Transcriptional repression in eukaryotic organisms is controlled by corepressor complexes that are recruited to target promoters by direct interactions with the DNA-bound repressors. The Cyc8–Tup1 corepressor complex was the first eukaryotic corepressor to be identified, and this study provides novel insight into the mechanism by which Cyc–Tup1 regulates transcriptional responses under stress conditions. The authors use a novel genetic approach and find that Cyc8–Tup1 is recruited to promoters by proteins that repress transcription when Cyc8–Tup1 is bound and activate transcription in Cyc8–Tup1's absence. The authors show that Cyc8–Tup1 functions primarily by masking the activation domain of the protein that recruited it under normal conditions. Thus, the critical mechanistic target of Cyc8–Tup1 is the activation domain of the recruiting protein, providing insight into a novel mechanism of the regulation of gene expression by Cyc8–Tup1.
Keywords: Cyc8–Tup1, transcriptional corepressor, gene regulation, stress responses, transcriptional activation, transcriptional repression
Abstract
The yeast Tup1–Cyc8 corepressor complex is recruited to promoters by DNA-binding repressors, but the mechanisms by which it inhibits expression of genes involved in various stress pathways are poorly understood. Conditional and rapid depletion of Tup1 from the nucleus leads to concurrent nucleosome depletion and histone acetylation, recruitment of coactivators (Swi/Snf, SAGA, and Mediator), and increased transcriptional activity. Conversely, coactivator dissociation occurs rapidly upon rerepression by Cyc8–Tup1, although coactivator association and transcription can be blocked even in the absence of nucleosomes. The coactivators are recruited to the sites where Tup1 was located prior to depletion, indicating that the repressor proteins that recruit Tup1 function as activators in its absence. Last, Cyc8–Tup1 can interact with activation domains in vivo. Thus, Cyc8–Tup1 regulates transcription primarily by masking and inhibiting the transcriptional activation domains of the recruiting proteins, not by acting as a corepressor. We suggest that the corepressor function of Cyc8–Tup1 makes only a modest contribution to expression of target genes, specifically to keep expression levels below the nonactivated state.
Prokaryotic repressor proteins bind promoter DNA sequences and inhibit transcription by steric hindrance of RNA polymerase. In contrast, transcriptional repression in eukaryotic organisms does not occur simply by binding of repressor proteins to their target sites. Instead, repression is mediated by corepressor complexes that are recruited to target promoters by direct interactions with the DNA-bound repressors. In many cases, corepressor complexes possess one or more histone-modifying enzymes (e.g., Rpd3 histone deacetylase, histone methylases for H3-K9 and K27, or histone demethylases for H3-K4 and K36) that are important for repression (Struhl 1998; Grewal and Jia 2007; Schwartz and Pirrotta 2007; Simon and Kingston 2009; Mosammaparast and Shi 2010). In some cases, corepressor complexes are recruited by nascent RNA or other RNA species (Buhler et al. 2006; Grewal and Elgin 2007; Rinn et al. 2007; Zhao et al. 2008). Some corepressor complexes are localized to discrete sites, whereas others are spread over larger genomic regions.
The Saccharomyces cerevisiae Cyc8–Tup1 complex, the first transcriptional corepressor to be described (Keleher et al. 1992; Tzamarias and Struhl 1994), is composed of four Tup1 subunits and one Cyc8 subunit (Varanasi et al. 1996). Tup1 is homologous to Groucho and TLE corepressors that play critical roles in multicellular eukaryotes. Cyc8–Tup1 is crucial to the biology of yeast cells, as it is required for repression of cell type-specific genes as well as genes expressed under a variety of environmental stress conditions, such as poor carbon source, hypoxia, DNA damage, mitochondrial dysfunction, and hyperosmolarity.
Cyc8–Tup1 is recruited to promoters via interactions with DNA-binding proteins, each of which represses genes in a specific biological pathway. These interactions with DNA-binding repressors are mediated mainly through different surfaces of Cyc8, although Tup1 also plays a role in some cases (Komachi et al. 1994; Tzamarias and Struhl 1995; Kobayashi et al. 2008). Tup1 contains a repression domain that is believed to confer the repressive function of the Cyc8–Tup1 complex via protein–protein interactions (Tzamarias and Struhl 1994; Zhang et al. 2002; Green and Johnson 2005). Interestingly, in response to osmotic or carbon source stress, Cyc8–Tup1 does not dissociate from target promoters, and in fact it contributes to recruitment of the Swi/Snf and SAGA coactivator complexes (Papamichos-Chronakis et al. 2002; Proft and Struhl 2002; Mennella et al. 2003). In these and other cases (Fragiadakis et al. 2004; Zhang and Reese 2005; Hickman and Winston 2007), the DNA-binding repressor also appears to function as an activator protein under appropriate stress conditions.
