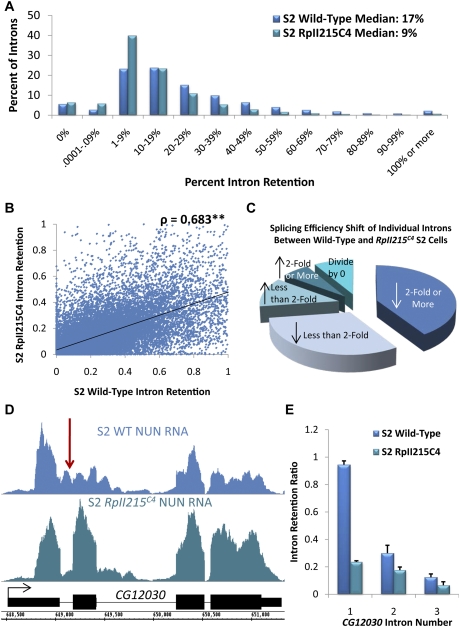
The data indicate a major effect of the RpII215C4 mutant on cotranscriptional splicing efficiency: Median intron retention is decreased by a factor of two compared with wild-type Pol II (Fig. 5A). In other words, introns are more efficiently spliced cotranscriptionally in the RpII215C4 mutant. The conclusion is based on the large number (25,892) of individual introns with sufficient reads in both wild-type and mutant nascent RNA for a comparison (Supplemental Fig. S14D). Although the data also indicate a statistically significant (ρ = 0.683, P < 0.01) correlation between the intron retention scores of the two samples (Fig. 5B), the correlation is inferior to those for the biological replicates (Supplemental Figs. S4A, S14B). Moreover, the individual intron retention scores of the RpII215C4 preparations are significantly different from those of the wild-type preparations (P < 0.001, Wilcoxon signed-rank test). Further analysis indicates that >72% of introns in the mutant strain have retention scores significantly lower than the wild-type strain (Fig. 5C).
Figure 5.
The slow-elongating RpII215C4 mutant increases cotranscriptional splicing efficiency in S2 cells. (A) A histogram of the percent of all introns of abundantly transcribed genes, grouped by intron retention. S2 cell wild-type NUN RNA is in blue, and RpII215C4 NUN RNA is in green. Retention = reads per base pair in introns/reads per base pair in all exons. (B) A scatter plot of intron retention values for individual introns in both wild-type and RpII215C4 S2 cells. There is a high correlation (Spearman's ρ = 0.683, P < 0.01) between the splicing of individual introns, and the slope of the regression line is flatter toward the wild type, indicating greater retention in that population. (C) A pie chart illustrating the percent of introns analyzed from both data sets that showed an increase or decrease in intron retention from wild-type to RpII215C4 S2 cells. (D) A visualization of CG12030 that has a high degree of retention in its first intron (red arrow) in the wild-type S2 cell NUN RNA sample (blue) and a dramatic decrease in the RpII215C4 (green) S2 cell sample. Gene structure is in black. (E) Quantification of intron retention for the introns of CG12030.
Preferentially retained introns represent a particularly important difference between wild-type and RpII215C4 data sets. The majority of the 509 introns retained at 50% or greater in the original wild-type data show a dramatic decrease in intron retention. For example, the first intron of CG12030 is inefficiently cotranscriptionally spliced in wild-type S2 cells, but is efficiently cotranscriptionally spliced in the RpII215C4 mutant S2 cells (Fig. 5D,E). In contrast, only 21 introns in this class show a twofold or greater increase in the RpII215C4 mutant samples. We conclude that elongating polymerase speed or some feature associated with RpII215C4 has a major impact on cotranscriptional splicing efficiency. Moreover, intron length, position, and annotation (alternative vs. constitutive) remain as predictors of cotranscriptional intron retention for the RpII215C4 data (Supplemental Fig. S15A–C), with no significant differences between alternative and constitutive first introns (Supplemental Fig. S7D).
Discussion
To address the extent of cotranscriptional splicing in Drosophila, we isolated and sequenced nascent RNA from S2 tissue culture cells and adult heads. Approximately 87% of analyzed introns are cotranscriptionally spliced 50% of the time or more. Cotranscriptional splicing is very similar between the two sources, with only a minor fraction of introns showing differences. Features such as intron length, intron location, and whether the intron is alternatively or constitutively spliced significantly impact cotranscriptional splicing efficiency. We also identified a select group of highly retained introns; they occur in ∼43% of analyzed genes, and other introns within the same transcripts are efficiently spliced. Use of the RpII215C4 mutant indicates that slowing the elongation rate of Pol II results in a substantial increase in global cotranscriptional splicing efficiency.
