Eukaryotic transcriptional control is mediated by regulatory DNA elements in enhancer regions and core promoter elements located within the promoter region. Martinez and colleagues identify and characterize factors that regulate the TFIID-dependent transcription stimulatory function of the Initiator (INR) core promoter element and the TATA box. The authors demonstrate that these factors are the protein HMGA1, which regulates transcription machinery, and the Mediator coregulator complex, which functions by stimulating the core promoter through the INR element in the presence of HMGA1. These studies significantly advance our understanding of eukaryotic gene regulation.
Keywords: gene regulation, transcription, RNA polymerase II, Initiator, Mediator, HMGA1
Abstract
The factors and mechanisms underlying the differential activity and regulation of eukaryotic RNA polymerase II on different types of core promoters have remained elusive. Here we show that the architectural factor HMGA1 and the Mediator coregulator complex cooperate to enhance basal transcription from core promoters containing both a TATA box and an Initiator (INR) element but not from “TATA-only” core promoters. INR-dependent activation by HMGA1 and Mediator requires the TATA-binding protein (TBP)-associated factors (TAFs) within the TFIID complex and counteracts negative regulators of TBP/TATA-dependent transcription such as NC2 and Topoisomerase I. HMGA1 interacts with TFIID and Mediator and is required for the synergy of TATA and INR elements in mammalian cells. Accordingly, natural HMGA1-activated genes in embryonic stem cells tend to have both TATA and INR elements in a synergistic configuration. Our results suggest a core promoter-specific regulation of Mediator and the basal transcription machinery by HMGA1.
Regulation of gene-specific transcription in eukaryotes is controlled by the combinatorial interplay of a variety of regulatory DNA elements located in promoter-proximal and -distal (e.g., enhancer) regions and core promoter elements located within the transcription initiation region (i.e., the core promoter). Regulatory elements are recognized by cognate sequence-specific DNA-binding regulators (activators or repressors), which in turn recruit a diversity of coregulators (i.e., coactivators or corepressors) (Roeder 2005). Activators often assemble cooperatively at enhancers to form stereo-specific activating complexes (e.g., enhanceosomes). Architectural DNA-binding proteins, such as HMGA1 (formerly HMGI/Y), have been shown to further assist in the formation of specific enhanceosomes (Thanos and Maniatis 1995; Reeves 2003). HMGA family proteins do not have an intrinsic transcription regulatory domain or a strict DNA sequence specificity but bind to the minor groove of AT-rich or structured DNA through “AT-hook” motifs and to numerous sequence-specific regulators. HMGA1 is thought to act as a chaperone to induce or stabilize DNA and/or protein conformations that facilitate cooperative binding of activators to specific enhancers (Reeves and Beckerbauer 2001; Reeves 2003; Panne 2008).
Once recruited by activators to regulatory DNA sequences, different classes of coactivators interplay to modify chromatin structure and/or directly interact with the general transcription machinery to enhance transcription by RNA polymerase II (Pol II) (Roeder 2005). The multiprotein Mediator complex belongs to the latter class of coactivators and has emerged as the prevalent “general coregulator” required for transcription of most, if not all, protein-coding genes in eukaryotes (Kornberg 2005; Malik and Roeder 2010). Mediator is recruited to regulatory DNA sequences by direct protein–protein interactions with a variety of activators, which further induce structural shifts in Mediator that may affect its functions (Malik and Roeder 2010; Meyer et al. 2010; Taatjes 2010). Mediator also interacts physically with Pol II and several general transcription factors (GTFs) and facilitates their assembly at the core promoter (Kornberg 2005; Malik and Roeder 2010). Accordingly, Mediator associates with both enhancers and core promoters in mammalian cells and has been shown to interact with cohesin in a complex that bridges enhancers to core promoters via DNA looping (Heintzman et al. 2009; Kagey et al. 2010). Besides facilitating activator-dependent recruitment of the general transcription machinery, Mediator also activates post-recruitment steps in transcription and stimulates phosphorylation of the C-terminal repeat domain (CTD) of Pol II (Kornberg 2005; Malik and Roeder 2010). These previous observations suggest that Mediator contributes to differential gene regulation by integrating signals emanating mostly from enhancers and gene-specific activators and may control the activity of the general transcription machinery at the core promoter of most genes. Consistent with this, Mediator is required for optimal activator-independent (i.e., basal) transcription from most target core promoters analyzed thus far in either yeast or metazoan cell-free transcription extracts in vitro (Kim et al. 1994; Mittler et al. 2001; Park et al. 2001; Baek et al. 2002; Reeves and Hahn 2003; Takagi and Kornberg 2006). Intriguingly, however, the stimulatory effect of Mediator on basal transcription is much less apparent in purified systems reconstituted with nonlimiting concentrations of the general Pol II transcription machinery (Mittler et al. 2001; Nair et al. 2005; Takagi and Kornberg 2006). This suggests that additional factors may be required for efficient Mediator-dependent stimulation of the general transcription machinery and/or that Mediator may antagonize inhibitory factors in cells and crude extracts (Malik and Roeder 2010).
The core promoter is the ultimate target of activators and Mediator and is defined as the DNA region where the GTFs (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) and Pol II assemble to form a functional preinitiation complex (PIC) (Juven-Gershon et al. 2008). Accordingly, this region is generally “nucleosome-free” or marked with unstable nucleosome variants at active (or poised) genes in vivo (Jin et al. 2009 and references therein). Core promoters have been most extensively studied in metazoans and are highly diverse in structure and intrinsic basal activity (Juven-Gershon et al. 2008). It has long been known that core promoter DNA sequences play an important regulatory role by influencing the transcriptional response of genes to distal activators and enhancers both in vivo and in vitro. However, how this is accomplished has remained obscure (Smale 2001; Juven-Gershon et al. 2008). Core promoters may contain different combinations of specific core promoter DNA elements that are recognized by distinct components of the PIC. The best-characterized core promoter elements include the TATA box, the TFIIB recognition elements (BREs) that flank the TATA box, the Initiator (INR) at the transcription start site, and the DPE, MTE, and DCE elements located downstream from the transcription start site (Juven-Gershon et al. 2008). Although the INR is the most prevalent element found in almost half of all core promoters from yeast to humans, none of these core promoter elements is universal, and some core promoters apparently lack all of the above elements (Ohler et al. 2002; Gershenzon and Ioshikhes 2005; Jin et al. 2006; Yang et al. 2007). Except for the BRE, core promoter elements are generally recognized by different subunits of the TFIID complex: The TATA box is bound by the TATA-binding protein (TBP), the INR is recognized by TBP-associated factor 1 (TAF1) in conjunction with TAF2, the DPE photo-cross-links to TAF6 and TAF9, and the DCE photo-cross-links to TAF1 (Juven-Gershon et al. 2008). Accordingly, specific combinations of core promoter elements synergize in basal and activated transcription in crude nuclear extracts and cultured cells and cooperatively recruit TFIID to the core promoter in vitro (O'Shea-Greenfield and Smale 1992; Colgan and Manley 1995; Burke and Kadonaga 1996; Emami et al. 1997; Juven-Gershon et al. 2008). Intriguingly, however, the intrinsic basal activities and synergistic functions of most core promoter elements cannot be recapitulated in systems reconstituted with purified GTFs and Pol II, suggesting that cooperative binding of TFIID to core elements can only partly explain their synergy and that additional factors may be required for optimal core promoter sequence-dependent Pol II activity. Indeed, INR function at mammalian TATA-less promoters and the strong synergy of TATA and INR elements were shown to require distinct TAF- and INR-dependent cofactors (TICs) whose identities have remained elusive (Martinez et al. 1998; Malecová et al. 2007). Similarly, TFIID-dependent DPE function and synergy with the INR require additional cofactors. These include the negative cofactor NC2, a negative regulator of TBP and TATA-dependent transcription, which stimulates DPE-dependent promoters in Drosophila cells and cell-free extracts (Willy et al. 2000; Hsu et al. 2008), and the protein kinase CK2, which enhances Sp1 activation of mammalian DPE-dependent promoters in a purified transcription system (Lewis et al. 2005). In yeast, the general transcription machinery may also require additional factors for efficient transcription from promoters with weak TATA boxes (Bjornsdottir and Myers 2008). Thus, the factors and mechanisms that regulate the general transcription machinery in a core promoter-specific manner may be diverse and remain poorly defined.