Repression by Cyc8–Tup1 inhibits TBP association and hence preinitiation complex formation (Kuras and Struhl 1999; Mennella et al. 2003). However, considerable work from a number of laboratories reveals a confusing picture about the repression mechanism, and several nonmutually exclusive models have been proposed (Smith and Johnson 2000; Malave and Dent 2006). First, Tup1 has genetic and biochemical interactions with several subunits of the Mediator complex (Kuchin and Carlson 1998; Gromoller and Lehming 2000; Papamichos-Chronakis et al. 2000; Han et al. 2001; Zhang and Emmons 2002), suggesting that it may interfere with Mediator function. However, a coherent picture of how this leads to repression as opposed to activation has yet to be elucidated. Second, Cyc8–Tup1 can interact with multiple histone deacetylases in certain in vitro conditions (Watson et al. 2000; Wu et al. 2001; Davie et al. 2003), and it has been proposed that repression is mediated by recruitment of the Hda1 histone deacetylase complex to target promoters (Wu et al. 2001; Robyr et al. 2002). Third, Tup1 interacts with hypoacetylated tails of histones H3 and H4 (Cooper et al. 1994; Edmondson et al. 1996; Huang et al. 1997) and appears to influence deposition of the Htz1 histone variant (Green and Johnson 2004; Gligoris et al. 2007; Morohashi et al. 2007). Fourth, Tup1 appears to stabilize nucleosome positioning, perhaps through the Isw2 nucleosome remodeling complex, although Tup1 and Isw2 independently associate with promoters (Zhang and Reese 2004b; Rizzo et al. 2011). Fifth, it has been suggested that Cyc8–Tup1 organizes long-range repressive chromatin structures (Fleming and Pennings 2001, 2007; Li and Reese 2001; Zhang and Reese 2004b). Last, in artificial situations, Tup1 may affect activator function (Gavin et al. 2000; Geisberg and Struhl 2000), although this effect may be mediated indirectly through chromatin.
A major problem with all of the mechanisms proposed above is that inactivation of any putative target results in minimal loss of repression as compared with the strong derepression observed in cyc8 or tup1 deletion mutants. Furthermore, while this observation has prompted the idea that these proposed mechanisms are redundant, strong repression is still observed in all attempts to inactivate multiple putative targets (Lee et al. 2000; Zhang and Reese 2004a). These observations strongly suggest that a critical aspect of regulation by Cyc8–Tup1 remains unknown. In addition, experimental and conceptual issues have complicated some previous conclusions leading to these models. Many studies rely on artificial, and often highly sensitized, constructs that are very different from native genes, and hence they may not reflect the true action of Cyc8–Tup1. Furthermore, most previous studies involved comparisons between repressing and nonrepressing conditions that involve environmental stress, thereby making it difficult to disentangle transcriptional repression mediated by Cyc8–Tup1 from stress-mediated activation, and this is further complicated by the role of Cyc8–Tup1 in recruiting coactivators (Papamichos-Chronakis et al. 2002; Proft and Struhl 2002; Mennella et al. 2003). Last, experiments involving cyc8 and tup1 deletion strains are complicated by their very poor growth and severe flocculation phenotype that could indirectly affect gene expression.
To circumvent these problems associated with previous analyses of Cyc8–Tup1 repression, we employed the anchor-away (AA) method (Haruki et al. 2008) to conditionally and rapidly remove Tup1 or Cyc8 out of the nucleus. A major advantage of this approach is the ability to kinetically follow the process of interest upon depletion of the protein, which is ideal for distinguishing direct versus indirect effects of a mutant allele (ts, null, or otherwise). Nuclear depletion by anchor-away occurs more rapidly and more effectively than obtained with inducible degron-based approaches (Moqtaderi et al. 1996; Dohmen and Varshavsky 2005), and the induction method for depletion is physiologically neutral, unlike the use of copper or heat in the degron-based methods. Using the approach, we demonstrate that Cyc8–Tup1 regulates transcription primarily by masking the activation domains of the diverse proteins that recruit Cyc8–Tup1 to target promoters. In this regard, Cyc8–Tup1 functions in a manner similar to that of Gal80, not as a corepressor. Our results suggest that the corepressor function of Cyc8–Tup1 only makes a modest contribution to regulation of target genes.