There are several indications that the NUN RNA fractionation reliably purifies nascent RNA with no more than a minor contamination by pA RNA. First, intron retention is 20-fold greater than in pA RNA. Second, RT–PCR assays of specific mRNAs after removal of pA RNA as well as comparisons between random priming and dT priming indicate that no more than a very small fraction of the NUN RNA is polyadenylated (Supplemental Figs. S1C, S2D). Third, the visible 5′-to-3′ gradient in the exon signal on many genes is consistent with a majority of nascent RNA. Fourth, the high retention of specific introns with a low retention of neighboring introns is also difficult to explain other than via nascent RNA.
Previous work looking at nascent splicing focused on the simpler yeast genome (Carrillo Oesterreich et al. 2010) or on a few select genes from metazoans (Kessler et al. 1993; Pandya-Jones and Black 2009; de la Mata et al. 2010). The PCR approach, detecting a pre-mRNA transcript in a population of total RNA by using PCR primers that span an intron–exon boundary (Kessler et al. 1993; Pandya-Jones and Black 2009; de la Mata et al. 2010), cannot address nascent splicing on a global scale but does avoid issues like rRNA contamination. Although the S2 cell NUN RNA also contained considerable rRNA, most likely nascent rRNA, the subtraction protocol reduced its representation to acceptable levels. There is considerably less rRNA in fly head nascent RNA, probably reflecting the much lower cell division rates than in S2 cells.
Although our analysis was restricted to the 35% of the genome with sufficiently abundant nascent RNA signal, efficient cotranscriptional splicing may reflect the general situation, with only exceptional introns showing high levels of retention. Because inefficiently spliced introns most often occur in transcripts with efficient introns, they may have features that mitigate against retention, although this possibility does not exclude an additional role for gene promoters or other features that specify cotranscriptional splicing efficiency. The vast majority of retained introns are not efficiently detected in pA RNA, which is contrary to previous work in the hamster (Kessler et al. 1993), and so are likely to be well spliced post-transcriptionally (although see nonsense-mediated decay [NMD] notion below). Indeed, the substantial reduction in retention in the slow elongation RpII215C4 mutant (Fig. 5D,E) suggests that these introns may just need more time to splice cotranscriptionally and therefore can be spliced either post-transcriptionally or cotranscriptionally. However, we cannot exclude the possibility that splicing of these introns is really poor and that these pre-mRNA transcripts are degraded within the nucleus or that they are transported to the cytoplasm and degraded there via NMD.
Many factors significantly correlate with these inefficiently spliced introns. For example, intron length is a robust predictor of cotranscriptional splicing efficiency. This is unlikely to be an artifact of more Pol II molecules collecting within longer introns. The metric that compares reads just before and just after a 3′SS also shows a robust increase in 3′SS ratio as a function of intron length, except for the infrequent case of very long introns of 10 kb or more. More efficient cotranscriptional splicing of these extra long introns may be due to recursive splicing (Burnette et al. 2005; data not shown).
Although there is some evidence for a processive model of intron excision in nascent transcripts (Wetterberg et al. 1996; Pandya-Jones and Black 2009), we found the opposite: First introns have less efficient cotranscriptional splicing than subsequent introns. The data are not simply because Drosophila first introns are longer than other introns and are not accounted for by alternative starts: Introns reflecting constitutive and alternative starts are about equally retained in S2 cells. Moreover, evidence for retention of first introns is not unique to this study (Kessler et al. 1993).
Previous work showed that the transcription initiation factor TFIIH associates with the U1 snRNA (Kwek et al. 2002). Furthermore, U1 snRNP has been implicated in the enhanced recruitment of the initiation factors TFIID, TFIIB, and TFIIH in mammalian cell lines (Damgaard et al. 2008). More recent work has identified a role for U1 snRNP in preventing premature cleavage and polyadenylation in HeLa cells (Kaida et al. 2010), suggesting another role for U1 snRNP that might interfere with the efficient cotranscriptional splicing of first introns. These considerations suggest a possible model for the delayed processing of 5′ introns: The U1 snRNP recruited to the 5′-most intron interacts with the transcription and polyadenylation machinery, which causes delays in subsequent steps in splicesome assembly; e.g., an interaction with the U2 snRNP.