Here, we present the biochemical identification of HMGA1 and Mediator as core promoter-selective cofactors required for the TFIID/TAF-dependent transcription stimulatory function of the INR element and its synergy with the TATA box (previously described as the TIC1 activity). HMGA1 functionally cooperates with Mediator and TFIID and elicits an INR-specific basal transcription stimulatory activity of Mediator, which requires TAFs and counteracts the negative regulation of TATA-dependent transcription by NC2. Consistent with their interdependent functions in vitro, HMGA1 specifically interacts with both Mediator and TFIID and is required for the synergy of TATA and INR elements in mammalian cells. Our results suggest a possible core promoter-dependent architectural or allosteric regulation of the general Pol II transcription machinery by HMGA1 and the possibility that HMGA1 and Mediator could act cooperatively at the interface between enhancers and core promoters to elicit gene-specific responses to regulatory stimuli.
Results
Biochemical identification of HMGA1 and Mediator as components of the TIC1 activity required for the synergy of TATA and INR core promoter elements
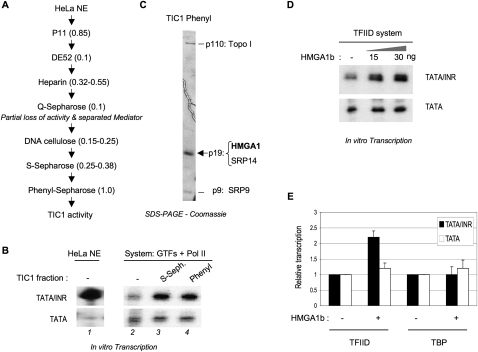
We previously partially purified a TFIID/TAF-dependent stimulatory activity (called TIC1) that restored INR function and the synergy of TATA and INR elements in a purified basal transcription system reconstituted with immunoaffinity-purified Flag-tagged TFIID; Ni2+ affinity-purified native TFIIA; recombinant 6His-tagged TFIIB, TFIIE, and TFIIF; and purified native TFIIH and Pol II (Martinez et al. 1998). To identify the active components of the crude TIC1 fractions, more extensive chromatographic fractionations were performed, and the TIC1 activity in chromatographic fractions was analyzed by complementation of the purified basal transcription system (see the Materials and Methods). We followed the ability of TIC1 to stimulate basal transcription selectively from a core promoter containing both TATA and INR consensus elements in a synergistic configuration (TATA/INR) but not from a derivative “TATA-only” core promoter (TATA) that differs only by point mutations that inactivate the INR (Supplemental Fig. S1A). The TIC1 activity was purified through seven chromatographic steps (Fig. 1A,B; Supplemental Fig. S1B–E), although fractionation on Q-Sepharose resulted in a significant loss of activity (see below; Supplemental Fig. S1B,C). A protein of ∼19 kDa (p19) consistently cofractionated with the TIC1 activity (Supplemental Fig. S1D,E; data not shown) and was enriched in the final TIC1 “Phenyl” fraction, which also contained two other protein bands: p110 and p9 (Fig. 1C). Tandem mass spectrometry analyses (LC-MS/MS) identified these proteins as DNA Topoisomerase I (Topo I) (p110), HMGA1 (p19), SRP14 (also in p19), and SRP9 (p9) (Fig. 1C; Supplemental Fig. S1F). SRP14/9 are abundant cytosolic (and nucleolar) proteins that heterodimerize and function within the signal recognition particle (SRP) in cotranslational targeting of proteins to the endoplasmic reticulum (Koch et al. 2003); hence, they were considered contaminants and were not investigated further.
Figure 1.
Purification and identification of HMGA1 as a component of the TIC1 activity. (A) Purification scheme used. Numbers in parentheses indicate the molar KCl concentration used to elute the TIC1 activity from each resin (see also Supplemental Fig. S1). (B) Representative in vitro transcription–primer extension analysis of transcripts from TATA and TATA/INR templates in HeLa nuclear extracts (NE, lane 1) and in the system reconstituted with purified GTFs and Pol II and complemented with TIC1 fractions from the last two purification steps. (C) Analysis of the purified TIC1 Phenyl fraction by SDS-PAGE and Coomassie staining. The proteins were excised from the gel and identified by LC/ESI/MS/MS. (D,E) In vitro transcription/primer extension assays with recombinant HMGA1b were performed with supercoiled templates in the purified system containing either TFIID or TBP. Autoradiograms shown for TATA and TATA/INR are from the same gel and exposure time. E shows a quantitation (mean ± SD) of more than three independent transcription experiments normalized to the promoter activities in the absence of HMGA1b.
Given their roles as architectural factors and transcription coregulators, Topo I and HMGA1 were further tested for TIC1 activity as purified recombinant proteins (Supplemental Fig. S2A,B). Purified recombinant Topo I did not have INR-dependent activity by itself (Supplemental Fig. S2D) and at higher concentrations repressed both TATA/INR and TATA promoters to a similar extent (data not shown). In contrast, recombinant HMGA1b selectively stimulated the TATA/INR core promoter without affecting the TATA template (Fig. 1D). Although modest, this INR-dependent stimulatory activity of recombinant HMGA1b was absolutely dependent on TAFs within TFIID (Fig. 1E). Notably, at higher concentrations, HMGA1b repressed transcription selectively from the TATA core promoter (Supplemental Fig. S2E), and TAFs were required to antagonize this repressive effect on the TATA/INR promoter (Supplemental Fig. S2F). The other major HMGA1 splicing isoform, HMGA1a, which only differs from HMGA1b by an extra 11 amino acids (Supplemental Fig. S1F), functioned similarly (see also below; Supplemental Fig. S2G; data not shown). For all subsequent experiments, we used recombinant HMGA1b at concentrations that activate TATA/INR but do not inhibit TATA-only transcription.