Results
Conditional and rapid depletion of the Cyc8–Tup1 corepressor results in concomitant and robust derepression of target genes
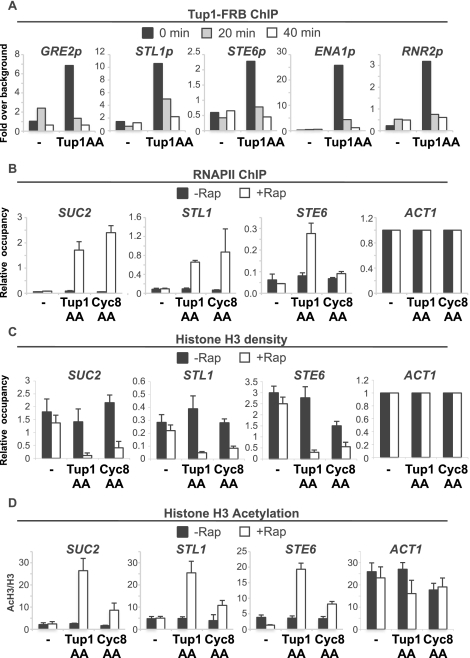
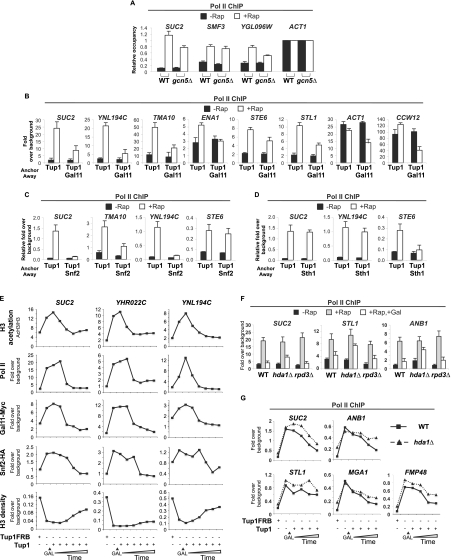
The anchor-away technique (Haruki et al. 2008) is based on the ability of rapamycin to rapidly form ternary complexes with the FKBP12 and FRB domains (Ho et al. 1996). Rapamycin addition causes rapid removal of the desired protein (tagged with FRB) from the nucleus into the cytoplasm, where it is stably anchored via FKBP-Rpl13A. As expected, strains expressing Tup1-FRB or Cyc8-FRB from their endogenous promoters grow normally in the absence of rapamycin, and rapamycin induces flocculation (data not shown). More importantly, chromatin immunoprecipitation (ChIP) experiments indicate that Tup1 association with target promoters is significantly reduced within 20 min of rapamycin treatment and are at background levels after 40 min (Fig. 1A).
Figure 1.
The anchor-away of Tup1 or Cyc8 triggers a conditional loss of Tup1–Cyc8 repression. (A) Tup1-FRB association with the indicated promoters in the control and Tup1-anchor-away strain treated with rapamycin for the indicated times. An ORF-free region of chromosome V was arbitrarily defined as background. (B) Pol II association with the indicated coding regions in the control, Tup1-anchor-away, and Cyc8-anchor-away strains that were or were not treated with rapamycin for 40 min. Association with the ACT1 coding region under the respective conditions served as a control. (C) H3 occupancy at the indicated promoters (relative to the ACT1 promoter) in the control, Tup1-anchor-away, and Cyc8-anchor-away strains that were or were not treated with rapamycin for 40 min. (D) H3 acetylation, expressed as a ratio to total H3 (AcH3/H3) at the indicated promoters in the control, Tup1-anchor-away, and Cyc8-anchor-away strains that were or were not treated with rapamycin for 40 min. Averages and standard errors of three individual experiments are shown.
Depletion of Tup1 or Cyc8 results in a dramatic and concomitant derepression of all tested Cyc8–Tup1 target genes as measured by RNA polymerase II (Pol II) occupancy at coding regions (Fig. 1B). In accord with previous results (Komachi et al. 1994; Tzamarias and Struhl 1995), depletion of Cyc8 has minimal, if any, effect on Tup1-dependent STE6 expression, indicating that nuclear export of Cyc8 does not affect Tup1, even though Cyc8–Tup1 can form a stable complex. Last, depletion of Tup1 or Cyc8 results in reduced histone H3 occupancy (Fig. 1C; Supplemental Fig. S1) and increased H3 acetylation (Fig. 1D) at Tup1 target promoters. Thus, the anchor-away approach reproduces all tested properties of tup1Δ and cyc8Δ mutants and hence can be used to study transcriptional repression mechanisms mediated by Cyc8–Tup1.