The issue of alternative splicing is more complex. A recent study in human cell lines found alternatively spliced introns of two genes to be more retained than constitutive ones (Pandya-Jones and Black 2009). This work comes to a similar genome-wide conclusion; the only exceptions are first introns of S2 cells. Perhaps they do not use many of their annotated alternative start sites, giving rise to a more constitutive situation than the more heterogeneous fly head RNA. This interpretation remains speculative, as different isoform contributions are not apparent in the nascent sequencing (nascent-seq) data.
There are also some differences in the splicing efficiency of individual introns between S2 cells and fly heads. Despite the similar picture of global cotranscriptional splicing (Fig. 3A), most introns manifest slight but statistically significant differences in retention values between the two tissues. Moreover, a few introns are dramatically different (Fig. 3C,D). Some of these differences may be related to growth rate, as genes related to translation and the ribosome are enriched for intron retention in S2 cells relative to heads (P = 8.7 × 10−8, DAVID functional annotation). Because rapidly dividing S2 cells undoubtedly require greater amounts of translational machinery than heads, the faster transcription of these genes may allow insufficient time for efficient cotranscriptional splicing. Indeed, kinetic coupling has been proposed to link polymerase speed and the efficiency of cotranscriptional splicing (Oesterreich et al. 2011).
To test the kinetic coupling model in vivo and on a genome-wide scale, Pol II elongation rate was altered by exploiting the RpII215C4 mutant (de la Mata et al. 2003, 2010; Boireau et al. 2007; Ip et al. 2011). Based on the global decrease in intron retention, we infer that the slower Pol II elongation rate of the mutant (Coulter and Greenleaf 1985; Boireau et al. 2007) dramatically increases cotranscriptional splicing efficiency (Fig. 5A). The data therefore link global cotranscriptional splicing to Pol II elongation rate and provide substantial evidence for the kinetic coupling model (Oesterreich et al. 2011). They also make it important to determine the effects of a faster Pol II. Interestingly, elongation rate does not have a universal effect on cotranscriptional splicing efficiency: Some introns show no change in splicing efficiency, whereas others even show a small decrease. This variability may reflect a nonuniform effect of the mutant polymerase on the elongation rate of different genes.
How will these Drosophila data compare with the cotranscriptional splicing efficiency of other species? Although the literature is rather sparse at present, it appears that yeast pre-mRNA cotranscriptional splicing is also quite efficient, with a median cotranscriptional splicing efficiency of 0.74, similar to the ratio of 0.83 shown here (Carrillo Oesterreich et al. 2010). In contrast, preliminary data indicate significantly less efficient cotranscriptional splicing in the mouse liver (YL Khodor, JS Menet, and J Rodriguez, unpubl.). One possibility is that the longer average intron size in mammals relative to Drosophila is relevant to this difference; i.e., that the relationship between intron length and cotranscriptional splicing described here for flies will also be true for mammals and will even account for a substantial fraction of intraspecific differences in cotranscriptional splicing efficiency.
Materials and methods
Plasmid construct
The pGB128 vector was cloned to contain a cDNA copy of the RPII215 gene with an N-terminal myc tag and the C4 mutation (G3973A) under the control of an actin promoter and a blastocidin resistance gene.
Tissue culture cells and stable lines
S2 tissue culture cells were obtained from Invitrogen and grown in Scheneider medium supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen) and 1% penicillin/streptomycin (MP Biomedicals). The RpII215C4 stable line was generated by transfection of the pGB128 vector using Effectene reagent (Qiagen). After 3 d, RpII215C4-containing cells were selected (Nawathean et al. 2005).
Fly strain
The fly strain used was yw.