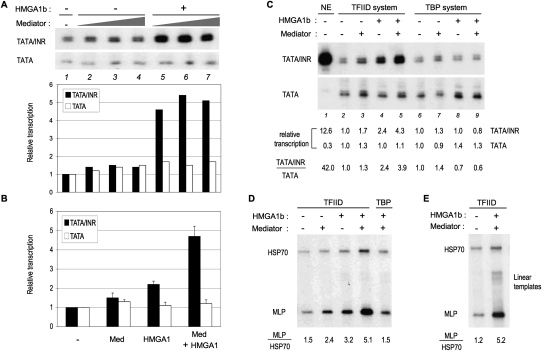
Since significant TIC1 activity was lost during the Q-Sepharose fractionation step, which also separated Mediator complexes from HMGA1 (Supplemental Fig. S1C), we considered the possibility that Mediator could be required for efficient HMGA1-mediated stimulation of INR-dependent transcription. As expected from numerous previous reports, a highly purified Mediator preparation that contains the various forms of Mediator, including CDK8-containing and CDK8-lacking complexes (Supplemental Fig. S2C), did not have core promoter selectivity in the reconstituted system in the absence of HMGA1 and only weakly stimulated basal transcription from both TATA/INR and TATA promoters (Fig. 2A, lanes 2–4). In contrast, a significant (about fivefold) preferential stimulation of TATA/INR was observed in the presence of HMGA1 (Fig. 2A,B). The INR-dependent basal stimulatory activity of HMGA1 and Mediator was not observed in the purified system reconstituted with TBP, but required TFIID/TAFs (Fig. 2C). Similarly, HMGA1 and Mediator stimulated basal transcription from the natural adenovirus major late core promoter (MLP), which is of the TATA/INR type, and had only a marginal effect on the natural core promoter of the human HSPA1A gene (HSP70), which has an identical consensus TATA box but no INR (Fig. 2D,E). Thus, the INR-dependent activity of HMGA1 and Mediator is observed with different DNA sequences flanking the consensus TATA and INR elements (Supplemental Fig. S1A). The core promoter-selective activity of HMGA1 and Mediator was similarly observed with the HMGA1a isoform (Supplemental Fig. S2G) and on linear templates (Fig. 2E; Supplemental Fig. S2H,I), indicating that a superhelical DNA structure is not required. The above results identify HMGA1 and Mediator as key positive components of the TIC1 activity and show that while Mediator (or TFIID) has no significant core promoter-selective transcription activity per se, in the presence of HMGA1, however, Mediator preferentially stimulates TATA/INR-containing core promoters by potentiating the TFIID/TAF-dependent synergy of TATA and INR elements.
Figure 2.
Cooperativity of HMGA1 and Mediator in TFIID/TAF-dependent INR function. (A) In vitro transcription experiment with supercoiled TATA and TATA/INR templates. Mediator (1, 2, and 4 μL) was titrated alone (lanes 2–4) or together with 40 ng of recombinant HMGA1b (lanes 5–7) in the purified TFIID system. The histogram shows the relative transcription activities for each template (normalized to lane 1). (B) The individual and combined effects of HMGA1b (40 ng) and Mediator (2 μL) on basal transcription in the purified system were quantitated from more than three independent experiments and plotted for each supercoiled TATA and TATA/INR promoter template as relative activities (mean ± SD) normalized to promoter activities in the absence of HMGA1 and Mediator. (C–E) In vitro transcription comparing the effects of HMGA1 and Mediator on the basal activities of different supercoiled (C,D) and linear (E) core promoters in the purified system containing either TFIID or TBP. (C) The relative transcription activities and “selectivity ratio” of TATA/INR to TATA are shown. HeLa nuclear extract (NE) was used as a reference. (D,E) Promoter activities were normalized to the activity of MLP in the TFIID system in the absence of HMGA1 and Mediator (left lanes), and only the selectivity ratios (MLP/HSP70) are shown. In D, the corresponding relative transcription activites (left to right) for MLP were 1.00, 2.24, 3.27, 8.24, 1.89; and for HSP70 were 0.67, 0.93, 1.02, 1.62, 1.26. See also Supplemental Figure S2H,I.
HMGA1 and Mediator counteract negative regulators of TBP/TATA-directed transcription in an INR-dependent manner
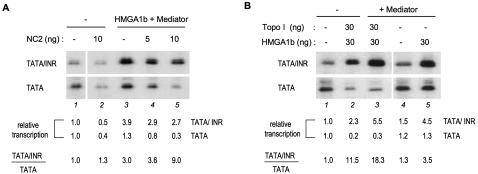
The above results suggested that the large (>40-fold) differential activity of TATA/INR versus TATA promoters observed in nuclear extracts is not solely the result of positive effects of HMGA1 and Mediator on TATA/INR, but also involves selective repression of TATA-only transcription by a nuclear extract component (e.g., see Fig. 2C, lane 1 vs. 5). Since NC2 (also known as DR1/DRAP1) inhibits TATA-dependent transcription, and its inhibitory activity is counteracted by the INR in a TAF-dependent manner in nuclear extracts but not in a purified system (Malecová et al. 2007), we tested whether the differential core promoter-selective repressive effect of NC2 is dependent on HMGA1 and Mediator. As expected, purified recombinant NC2 (Supplemental Fig. S3) repressed both TATA/INR and TATA core promoters similarly in the purified system (Fig. 3A, lane 1 vs. 2). However, in the presence of HMGA1 and Mediator, the TATA/INR promoter became more resistant to NC2 repression, while the TATA promoter was efficiently repressed (Fig. 3A, lanes 3–5). Thus, besides potentiating TATA/INR synergy, HMGA1 and Mediator also antagonize NC2-mediated repression on TATA in an INR-dependent manner, leading to an increased differential activity of TATA/INR versus TATA in the presence of NC2 (about ninefold).
Figure 3.
HMGA1 and Mediator counteract the negative functions of NC2 and Topo I in an INR-dependent manner. Recombinant NC2 (A) (see Supplemental Fig. S3) or Topo I (B) was added to the purified TFIID-based system in the presence or absence of HMGA1 and Mediator, as indicated. Basal transcription was analyzed from supercoiled TATA and TATA/INR promoters. Autoradiograms in each of the two panels are from the same gel and film exposure. The relative transcription signals (normalized to lane 1) and the ratio of TATA/INR to TATA signals are shown.
Similar to NC2 (Malecová et al. 2007), Topo I (also known as PC3 or Dr2) was shown to repress basal TATA-dependent but not TATA-less INR-dependent transcription (Kretzschmar et al. 1993; Merino et al. 1993). Although at low concentrations Topo I did not have this repressive effect (Supplemental Fig. S2D), in the presence of HMGA1, however, a selective repression of the TATA core promoter was observed both in the absence and presence of Mediator (Fig. 3B, lanes 2,3). In contrast, the TATA/INR promoter was stimulated in the presence of Topo I, HMGA1, and Mediator, leading to a high (∼18-fold) differential core promoter activity (Fig. 3B, lane 3).