Tup1 inhibits H3 acetylation by blocking recruitment of Gcn5 histone acetylase, not by recruiting Hda1 histone deacetylase
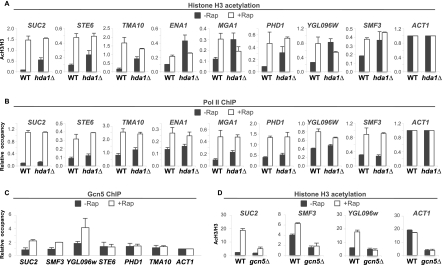
In principle, Tup1 could inhibit histone acetylation by recruiting a histone deacetylase or blocking the recruitment of a histone acetylase. Although one proposed model invokes Tup1-mediated recruitment of the Hda1 histone deacetylase complex to target promoters, we were unable to detect association of HA-tagged Hda1, Hda2, or Hda3 binding to Tup1-regulated promoters by ChIP or whole-genome ChIP sequencing (ChIP-seq) experiments (data not shown). We therefore reinvestigated the contribution of Hda1 to Tup1 repression by deleting the HDA1 gene in the Tup1-anchor-away strain. In agreement with previous results (Green and Johnson 2004; Zhang and Reese 2004a), deletion of HDA1 causes increased H3 acetylation at Tup1 targets, with the magnitude of the effect depending on the promoter (Fig. 2A). In all cases, there is little or no derepression observed in the HDA1 deletion strain (Fig. 2B), indicating that Hda1 and H3 acetylation status per se plays a very limited role in Tup1 repression.
Figure 2.
Elevated histone H3 acetylation in the absence of Tup1 is caused by the loss of Hda1 and the increased recruitment of Gcn5. (A) Histone H3 acetylation at the indicated promoters (relative to the ACT1 promoter) in the Tup1-anchor-away strains with (hda1Δ) or without (WT) HDA1 deletion that were or were not treated with rapamycin for 40 min. (B) Pol II association with the indicated coding regions (relative to the ACT1 coding region) in the Tup1-anchor-away strains with (hda1Δ) or without (WT) HDA1 deletion that were or were not treated with rapamycin for 40 min. (C) Gcn5 association with the indicated promoters in the Tup1-anchor-away strain that was or was not treated with rapamycin for 40 min. Association with the ACT1 promoter under the respective conditions served as a control. (D) Histone H3 acetylation at the indicated promoters in the Tup1-anchor-away strains with (gcn5Δ) or without (WT) GCN5 deletion that were or were not treated with rapamycin for 40 min. Histone H3 acetylation level was expressed as a ratio to the total histone H3 density (AcH3/H3). Averages and standard errors of three individual experiments are shown.
In contrast, at some Tup1-regulated promoters, association of Gcn5, the catalytic subunit of the SAGA histone acetylase coactivator complex, is rapidly increased upon depletion of Tup1 (Fig. 2C). Furthermore, H3 acetylation at the promoters showing elevated Gcn5 binding after anchor-away of Tup1 was inhibited (to various extents) in a gcn5Δ mutant (Fig. 2D). At other promoters, increased association of Gcn5 is not observed, suggesting the presence of a distinct histone acetylase. Thus, Tup1 blocks Gcn5 recruitment to target promoters and is responsible for some or most of the increased H3 acetylation that occurs upon depletion of Tup1.
Tup1 blocks recruitment of nucleosome remodeling complexes and nucleosome eviction at target promoters
Relief of Tup1 repression is associated with a loss of nucleosomes at promoters (Figs. 1C; Supplemental Fig. S1; Hirschhorn et al. 1992; Matallana et al. 1992; Cooper et al. 1994), and it has been proposed that Cyc8–Tup1 physically maintains a repressive chromatin structure. However, it is also possible that the effect of Cyc8–Tup1 on nucleosome occupancy is due to blocking the recruitment and/or function of nucleosome remodeling complexes or is an indirect effect due to blocking transcription.