Nascent RNA isolation in S2 cells
The NUN RNA isolation protocol was adapted from Nechaev et al. (2010) and Wuarin and Schibler (1994): S2 cells grown in a monolayer in T-75 flasks 2 d after passage were harvested via centrifugation at 1000g and washed twice in ice-cold 1× PBS. Cells were resuspended in ice-cold buffer AT (15 mM HEPES-KOH at pH 7.6, 10 mM KCl, 5 mM MgOAc, 3 mM CaCl2, 300 mM sucrose, 0.1% Triton X-100, 1 mM DTT, 1× Complete protease inhibitors [Roche]) and dounced 30 times in a dounce homogenizer with tight pestle to lyse. The resulting lysate was divided into 0.5-mL aliquots and layered over 1 mL of buffer B (15 mM HEPES-KOH at pH 7.6, 10 mM KCl, 5 mM MgOAc, 3 mM CaCl2, 1 M sucrose, 1 mM DTT, 1× Complete protease inhibitors), then centrifuged at 8000 rpm for 15 min at 4°C. The supernatant was removed and the pellet was resuspended in 5 vol of nuclear lysis buffer (10 mM HEPES-KOH at pH 7.6, 100 mM KCl, 0.1 mM EDTA, 10% glycerol, 0.15 mM spermine, 0.5 mM spermidine, 0.1 M NaF, 0.1 M Na3VO4, 0.1 mM ZnCl2, 1 mM DTT, 1× Complete protease inhibitors, 1 U/μL RNasin Plus [Promega]) and dounced three times with loose pestle and twice with tight pestle to resuspend. We added 2× NUN buffer (25 mM HEPES-KOH at pH 7.6, 300 mM NaCl, 1 M Urea, 1% NP-40, 1 mM × Complete protease inhibitors) 1:1 to the nuclear suspension drop by drop while vortexing, and the suspension was placed on ice for 20 min prior to spinning at 14,000 rpm for 30 min at 4°C. The supernatant was removed and TRIzol reagent (Invitrogen) was added to DNA–Histone–Pol II-RNA pellets. The TRIzol–pellet tube was heated to 65°C to dissolve the pellet, and RNA was extracted following the manufacturer's protocol. The resulting RNA was subjected to rRNA removal (see below) and pA depletion with Oligo(dT) magnetic beads (Invitrogen). For the stable RpII215C4 cell line, this protocol was performed 24 h after blastocidin removal and the addition of 5 μg/mL α-amanitin.
Nascent RNA isolation in fly heads
Flies were harvested and frozen on dry ice. Fly heads were collected to 1 mL and ground on dry ice with a mortar and pestle for 1 min before being transferred into a dounce homogenizer. Five volumes of homogenization buffer (10 mM HEPES-KOH at pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 0.8 M sucrose, 0.5 mM EDTA, 1 mM DTT, 1× Complete protease inhibitor) was added to the ground fly heads, and the mixture was dounced 15 times for 1 min with the loose pestle. The resulting lysate was filtered through 100-μm mesh into a 50-mL Falcon tube and centrifuged for 2 min at 300 rpm. The flow-through was layered onto 5 mL of sucrose cushion buffer (10 mM HEPES-KOH at pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 M sucrose, 10% glycerol, 0.5 mM EDTA, 1 mM DTT, Complete protease inhibitor) in glass Kimble centrifuge tubes. The samples were spun at 11,000 rpm for 10 min at 4°C, and pellets were resuspended in 5 vol of nuclear lysis buffer as above and processed as above.
Total pA isolation in S2 cells
Medium was removed from, and 1 mL of TRIzol reagent (Invitrogen) was added to, S2 cells grown in a monolayer in one well of a six-well plate 2 d after passage. RNA was extracted following the manufacturer's protocol. Once resuspended, pA RNA was doubly selected by use of Oligo(dT) magnetic beads, following the manufacturer's protocol (Invitrogen).
RNA signal analysis by quantitative PCR (qPCR)
Nascent and total RNA were reverse-transcribed with SuperScript II reverse transcriptase (Invitrogen), and qPCR was performed on selected genes as described previously (Kadener et al. 2009). Primers used are listed in Supplemental Table S1.
rRNA removal
Six micrograms of NUN RNA was reverse-transcribed with a mix of biotinylated antisense oligos (Supplemental Table S1) spaced ∼500 base pairs (bp) apart on the rRNA primary transcript and the 5S rRNA primary transcript. The resulting RNA:DNA hybrid was subjected to pull-down using two aliquots of 400 μL of streptavidin magnetic beads (Dynabead M-270, Invitrogen) and precipitated with ethanol.
RNA sequencing and alignment
Sequencing library preparation for both nascent and pA RNA samples was performed according to the manufacturer's protocol (Illumina), and libraries were assayed on the Agilent Bioanalyzer prior to being loaded onto the Illumina Genome Analyzer. Seventy-six-base-pair reads were sequenced for one S2 cell replicate trimmed to 64 bp and mapped to the Drosophila dm3 genome alignment obtained from the University of California at Santa Cruz Genome Browser (http://www.genome.ucsc.edu/cgi-bin/hgTables?command=start) using TopHat (http://www.tophat.cbcb.umd.edu) with Bowtie. The parameters used were “−m 1 −F 0 −g 1–microexon-search–no-closure-search −I 50000.” The unique mappable reads for each lane are listed in Supplemental Table S2. The .WIG format files were used for visualization on the Affymetrix Integrated Genome Browser and further analysis.