These results suggest that the differential activity of the general Pol II transcription machinery on TATA and TATA/INR core promoters is the result of not only positive cooperative effects of HMGA1 and Mediator on TFIID/TAF-dependent INR function, but also antagonistic INR-dependent effects of HMGA1, Mediator, and TAFs on negative regulators of TBP/TATA-directed transcription, such as NC2 and Topo I. Interestingly, HMGA1 itself has both positive effects (in concert with TAFs and Mediator) and negative effects (in concert with Topo I, or by itself at high concentrations) on TATA-dependent transcription, which depend on the presence or absence of a synergistic INR element.
Role of HMGA1 in the synergy of TATA and INR elements in mammalian cells
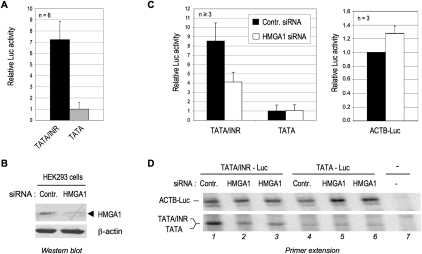
To test the possible INR-dependent function of endogenous HMGA1 in mammalian cells, we analyzed the activity of the TATA/INR and TATA core promoters fused to a luciferase reporter gene in transfected HEK293 cells. As expected, the basal TATA/INR-dependent luciferase activity was significantly higher than that of the TATA promoter (Fig. 4A). Depletion of endogenous cellular HMGA1 by RNAi using a specific siRNA (Fig. 4B) selectively inhibited TATA/INR but not TATA reporter activity or the activity of the TATA-only β-actin promoter–luciferase (ACTB-Luc) reporter (Fig. 4C). Importantly, the TATA/INR promoter-selective effect of HMGA1 knockdown was confirmed by primer extension analyses of correctly initiated luciferase mRNA transcripts (Fig. 4D). Moreover, the HMGA1 requirement for TATA/INR-dependent transcription was also observed with a different construct having different DNA sequences flanking the consensus TATA and INR elements (Supplemental Fig. S4), suggesting that specific flanking sequences are not required for the TATA/INR-specific basal activity of HMGA1 either in vitro (see above) or in vivo.
Figure 4.
HMGA1 is required selectively for TATA/INR but not TATA-mediated transcription in vivo. (A) TATA/INR-Luc and TATA-Luc were transfected in HEK293 cells, and the relative luciferase activities (mean ± SD) from six independent experiments (each in duplicate) are shown. The luciferase activity of TATA-Luc was arbitrarily set to 1. (B) HEK293 cells were transfected with either control (Contr.) or specific siRNA against HMGA1. Endogenous HMGA1 in whole-cell extracts was analyzed by Western blot. (C) TATA/INR-Luc, TATA-Luc, or ACTB-Luc reporters were transfected in HEK293 cells with a control siRNA (black bars) or the HMGA1-specific siRNA (open bars). The relative luciferase activities are shown (as in A). TATA-Luc activity (left panel) and ACTB-Luc activity (right panel) in cells transfected with the control siRNA were set arbitrarily to 1. (D) Total mRNA from HEK293 cells transfected with ACTB-Luc (lanes 1–6) and either TATA/INR-Luc (lanes 1–3) or TATA-Luc (lanes 4–6) and the indicated siRNAs was analyzed by primer extension with the Luc 24mer primer. Lane 7 is a control reaction with mRNA from mock-transfected cells. The position of correctly initiated transcripts is indicated for each promoter construct (length of 172 nucleotides [nt] for ACTB-Luc and 128 nt for TATA and TATA/INR-Luc). The top and bottom autoradiograms are from the top and bottom parts of the same gel; the bottom autoradiogram is from a longer X-ray film exposure time. See also Supplemental Figure S4 for an analysis of a different TATA/INR core promoter.
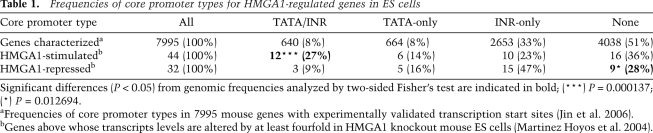
To further investigate the possible core promoter-specific regulation of natural target genes by HMGA1, we performed a statistical analysis of published differential mRNA expression data obtained from mouse embryonic stem (ES) cells after the knockout of the Hmga1 gene (Martinez-Hoyos et al. 2004). Of 13,059 murine transcripts that were analyzed by Affymetrix oligo array, a total of 1863 (14.3%) were differentially expressed by at least twofold in Hmga1 knockout ES cells. To minimize indirect effects and false positives, we focused on the 250 differentially expressed gene transcripts (1.9%) that were most highly dependent on HMGA1; i.e., affected by at least fourfold and validated by RT–PCR (Martinez Hoyos et al. 2004). These included 103 transcripts from known mouse genes, of which 76 had experimentally validated transcription initiation sites and well-annotated core promoter elements from genome-wide bioinformatics studies (Jin et al. 2006). Of these 76 HMGA1-dependent transcripts, 44 were down-regulated and 32 were up-regulated (i.e., fourfold or more) in Hmga1 knockout ES cells. We separated the genes in these two groups according to the reported presence or absence of TATA and/or INR elements in their core promoters (Jin et al. 2006) and compared the frequencies of specific core promoter types in HMGA1-activated and HMGA1-repressed groups with the global frequencies of promoter types in the mouse genome (Table 1). Interestingly, the group of HMGA1-stimulated genes was significantly enriched in core promoters having both TATA and INR elements (TATA/INR type), while other promoter types in this group did not significantly differ from their global frequencies in the genome. In contrast, the frequencies of most promoter types, including TATA/INR, in the HMGA1-repressed group of genes did not significantly differ from their global genomic frequencies. We note, however, that the “none” category of promoters lacking both TATA and INR is underrepresented in the HMGA1-repressed group of genes, consistent with the fact that these promoters are generally GC-rich (AT-poor). Altogether, these results are consistent with our in vitro transcription analyses and suggest a novel core promoter-dependent role of HMGA1 in gene-specific regulation in mammalian cells involving potentiation of the transcription synergy of TATA and INR elements.
Table 1.
Frequencies of core promoter types for HMGA1-regulated genes in ES cells
HMGA1 C-terminal acidic tail domain has core promoter-specific functions and is required for HMGA1 interaction with TFIID and Mediator in mammalian cells
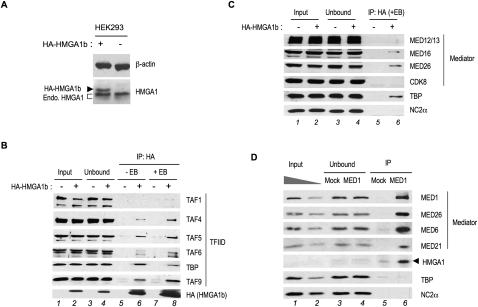
To investigate the possible mechanisms for the functional cooperativity of HMGA1 with TFIID and Mediator, we tested their possible interaction in HEK293 cells. We first used a HEK293 cell line that expresses low levels of ectopic HA epitope-tagged HMGA1b (HA-HMGA1b) (Fig. 5A). Immunoprecipitation with an anti-HA antibody and Western blot analyses demonstrated a specific interaction of HA-HMGA1b with all TFIID subunits tested (Fig. 5B) and several subunits of Mediator, but not the CDK8 subunit (Fig. 5C). We further confirmed the specific interaction of the Mediator complex with endogenous HMGA1 in HeLa and HEK293 cells by coimmunoprecipitation with an anti-MED1 antibody (Fig. 5D; Supplemental Fig. S5). Notably, endogenous HMGA1b was significantly enriched in the anti-MED1 immunoprecipitate (Fig. 5D, lane 6; Supplemental Fig. S5). Moreover, the interactions observed were not affected by the presence of ethidium bromide, suggesting that they were not indirect effects of DNA binding.