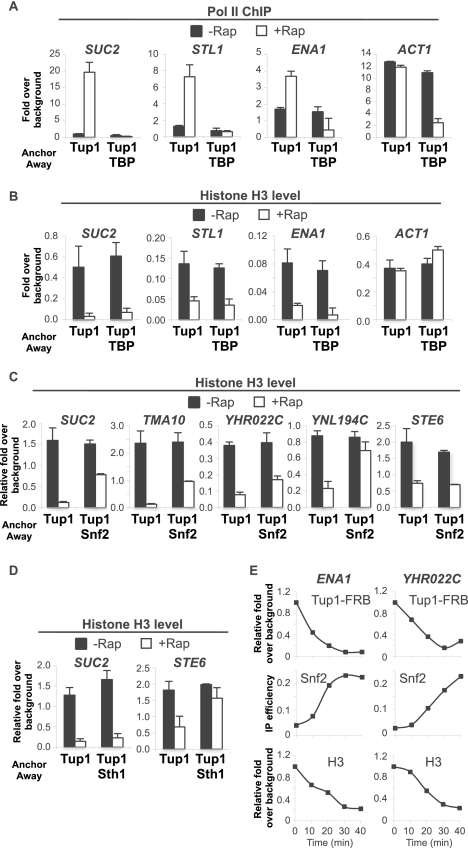
To address this issue, we generated double anchor-away strains to simultaneously deplete Tup1 with TBP, Snf2 (catalytic subunit of the Swi/Snf nucleosome remodeling complex), or Sth1 (catalytic subunit of the RSC nucleosome remodeling complex). Simultaneous depletion of Tup1 and TBP completely blocks derepression for all Tup1 targets analyzed (Fig. 3A), but histone eviction is still observed (Fig. 3B), indicating that the histone eviction is independent of transcription. In contrast, histone eviction at Tup1-repressed promoters is significantly reduced when Tup1 was simultaneously depleted with Snf2 or Sth1 (Fig. 3C,D). Notably, the two remodelers affected the histone eviction to different extents at different promoters, presumably reflecting promoter-specific dependence on and/or functional redundancy between histone remodelers.
Figure 3.
Tup1 maintains high nucleosome density at the UPR of its targets by preventing recruitment of histone remodelers. (A) Pol II association with the indicated coding regions in the Tup1-anchor-away or Tup1/TBP-anchor-away strains that were or were not treated with rapamycin for 1 h. (B) H3 occupancy with the indicated promoters in the Tup1-anchor-away or Tup1/TBP-anchor-away strains that were or were not treated with rapamycin for 1 h. (C) H3 occupancy with the indicated promoters in the Tup1-anchor-away and Tup1/Snf2-anchor-away strains that were or were not treated with rapamycin for 1 h. Averages and standard errors of three individual experiments are shown. (D) H3 occupancy with the indicated promoters in the Tup1-anchor-away and Tup1/Sth1-anchor-away strains that were or were not treated with rapamycin for 1 h. (E) Time-course ChIP experiments measuring occupancy of Tup1-FRB, Snf2-HA, and H3 with the indicated promoters in the Tup1-anchor-away strain upon addition of rapamycin. A representative result is shown. (A–E) An ORF-free region of chromosome V was arbitrarily defined as background. Averages and standard errors of three individual experiments are shown.
To address whether Tup1 prevents the activity or the recruitment of histone remodelers, we investigated the association of Snf2. In the presence of Tup1, there was a low or background level of Snf2-HA at all Tup1-repressed promoters (Supplemental Fig. S2). After Tup1 was anchored away, elevated Snf2 binding was observed at all target promoters, with the exception of STE6, whose histone eviction is not dependent on Snf2 (see Fig. 3C). Furthermore, time-course experiments show that Snf2 recruitment, H3 depletion, and Tup1 depletion occur concurrently (Fig. 3E). Taken together, these results show that Cyc8–Tup1 affects nucleosome occupancy indirectly by blocking recruitment of nucleosome remodeling complexes.
Tup1 blocks Mediator binding to promoters
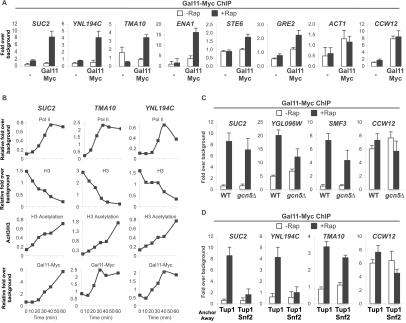
We also examined the association of the Gal11 subunit of Mediator, another coactivator recruited by activator proteins. As observed with Swi/Snf and SAGA, association of Gal11 at all target promoters tested is very low under conditions of Tup1 repression and increases dramatically upon removal of Tup1 (Fig. 4A), indicating that Cyc8–Tup1 also blocks the recruitment of Mediator. Interestingly, kinetic analysis indicates that initial Gal11 recruitment is slightly slower than those of histone H3 acetylation and eviction (cf. the period between 0 and 20 min) but is concurrent with transcriptional derepression (Fig. 4B). In addition, Gal11 recruitment upon Tup1 depletion is slightly compromised in the gcn5Δ mutant and is markedly reduced at some promoters in the absence of Snf2 (Fig. 4C,D). Therefore, robust Gal11 recruitment to target promoters depends on the loss of Tup1 as well as chromatin modification and/or remodeling, which presumably depend on the recruitment of other coactivators (e.g., Swi/Snf and SAGA).