Intron retention determination
Custom scripts were used to calculate intron retention, which was calculated by averaging mapped reads per base pair in a given intron over the average mapped reads per base pair in all exons of the isoform, or the average of the isoforms, of the gene in which the intron appears. Constitutive introns were defined as introns appearing in all isoforms of the gene, and their retention was calculated as an average retention for all isoforms of the gene.
Alternative intron retention determination
To verify the validity of the first metric, a second metric was used: All isoforms of a gene were condensed down into one isoform, with introns being defined as regions between the 5′ and 3′ ends that did not contain an exon in any isoform. Retention was then calculated for these nonexonic regions as above (Supplemental Fig. S5B).
3′SS ratio determination
To determine splicing efficiency by the ratio of reads about the 3′SS, we determined the number of reads at each base pair for the last 25 bp of a given intron and the first 25 bp of the 3′ exon. The numbers were then divided. Alternative introns with overlapping exons in this region were excluded from this analysis (Supplemental Fig. S5C).
Statistical analysis
Selected introns meeting our criteria (see the Results) were analyzed by length, position, and prior annotation as alternative or constitutive using the PASW Statistics 18 software (IBM). Nonparametric analysis was used, as the distributions were not normal.
Data availability
Raw and processed sequencing data used in this work are available for download from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo), accession number GSE32950.
Acknowledgments
We thank Michael Tolan for assistance with custom analytical code, Kristyna Palm Danish for administrative assistance, and our colleagues in the Rosbash laboratory for helpful suggestions. We also thank Douglas Black, Sebastian Kadener, Nelson Lau, and Amy Pandya-Jones for their feedback and advice. This research was supported by the Howard Hughes Medical Institute, NIH grants GM023549 and NS044232 (to M.R.), the NIH Genetics Training Grant (to Y.L.K.), and NSF IGERT fellowship DGE-0549390 (for J.R.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.178962.111.
Note added in proof
Subsequent to acceptance of this manuscript, we noted a small but significant increase in intron retention of final introns. This increase is less dramatic than the difference between the retention of the first and all other introns.
References
- Abruzzi KC, Lacadie S, Rosbash M 2004. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J 23: 2620–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RD, Innocente SA, Barrass JD, Beggs JD 2010. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell 40: 582–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauren G, Wieslander L 1994. Splicing of balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell 76: 183–192 [DOI] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev 2: 754–765 [DOI] [PubMed] [Google Scholar]
- Bird G, Zorio DA, Bentley DL 2004. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol Cell Biol 24: 8963–8969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boireau S, Maiuri P, Basyuk E, de la Mata M, Knezevich A, Pradet-Balade B, Backer V, Kornblihtt A, Marcello A, Bertrand E 2007. The transcriptional cycle of HIV-1 in real-time and live cells. J Cell Biol 179: 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody Y, Neufeld N, Bieberstein N, Causse SZ, Bohnlein EM, Neugebauer KM, Darzacq X, Shav-Tal Y 2011. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol 9: e1000573 doi: 10.1371/journal.pbio.1000573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette JM, Miyamoto-Sato E, Schaub MA, Conklin J, Lopez AJ 2005. Subdivision of large introns in Drosophila by recursive splicing at nonexonic elements. Genetics 170: 661–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo Oesterreich F, Preibisch S, Neugebauer KM 2010. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell 40: 571–581 [DOI] [PubMed] [Google Scholar]
- Coulter DE, Greenleaf AL 1985. A mutation in the largest subunit of RNA polymerase II alters RNA chain elongation in vitro. J Biol Chem 260: 13190–13198 [PubMed] [Google Scholar]
- Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J 2008. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol Cell 29: 271–278 [DOI] [PubMed] [Google Scholar]
- Das R, Dufu K, Romney B, Feldt M, Elenko M, Reed R 2006. Functional coupling of RNAP II transcription to spliceosome assembly. Genes Dev 20: 1100–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R 2007. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell 26: 867–881 [DOI] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 12: 525–532 [DOI] [PubMed] [Google Scholar]