Figure 5.
HMGA1 interacts with TFIID and Mediator, but not CDK8, in human cells. (A) Western blot analysis of total HMGA1 in normal HEK293 cells (−) and a derivative clonal cell line that was stably transfected with HA-HMGA1b (+). Positions of endogenous HMGA1 and ectopic HA-HMGA1b proteins are indicated. An antibody to β-actin was used as loading control. (B,C) Whole-cell extracts from HEK293 cells and HEK293 cells stably expressing HA-HMGA1b (described above) were adjusted to 175 mM KCl and immunoprecipitated with anti-HA antibody resin in the presence or absence of ethidium bromide (EB), as indicated. The Western blot was probed with antibodies to the indicated TFIID and Mediator subunits. NC2α served as the negative control. (D) HeLa cell nuclear extracts were immunoprecipitated with a MED1 antibody or mock-immunoprecipitated with goat IgG (Mock), and associated proteins were analyzed by Western blot with the indicated antibodies. Similar results were obtained with HEK293 cells (Supplemental Fig. S5).
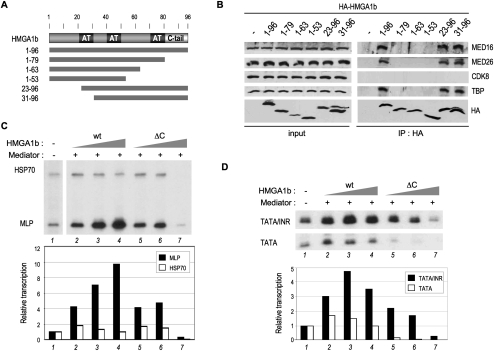
To map the domains of HMGA1b required for interaction with TFIID and Mediator, HA-HMGA1b wild type and several deletion mutants were transiently transfected in HEK293 cells and analyzed by coimmunoprecipitation and Western blotting, as above. Interestingly, we found that the C-terminal acidic tail domain is important for HMGA1b interaction with both TFIID and Mediator (Fig. 6A,B). To our knowledge, this represents the first identification of HMGA1-interacting factors that depend on this conserved domain of HMGA proteins. We further tested the possible contribution of the C-terminal tail in the core promoter-specific basal stimulatory activity of HMGA1b by titrating purified recombinant HMGA1b wild type and a tail-deleted (ΔC) mutant (Supplemental Fig. S6) in the purified transcription system in the presence of Mediator. While HMGA1b wild type significantly stimulated the MLP core promoter, but not the HSP70 promoter, at all concentrations tested (up to 10-fold) (Fig. 6C, lanes 2–4), the ΔC mutant had a drastically reduced activity and repressed transcription from both promoters at the highest concentration (Fig. 6C, lanes 5–7). Similarly, the HMGA1b ΔC mutant had a reduced INR-specific stimulatory activity on the TATA/INR core promoter and an increased repressive function at the highest concentration on both promoters (Fig. 6D). However, the TATA/INR promoter was less sensitive to this repressive effect, consistent with the INR-dependent activity of TAFs and Mediator in antagonizing the negative function of HMGA1 and other negative cofactors, described previously (Fig. 3; Supplemental Fig. S2E,F). Thus, HMGA1 has both (1) a core promoter/INR-selective basal stimulatory function that cooperates with Mediator and TFIID/TAFs and requires the C-tail domain (and possibly the N-terminal AT hooks), and (2) a negative function at the N terminus (containing the three AT hooks) that is suppressed by the C-tail domain and antagonized in an INR-dependent manner by Mediator and TFIID/TAFs. Hence, these results suggest that the interaction of TFIID and Mediator with HMGA1 is dependent on the acidic C-tail domain and important for their concerted core promoter-selective basal activities.
Figure 6.
The HMGA1 acidic C-tail domain is required for interaction with TFIID and Mediator and for stimulation of INR-dependent transcription. (A) Scheme of HMGA1b wild-type structure, including AT hooks (AT) and acidic tail (C tail), and deletion mutants used for immunoprecipitation experiments below. (B) HEK293 cells were mock-transfected (−) or transiently transfected with HA-HMGA1b wild type (1–96) or the indicated deletion mutants. Whole-cell extracts (input) were immunoprecipitated with anti-HA antibody (IP: HA) and analyzed by Western blot with the indicated antibodies. (C,D) In vitro transcription was performed with the indicated core promoter constructs (linear form) in the purified TFIID-based system complemented with Mediator and different amounts (20, 40, and 60 ng in C; and 35, 40, and 45 ng in D) of either wild-type HMGA1b (wt) or a deletion mutant 1–81 (ΔC) that lacks the acidic C-tail domain (purified proteins are shown in Supplemental Fig. S6). Relative transcription levels were normalized to the signals in the absence of HMGA1/Mediator (shown in lanes 1).
Discussion
The factors and mechanisms responsible for the strong synergistic stimulation of Pol II-dependent transcription by TATA and INR core promoter elements have remained poorly understood. Previous results indicated that TFIID/TAF-dependent INR function in synergy with the TATA box not only entails a TFIIA-dependent cooperative recruitment of TFIID to core promoters containing both elements in a synergistic configuration (Emami et al. 1997), but also involves TAF-dependent cofactors that are distinct from GTFs and have remained elusive (Martinez et al. 1998). Here we identified these cofactors as the architectural protein HMGA1 and the Mediator coregulator complex. Significantly, we found that the basal transcription stimulatory function of Mediator, which up to now has been considered “general” or invariant on all core promoters, can be stimulated by HMGA1 and TAFs in an INR-dependent manner (Fig. 2). Our results thus unveil a “facultative” core promoter-dependent activity of Mediator and HMGA1 and a functional core promoter-selective cooperativity of HMGA1, Mediator, and TFIID/TAFs, as none of these factors alone (or in pairs) can significantly stimulate INR-dependent transcription by the purified Pol II transcription machinery. We note, however, that the maximal level of INR-dependent activation observed with crude nuclear extracts has yet to be reached in the purified system, which may suggest the involvement of additional cofactors or post-translational modifications that may be missing in the purified reconstituted system. For instance, HMGA1 is a substrate for multiple post-translational modifications in vivo, which influence its DNA binding and transcription functions (Reeves 2003).