Figure 4.
Gal11 binding is prevented by Tup1 and nucleosomes at UPRs. (A) Gal11-Myc association with the indicated promoters in the Tup1-anchor-away strains containing untagged (−) or Myc-epitope-tagged (Gal11Myc) Gal11 that were or were not treated with rapamycin. (B) Time-course ChIP experiments measuring association of Pol II at the indicated coding regions, occupancy of H3 and Gal11-Myc, and acetylation of H3 at the indicated promoter regions in the Tup1-anchor-away strain upon rapamycin treatment. The results of a representative experiment are shown. (C) Gal11-Myc association with the indicated promoters in the Tup1-anchor-away strains containing wild type or gcn5Δ mutation that were or were not treated with rapamycin. (D) Gal11-Myc association with the indicated promoters in the Tup1-anchor-away and Tup1/Snf2-anchor-away strains that were or were not treated with rapamycin. (A–D) An ORF-free region of chromosome V was arbitrarily defined as background. Averages and standard errors of three individual experiments are shown.
Recruitment of SAGA, Swi/Snf, and Mediator is necessary for full derepression of Tup1-repressed targets
Our findings strongly suggest that the primary mechanism of Tup1 repression is to block the recruitment of coactivators. This model predicts that derepression of Cyc8–Tup1 targets would be reduced when coactivator functions are impaired. While deletion of GCN5 has no effect on transcription before Tup1 is removed, the level of derepression for some targets is slightly affected (Fig. 5A), suggesting that the recruitment of Gcn5 is necessary for full derepression. On the other hand, simultaneous depletion of Gal11 and Tup1 drastically reduces the levels of derepression of all Tup1-repressed genes analyzed (Fig. 5B). Similarly, transcription of the constitutively active genes ACT1 and CCW12 were also affected. Consistent with the dependence of Gal11 recruitment on nucleosome eviction, the lack of the nucleosome remodeler Snf2 or Sth1 function also significantly impacts derepression of their respective Cyc8–Tup1 targets (Fig. 5C,D), and the effect is comparable with that observed in the absence of Gal11. Taken together, these results reinforce that transcriptional activation is the main target of Tup1 repression.
Figure 5.
Derepression of Tup1 targets depends on the recruitment of histone remodelers and Mediator. (A) Pol II association with the indicated promoters in the Tup1-anchor-away strains with (WT) or without Gcn5 (gcn5Δ) that were or were not treated with rapamycin for 1 h. (B) Pol II association with the indicated promoters in the Tup1-anchor-away and Tup1/Gal11-anchor-away strains that were or were not treated with rapamycin for 1 h. (C) Pol II association with the indicated promoters in the Tup1-anchor-away and Tup1/Snf2-anchor-away strains that were or were not treated with rapamycin for 1 h. (D) Pol II association with the indicated promoters in the Tup1-anchor-away and Tup1/Sth1-anchor-away strains that were or were not treated with rapamycin for 1 h. (E) Kinetics of H3 acetylation and occupancy of Gal11-Myc, Snf2-HA, and H3 at the indicated promoters and coding regions in the Tup1-anchor-away strain carrying a galactose-inducible GAL(p)TUP1 plasmid before and after anchoring away of Tup1FRB by rapamycin treatment for 1 h and at the indicated times after galactose induction. (F) Pol II ChIP experiments measuring transcription of the indicated coding regions in the Tup1-anchor-away strains of wild-type, hda1Δ, or rpd3Δ background before (−Rap) and after (+Rap) depletion of Tup1-FRB by rapamycin treatment, and after reintroduction of Tup1 by galactose induction of GAL(p)TUP1 for 1 h (+Rap, +Gal). (G) Pol II association of the indicated coding regions were monitored as described in A, except the analysis was done in the Tup1-anchor-away strains of wild-type or hda1Δ background. (A–G) An ORF-free region of chromosome V was arbitrarily defined as background. Averages and standard errors of three individual experiments are shown.