- de la Mata M, Lafaille C, Kornblihtt AR 2010. First come, first served revisited: Factors affecting the same alternative splicing event have different effects on the relative rates of intron removal. RNA 16: 904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Bentley DL 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: Different functions for different segments of the CTD. Genes Dev 15: 1783–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornemann J, Kotovic KM, Hujer K, Neugebauer KM 2005. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell 19: 53–63 [DOI] [PubMed] [Google Scholar]
- Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, Blencowe BJ 2011. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res 21: 390–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Cramer P, Nogues G, Cazalla D, de la Mata M, Fededa JP, Werbajh SE, Srebrow A, Kornblihtt AR 2001. Antagonistic effects of T-Ag and VP16 reveal a role for RNA pol II elongation on alternative splicing. EMBO J 20: 5759–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Rodriguez J, Abruzzi KC, Khodor YL, Sugino K, Marr MT II, Nelson S, Rosbash M 2009. Genome-wide identification of targets of the drosha-pasha/DGCR8 complex. RNA 15: 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G 2010. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler O, Jiang Y, Chasin LA 1993. Order of intron removal during splicing of endogenous adenine phosphoribosyltransferase and dihydrofolate reductase pre-mRNA. Mol Cell Biol 13: 6211–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva E, Wurtz T, Visa N, Daneholt B 1994. Assembly and disassembly of spliceosomes along a specific pre-messenger RNP fiber. EMBO J 13: 6052–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwek KY, Murphy S, Furger A, Thomas B, O'Gorman W, Kimura H, Proudfoot NJ, Akoulitchev A 2002. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat Struct Biol 9: 800–805 [DOI] [PubMed] [Google Scholar]
- Lacadie SA, Rosbash M 2005. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol Cell 19: 65–75 [DOI] [PubMed] [Google Scholar]
- Lacadie SA, Tardiff DF, Kadener S, Rosbash M 2006. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes Dev 20: 2055–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMaire MF, Thummel CS 1990. Splicing precedes polyadenylation during Drosophila E74A transcription. Mol Cell Biol 10: 6059–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell 9: 1101–1111 [DOI] [PubMed] [Google Scholar]
- Listerman I, Sapra AK, Neugebauer KM 2006. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol 13: 815–822 [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361 [DOI] [PubMed] [Google Scholar]
- Morris DP, Greenleaf AL 2000. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 275: 39935–39943 [DOI] [PubMed] [Google Scholar]
- Nawathean P, Menet JS, Rosbash M 2005. Assaying the Drosophila negative feedback loop with RNA interference in S2 cells. Methods Enzymol 393: 610–622 [DOI] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K 2010. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327: 335–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer KM 2002. On the importance of being co-transcriptional. J Cell Sci 115: 3865–3871 [DOI] [PubMed] [Google Scholar]
- Oesterreich FC, Bieberstein N, Neugebauer KM 2011. Pause locally, splice globally. Trends Cell Biol 21: 328–335 [DOI] [PubMed] [Google Scholar]
- Pandya-Jones A, Black DL 2009. Co-transcriptional splicing of constitutive and alternative exons. RNA 15: 1896–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R, Bentley D 2009. ‘Cotranscriptionality’: The transcription elongation complex as a nexus for nuclear transactions. Mol Cell 36: 178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ 2003. Dawdling polymerases allow introns time to splice. Nat Struct Biol 10: 876–878 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ 2002. Integrating mRNA processing with transcription. Cell 108: 501–512 [DOI] [PubMed] [Google Scholar]
- Schwarze U, Starman BJ, Byers PH 1999. Redefinition of exon 7 in the COL1A1 gene of type I collagen by an intron 8 splice-donor-site mutation in a form of osteogenesis imperfecta: Influence of intron splice order on outcome of splice-site mutation. Am J Hum Genet 65: 336–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff DF, Lacadie SA, Rosbash M 2006. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol Cell 24: 917–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetterberg I, Bauren G, Wieslander L 1996. The intranuclear site of excision of each intron in Balbiani ring 3 pre-mRNA is influenced by the time remaining to transcription termination and different excision efficiencies for the various introns. RNA 2: 641–651 [PMC free article] [PubMed] [Google Scholar]
- Wetterberg I, Zhao J, Masich S, Wieslander L, Skoglund U 2001. In situ transcription and splicing in the Balbiani ring 3 gene. EMBO J 20: 2564–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J, Schibler U 1994. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: Evidence for cotranscriptional splicing. Mol Cell Biol 14: 7219–7225 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw and processed sequencing data used in this work are available for download from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo), accession number GSE32950.