In addition to their TAF- and INR-dependent stimulatory activities, HMGA1 and Mediator also antagonize repression of the basal transcription machinery by NC2 and Topo I in an INR-dependent manner (Fig. 3). While not addressed here, this concerted anti-repressive activity of HMGA1 and Mediator could also more broadly antagonize the inhibitory effects of general chromatin components at specific promoters in vivo. Indeed, HMGA1 was shown to dynamically compete with histone H1 binding to chromatin in live mammalian cells (Catez et al. 2004). Hence, the combined stimulatory and anti-repressive effects of HMGA1, Mediator, and TAFs may account for the large differential activity of TATA and TATA/INR core promoters observed in more physiological cell-free extracts and in live cells (O'Shea-Greenfield and Smale 1992; Colgan and Manley 1995; Malecová et al. 2007). In support of a core promoter-selective stimulatory function of HMGA1 in vivo, we further showed that endogenous HMGA1 in mammalian cells contributes to the transcription stimulatory activity of the INR at TATA- and INR-containing (TATA/INR) core promoters and that physiological HMGA1-activated (but not HMGA1-repressed) target genes in ES cells often have core promoters with TATA and INR elements in a synergistic configuration (Fig. 4; Table 1). Given the multiple post-translational modifications and diverse chromatin and gene regulatory roles of HMGA1 (Reeves 2003), including the novel core promoter-specific functions described here, the particular contributions of HMGA1 in regulation of specific genes are likely to be cell type- and context-dependent, consistent with the observed tissue specificity of HMGA1-dependent gene regulation (Martinez Hoyos et al. 2004).
While the detailed molecular mechanisms underlying the core promoter-selective cooperativity of HMGA1, Mediator, and TFIID/TAFs in INR-dependent transcription remain to be fully characterized, our results point to an important regulatory role of the conserved acidic C-tail domain of HMGA1. This acidic C-tail domain is conserved in HMGA proteins (HMGA1a/b and HMGA2) and appears to have important biological functions in regulation of cell proliferation and oncogenic transformation by HMGA proteins, whose overexpression is a hallmark of malignant tumors (Pierantoni et al. 2003; Fusco and Fedele 2007; Li et al. 2007). However, the molecular functions of this C-tail domain have remained unclear. The acidic C-tail domain of HMGA1 does not have an intrinsic transactivating function when fused to the DNA-binding domain of GAL4 (Thanos and Maniatis 1992; Zhou et al. 1996), although it appears to be required for HMGA1 coactivation of some, but not all, HMGA1-dependent activators and target promoters in transfected cells (Yie et al. 1997; Chin et al. 1998). Here, we showed that the C-tail domain of HMGA1 is required for both HMGA1 interaction with Mediator and TFIID in human cells (Fig. 6B) and maximal stimulation of INR-dependent basal transcription by Mediator and TFIID/TAFs in vitro (Fig. 6C,D). In addition, the C-tail domain antagonizes the repressive function of the N-terminal region containing the three AT hooks (Fig. 6C,D). This anti-repressive function of the C tail correlates with the reported roles of the acidic C-tail domains of HMGA1 and HMGA2 in restricting the DNA-binding and self-association activities of the N-terminal region containing the AT hooks (Nissen and Reeves 1995; Yie et al. 1997; Noro et al. 2003). Thus, the DNA-binding activity of HMGA1, which recognizes AT-rich and structured sequences including TATA elements and nucleosomal DNA, could be altered in association with TFIID and/or Mediator. Alternatively (or in addition), HMGA1 could selectively interact with specific variants of Mediator or TFIID complexes or could affect their structure at enhancers and/or core promoters. Indeed, HMGA1 interacts selectively with a form of Mediator that lacks the CDK8 subunit (Fig. 5C). In addition, structural effects of activators on Mediator conformation (Taatjes et al. 2002; Meyer et al. 2010; Taatjes 2010) and on isomerization of TFIID on core promoter DNA (Horikoshi et al. 1988; Lieberman and Berk 1994; Chi and Carey 1996) have been reported, which suggests a possible malleability of these complexes. In this context, it is interesting to note that the C-tail domain of HMGA1 is phosphorylated by CK2 (for review, see Reeves 2003), a protein kinase with reported core promoter-specific regulatory activities (Lewis et al. 2005). Thus, post-translational modifications of HMGA1 could modulate its interactions with Mediator or TFIID and hence regulate its core promoter-selective functions.
It has been shown that a heterodimeric complex composed of human TAF1 and Drosophila TAF2 subunits of TFIID, but not either subunit alone, can bind specifically to INR sequences (Chalkley and Verrijzer 1999). Interestingly, Drosophila TAF1 has HMGA-like AT hooks—one or two motifs, depending on the TAF1 isoform—and only TAF1 isoforms with both AT hooks can directly bind, independently of TAF2, to the transcription start site of several Drosophila core promoters (Metcalf and Wassarman 2006). Furthermore, TAF1 is the only subunit within Drosophila TFIID that contacts the INR, as indicated by short-range protein–DNA cross-linking, suggesting that TAF2 plays only an accessory role (Wu et al. 2001). In contrast, human TAF1 (and TAF1 in many/most other organisms) does not have any AT hook, but instead has an HMG box (Sekiguchi et al. 1991). Thus, it is tempting to speculate that the interaction of HMGA1 with TFIID complexes that lack one or both AT hooks could compensate for the missing AT hooks and facilitate specific TFIID–INR interactions or stereo-specific conformations at TATA/INR core promoters. For instance, HMGA1 is able to bind DNA on the surface of nucleosomes, inducing localized changes in the rotational setting of the DNA (for review, see Reeves 2003), and could perhaps similarly bind to TATA/INR core promoter DNA that is bent/wrapped by TFIID/TAFs (Oelgeschläger et al. 1996). Thus, HMGA1 could have architectural or DNA chaperone functions at certain core promoters, similar to its proposed role in facilitating the cooperative assembly of nucleoprotein enhanceosome complexes (Reeves 2003; Panne 2008). The recruitment of both HMGA1 and Mediator to regulatory DNA/enhancer sequences and the reported involvement of both HMGA1 and Mediator in long-range enhancer function via DNA/chromatin looping (Bagga et al. 2000; Kagey et al. 2010) and in core promoter-selective stimulation (as shown here) suggest possible concerted functions of HMGA1 and Mediator at the interface between distal regulatory elements and specific core promoters. Such cooperative interactions could not only facilitate the long-range communication between activators/enhancers and the basal transcription machinery, but also mediate the core promoter-selective function of certain activators and enhancers.
In summary, our results uncovered a novel core promoter-selective cooperative function of HMGA1 and Mediator through the INR that depends on TAFs and the C-terminal acidic tail domain of HMGA1. From the dual roles of HMGA1 and Mediator at enhancers and core promoters, we propose that distal regulators and their recruited coregulators could, beyond a simple recruitment model, coordinate the assembly of stereo-specific PICs at core promoters and dictate the productive utilization of specific core promoter DNA elements by the basal transcription machinery and hence the levels of transcription in response to specific stimuli. The reconstitution of alternative core promoter element-dependent transcription pathways in vitro with purified factors is an important first step to further test this model.