Tup1 can block Mediator binding and repress transcription in the absence of nucleosomes
As the above finding implies that high nucleosome density is critical for Cyc8–Tup1 repression, we monitored nucleosome density and other promoter activities during re-establishment of the repression. Upon galactose induction of untagged Tup1 that is not subject to rapamycin-induced depletion, re-establishment of Cyc8–Tup1 repression is clearly evident from the loss of the rapamycin-induced derepression (Pol II occupancy) (Fig. 5E). Interestingly, kinetic analysis reveals that H3 deacetylation precedes the establishment of repression (Fig. 5E). However, the repression was not affected in the absence of Rpd3 function and was only slightly reduced in the hda1Δ mutant (Fig. 5F). The kinetics of the repression might be slower for some genes (SUC2 and ANB1) (Figure 5G) in the absence of Hda1 as compared with wild type, although the effect is subtle.
Interestingly, repression is observed prior to the deposition of nucleosomes back to the respective target promoters (Fig. 5E), and similar results have been observed for Tup1-mediated repression in another context (Bryant et al. 2008). In contrast, the kinetics of the loss of Mediator is highly correlated with the onset of the repression, while the disappearance of Snf2 from the UPRs (upstream promoter regions) is slightly slower. Taken together, these results reveal that repressive chromatin structure contributes to, but is not necessary for, Tup1 repression and that Tup1 repression is linked to dissociation of coactivators from target promoter regions.
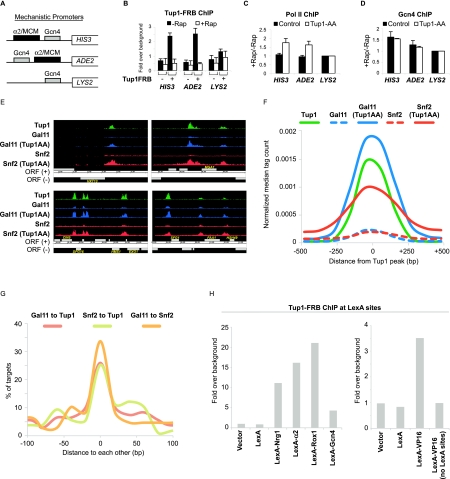
Tup1 does not affect activator binding to promoter
In principle, Cyc8–Tup1 could block activator-mediated recruitment of coactivator complexes or block binding of the activator to target promoters, although previous studies suggest that activator binding is not inhibited (Redd et al. 1996; Papamichos-Chronakis et al. 2002, 2004; Proft and Struhl 2002; Zhang and Reese 2005). As an independent approach to address the effect of Cyc8–Tup1 on activator binding, we generated a “mechanistic” anchor-away strain that contains three artificial Gcn4-activated promoters (Fig. 6A). The control promoter driving LYS2 contains a Gcn4-binding site upstream of the TATA and initiator elements of the HIS3 gene. The other two promoters controlling expression of HIS3 and ADE2 are identical to the control promoter except that they contain a Cyc8–Tup1 recruitment site (derived from the α2/Mcm1-binding site in the STE6 promoter) upstream of or downstream from, respectively, the Gcn4-binding site.
Figure 6.
Tup1 represses transcription by masking the activating potential of a diverse class of repressor–activator proteins. (A) Diagram illustrating the arrangement of binding sites (α2/MCM, which is bound by a Tup1-recruiting DNA-binding repressor complex, and Gcn4) at three artificial promoters fused to the HIS3, ADE2, and LYS2 genes. (B) Tup1 association with the indicated artificial promoters in the control and the Tup1-anchor-away strains that contain the mechanistic promoters shown in A before and after rapamycin treatment for 1 h. (C) Changes in Pol II association with the indicated artificial promoters in the control and the Tup1-anchor-away after rapamycin treatment for 1 h. (D) Changes in Gcn4 binding in the control and the Tup1-anchor-away that contain the mechanistic promoters.. (A–D) An ORF-free region of chromosome V was arbitrarily defined as background. Averages and standard errors of three individual experiments are shown. (E) Representative genome browser (Affymetrix Integrated Genome Browser) screenshots showing binding regions of Tup1, Snf2-HA, and Gal11-Myc in the presence or absence of Tup1 (after Tup1 depletion) around the indicated ORFs. (F) Binding profile of Tup1 (black line), Gal11-Myc (blue line), and Snf2-HA (red line) at Tup1-repressed promoters before (broken line) and after (solid line) Tup1 depletion (Tup1AA). (G) Distances between peak summits of Gal11 and Snf2 relative to Tup1 peak summits, and between Gal11 and Snf2 peak summits at all Tup1-repressed promoters. (H) Tup1 association with four integrated tandem LexA sites in strains expressing the indicated LexA derivatives.