Materials and methods
Protein purification and in vitro transcription
TIC1 purification is described in the Supplemental Material. Purification of Pol II and GTFs was described previously (Martinez et al. 1998). The purified basal transcription system consisted of 0.7 μL of Pol II (DE), 1 μL of TFIIA (Ni2+-NTA-agarose), 15 ng of recombinant 6His-TFIIB, 1 μL of Flag-tagged TFIID (∼5 ng of f:TBP per microliter), 20 ng of recombinant 6His-TFIIF, and 2 μL of TFIIE/H fraction (see the Supplemental Material). Alternatively, 20 ng of recombinant 6His-TFIIE and 0.15 μL of highly purified TFIIH (Q2) were used instead of the TFIIE/H fraction, with similar results (Supplemental Fig. S2H,I). When indicated, 5–10 ng of recombinant 6His-TBP was used instead of Flag-tagged TFIID. Purification of Mediator, reporter plasmids, in vitro transcription, and primer extension are described in the Supplemental Material.
Cell culture, transient transfection, luciferase assay, and RNAi
HeLa and HEK293 cells and a HEK293 cell line stably transfected with pHA-HMGA1b and expressing low levels of HA-tagged mouse HMGA1b were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum at 37°C with 5% CO2. Transient transfections of HEK293 cells were performed with Lipofectamine 2000 (Invitrogen), and luciferase assays were performed as described previously (Faiola et al. 2005). RNAi analyses are described in the Supplemental Material.
Antibodies, Western blotting, and immunoprecipitation
The antibodies obtained from commercial sources were HMGA1 (sc-1564), MED1/TRAP220 (sc-5334x), MED12/TRAP230 (sc-5374x), MED13/TRAP240 (sc-12013x), MED16/TRAP95 (sc-5363x), MED26/CRSP70 (sc-48776x), CDK8 (sc-1521), TAF1 (sc-735x), TAF9 (sc-1247x), and β-actin (sc-1616R), all from Santa Cruz Biotechnology; and anti-HA antibody (12CA5) and anti-HA resin from Covance. The NC2α antibody was a kind gift from Dr. Thomas Oelgeschläger. Whole-cell extract preparation, immunoprecipitation, and Western blotting were essentially as previously described (Faiola et al. 2005). Where indicated, 50 μg/mL ethidium bromide was added to cell extracts before immunoprecipitation.
Core promoter statistics
For the comparisons of core promoter frequencies in Table 1, significant differences at P < 0.05 were determined by using the Fisher's exact test and two-sided P-values (sum of small p's method).
Acknowledgments
We thank Drs. Aram Akopian, Alfredo Fusco, Thomas Oelgeschläger, Stephen Smale, and Yinsheng Wang for generous gifts of reagents; Dr. Subir Ghosh for advice on statistical methods; and Hsiao-LIng Yeh and other members of the laboratory for advice and discussions. This work was supported by grants from NIH (CA129325 to R.G.R.) and NSF (MCB-0448488 and MCB-1021696 to E.M.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.177360.111.
References
- Baek HJ, Malik S, Qin J, Roeder RG 2002. Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol Cell Biol 22: 2842–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga R, Michalowski S, Sabnis R, Griffith JD, Emerson BM 2000. HMG I/Y regulates long-range enhancer-dependent transcription on DNA and chromatin by changes in DNA topology. Nucleic Acids Res 28: 2541–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsdottir G, Myers LC 2008. Minimal components of the RNA polymerase II transcription apparatus determine the consensus TATA box. Nucleic Acids Res 36: 2906–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TW, Kadonaga JT 1996. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev 10: 711–724 [DOI] [PubMed] [Google Scholar]
- Catez F, Yang H, Tracey KJ, Reeves R, Misteli T, Bustin M 2004. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol Cell Biol 24: 4321–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkley GE, Verrijzer CP 1999. DNA binding site selection by RNA polymerase II TAFs: A TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J 18: 4835–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi T, Carey M 1996. Assembly of the isomerized TFIIA–TFIID–TATA ternary complex is necessary and sufficient for gene activation. Genes Dev 10: 2540–2550 [DOI] [PubMed] [Google Scholar]
- Chin MT, Pellacani A, Wang H, Lin SS, Jain MK, Perrella MA, Lee ME 1998. Enhancement of serum-response factor-dependent transcription and DNA binding by the architectural transcription factor HMG-I(Y). J Biol Chem 273: 9755–9760 [DOI] [PubMed] [Google Scholar]
- Colgan J, Manley JL 1995. Cooperation between core promoter elements influences transcriptional activity in vivo. Proc Natl Acad Sci 92: 1955–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami KH, Jain A, Smale ST 1997. Mechanism of synergy between TATA and initiator: Synergistic binding of TFIID following a putative TFIIA-induced isomerization. Genes Dev 11: 3007–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faiola F, Liu X, Lo S, Pan S, Zhang K, Lymar E, Farina A, Martinez E 2005. Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol Cell Biol 25: 10220–10234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco A, Fedele M 2007. Roles of HMGA proteins in cancer. Nat Rev Cancer 7: 899–910 [DOI] [PubMed] [Google Scholar]
- Gershenzon NI, Ioshikhes IP 2005. Synergy of human Pol II core promoter elements revealed by statistical sequence analysis. Bioinformatics 21: 1295–1300 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi M, Carey MF, Kakidani H, Roeder RG 1988. Mechanism of action of a yeast activator: Direct effect of GAL4 derivatives on mammalian TFIID–promoter interactions. Cell 54: 665–669 [DOI] [PubMed] [Google Scholar]
- Hsu JY, Juven-Gershon T, Marr MT II, Wright KJ, Tjian R, Kadonaga JT 2008. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev 22: 2353–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin VX, Singer GA, Agosto-Pérez FJ, Liyanarachchi S, Davuluri RV 2006. Genome-wide analysis of core promoter elements from conserved human and mouse orthologous pairs. BMC Bioinformatics 7: 114 doi: 10.1186/1471-2105-7-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G 2009. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 41: 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT 2008. The RNA polymerase II core promoter—The gateway to transcription. Curr Opin Cell Biol 20: 253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Björklund S, Li Y, Sayre MH, Kornberg RD 1994. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77: 599–608 [DOI] [PubMed] [Google Scholar]
- Koch HG, Moser M, Müller M 2003. Signal recognition particle-dependent protein targeting, universal to all kingdoms of life. Rev Physiol Biochem Pharmacol 146: 55–94 [DOI] [PubMed] [Google Scholar]
- Kornberg RD 2005. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci 30: 235–239 [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Meisterernst M, Roeder RG 1993. Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proc Natl Acad Sci 90: 11508–11512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BA, Sims RJ III, Lane WS, Reinberg D 2005. Functional characterization of core promoter elements: DPE-specific transcription requires the protein kinase CK2 and the PC4 coactivator. Mol Cell 18: 471–481 [DOI] [PubMed] [Google Scholar]
- Li Y, Lu J, Prochownik EV 2007. Dual role for SUMO E2 conjugase Ubc9 in modulating the transforming and growth-promoting properties of the HMGA1b architectural transcription factor. J Biol Chem 282: 13363–13371 [DOI] [PubMed] [Google Scholar]
- Lieberman PM, Berk AJ 1994. A mechanism for TAFs in transcriptional activation: Activation domain enhancement of TFIID–TFIIA–promoter DNA complex formation. Genes Dev 8: 995–1006 [DOI] [PubMed] [Google Scholar]
- Malecová B, Gross P, Boyer-Guittaut M, Yavuz S, Oelgeschläger T 2007. The initiator core promoter element antagonizes repression of TATA-directed transcription by negative cofactor NC2. J Biol Chem 282: 24767–24776 [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG 2010. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 11: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Ge H, Tao Y, Yuan CX, Palhan V, Roeder RG 1998. Novel cofactors and TFIIA mediate functional core promoter selectivity by the human TAFII150-containing TFIID complex. Mol Cell Biol 18: 6571–6583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Hoyos J, Fedele M, Battista S, Pentimalli F, Kruhoffer M, Arra C, Orntoft TF, Croce CM, Fusco A 2004. Identification of the genes up- and down-regulated by the high mobility group A1 (HMGA1) proteins: Tissue specificity of the HMGA1-dependent gene regulation. Cancer Res 64: 5728–5735 [DOI] [PubMed] [Google Scholar]
- Merino A, Madden KR, Lane WS, Champoux JJ, Reinberg D 1993. DNA topoisomerase I is involved in both repression and activation of transcription. Nature 365: 227–232 [DOI] [PubMed] [Google Scholar]
- Metcalf CE, Wassarman DA 2006. DNA binding properties of TAF1 isoforms with two AT-hooks. J Biol Chem 281: 30015–30023 [DOI] [PubMed] [Google Scholar]
- Meyer KD, Lin SC, Bernecky C, Gao Y, Taatjes DJ 2010. p53 activates transcription by directing structural shifts in Mediator. Nat Struct Mol Biol 17: 753–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler G, Kremmer E, Timmers HT, Meisterernst M 2001. Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep 2: 808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair D, Kim Y, Myers LC 2005. Mediator and TFIIH govern carboxyl-terminal domain-dependent transcription in yeast extracts. J Biol Chem 280: 33739–33748 [DOI] [PubMed] [Google Scholar]
- Nissen MS, Reeves R 1995. Changes in superhelicity are introduced into closed circular DNA by binding of high mobility group protein I/Y. J Biol Chem 270: 4355–4360 [DOI] [PubMed] [Google Scholar]
- Noro B, Licheri B, Sgarra R, Rustighi A, Tessari MA, Chau KY, Ono SJ, Giancotti V, Manfioletti G 2003. Molecular dissection of the architectural transcription factor HMGA2. Biochemistry 42: 4569–4577 [DOI] [PubMed] [Google Scholar]
- Oelgeschläger T, Chiang CM, Roeder RG 1996. Topology and reorganization of a human TFIID–promoter complex. Nature 382: 735–738 [DOI] [PubMed] [Google Scholar]
- Ohler U, Liao GC, Niemann H, Rubin GM 2002. Computational analysis of core promoters in the Drosophila genome. Genome Biol 3: research0087.1–research0087.12 doi: 10.1186/gb-2002-3-12-research0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea-Greenfield A, Smale ST 1992. Roles of TATA and initiator elements in determining the start site location and direction of RNA polymerase II transcription. J Biol Chem 267: 1391–1402 [PubMed] [Google Scholar]
- Panne D 2008. The enhanceosome. Curr Opin Struct Biol 18: 236–242 [DOI] [PubMed] [Google Scholar]
- Park JM, Gim BS, Kim JM, Yoon JH, Kim HS, Kang JG, Kim YJ 2001. Drosophila Mediator complex is broadly utilized by diverse gene-specific transcription factors at different types of core promoters. Mol Cell Biol 21: 2312–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierantoni GM, Battista S, Pentimalli F, Fedele M, Visone R, Federico A, Santoro M, Viglietto G, Fusco A 2003. A truncated HMGA1 gene induces proliferation of the 3T3-L1 pre-adipocytic cells: A model of human lipomas. Carcinogenesis 24: 1861–1869 [DOI] [PubMed] [Google Scholar]
- Reeves R 2003. HMGA proteins: Flexibility finds a nuclear niche? Biochem Cell Biol 81: 185–195 [DOI] [PubMed] [Google Scholar]
- Reeves R, Beckerbauer L 2001. HMGI/Y proteins: Flexible regulators of transcription and chromatin structure. Biochim Biophys Acta 1519: 13–29 [DOI] [PubMed] [Google Scholar]
- Reeves WM, Hahn S 2003. Activator-independent functions of the yeast mediator sin4 complex in preinitiation complex formation and transcription reinitiation. Mol Cell Biol 23: 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG 2005. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett 579: 909–915 [DOI] [PubMed] [Google Scholar]
- Sekiguchi T, Nohiro Y, Nakamura Y, Hisamoto N, Nishimoto T 1991. The human CCG1 gene, essential for progression of the G1 phase, encodes a 210-kilodalton nuclear DNA-binding protein. Mol Cell Biol 11: 3317–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST 2001. Core promoters: Active contributors to combinatorial gene regulation. Genes Dev 15: 2503–2508 [DOI] [PubMed] [Google Scholar]
- Taatjes DJ 2010. The human Mediator complex: A versatile, genome-wide regulator of transcription. Trends Biochem Sci 35: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taatjes DJ, Näär AM, Andel F III, Nogales E, Tjian R 2002. Structure, function, and activator-induced conformations of the CRSP coactivator. Science 295: 1058–1062 [DOI] [PubMed] [Google Scholar]
- Takagi Y, Kornberg RD 2006. Mediator as a general transcription factor. J Biol Chem 281: 80–89 [DOI] [PubMed] [Google Scholar]
- Thanos D, Maniatis T 1992. The high mobility group protein HMG I(Y) is required for NF-κB-dependent virus induction of the human IFN-β gene. Cell 71: 777–789 [DOI] [PubMed] [Google Scholar]
- Thanos D, Maniatis T 1995. Virus induction of human IFN β gene expression requires the assembly of an enhanceosome. Cell 83: 1091–1100 [DOI] [PubMed] [Google Scholar]
- Willy PJ, Kobayashi R, Kadonaga JT 2000. A basal transcription factor that activates or represses transcription. Science 290: 982–985 [DOI] [PubMed] [Google Scholar]
- Wu CH, Madabusi L, Nishioka H, Emanuel P, Sypes M, Arkhipova I, Gilmour DS 2001. Analysis of core promoter sequences located downstream from the TATA element in the hsp70 promoter from Drosophila melanogaster. Mol Cell Biol 21: 1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E 2007. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene 389: 52–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yie J, Liang S, Merika M, Thanos D 1997. Intra- and intermolecular cooperative binding of high-mobility-group protein I(Y) to the β-interferon promoter. Mol Cell Biol 17: 3649–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Benson KF, Przybysz K, Liu J, Hou Y, Cherath L, Chada K 1996. Genomic structure and expression of the murine Hmgi-c gene. Nucleic Acids Res 24: 4071–4077 [DOI] [PMC free article] [PubMed] [Google Scholar]