As expected, Tup1-FRB binding to its endogenous targets and the artificial promoters driving HIS3 and ADE2 is lost after rapamycin treatment (Fig. 6B), and expression of these genes, but not the control LYS2 gene, is derepressed (Fig. 6C). Interestingly, Tup1 repression of ADE2 and HIS3 occurs to a comparable extent even though the arrangements of Tup1 recruitment and the Gcn4-binding site relative to the core promoter are different, suggesting that Tup1 repression is unlikely to be caused by steric hindrance. Importantly, binding of Gcn4 to these artificial Tup1-repressed promoters is unchanged after removal of Tup1 (Fig. 6D), indicating that Tup1 does not interfere with activator binding.
Coactivators are recruited to sites previously occupied by Tup1
Recruitment of Swi/Snf, SAGA, and Mediator to Tup1-regulated genes upon loss of Tup1 presumably requires activators bound to target sites. As a consequence, activator-binding sites necessary for derepression can be mapped by determining the location of the recruited coactivators on a genome-wide level. Specifically, we used ChIP-seq to map Snf2 and Gal11 under conditions in which Tup1 was or was not depleted and compared the coactivator recruitment sites with the location of Tup1.
In accord with previous results (Venters et al. 2011), Tup1-binding sites are found mostly at distal promoter regions and are associated with motifs for the known DNA-binding repressors (data not shown). To determine whether Tup1 binds to specific sites or spreads over broad regions as previously suggested (Ducker and Simpson 2000), we compared the binding profiles of Tup1 and the activator Gcn4 that binds to a well-defined TGACTCA motif (Hill et al. 1986; Oliphant et al. 1989). The majority of Gcn4 motifs are located within 10–20 base pairs (bp) from the Gcn4 peak summits (Supplemental Fig. S3), indicating that our ChIP-seq data can map binding sites in vivo to high resolution. Importantly, the ChIP signal profile of Tup1 is comparable with that of Gcn4 ChIP binding profile (Supplemental Fig. S3), indicating that Tup1 is recruited to highly localized sites at target promoters.
Strikingly, the coactivators are recruited to all Tup1-repressed promoters when Tup1 is removed, indicating that the block of transcriptional activation is a general mode of Cyc8–Tup1 repression. Importantly, Snf2 and Gal11 are recruited to regions that coincide with the Tup1 sites (Fig. 6E; Supplemental Fig. S4), and these associations are also seen in the presence of Tup1, albeit at much lower levels (Figs. 6E,F; Supplemental Fig. S4). Most Tup1 peaks are within 20 bp of the Gal11 and Snf2 peaks, and the distances between the Tup1 and the coactivators peaks are as close as those between the two recruited coactivators themselves (Fig. 6G). Furthermore, Tup1 and the recruited coactivators share extremely similar binding profiles over all Tup1-repressed promoters (Fig. 6F). A simulation in which coactivator ChIP signals are artificially moved 25 bp or 50 bp away from their true location shows that the simulated coactivator peak is clearly distinguishable from the Tup1 peak (Supplemental Fig. S5), suggesting that the Tup1 and the coactivators target sites must be within 25 bp of each other.
The observation that Swi/Snf and Mediator are recruited to the same site previously occupied by Tup1 strongly suggests that the DNA-binding repressor and the DNA-binding activator for a given gene are bound to the same site and hence that the same protein functions as both a repressor and activator. Therefore, we suggest that most, if not all, of the DNA-binding repressors have the ability to activate transcription in the absence of Tup1. More importantly, this implies that, under nonstressed conditions, Tup1 masks the activating potential of these repressor–activator proteins.
Tup1 interacts with activation domain in vivo
As transcriptional activation domains directly and independently interact with Swi/Snf, SAGA, and Mediator, the ability of Cyc8–Tup1 to block the recruitment of these three different coactivators at many different promoters strongly suggests that Tup1 interacts with and masks the activation domain. Indeed, LexA-Gcn4 and LexA-VP16 (a mammalian activation domain), but not LexA alone, recruit Tup1 to a genomic region with four LexA sites, and such Tup1 recruitment is not observed in a control strain lacking the LexA sites (Fig. 6H). Interestingly, the level of Tup1 recruitment mediated by Gcn4 and the VP16 activation domain is noticeably lower than that mediated by LexA fusions to the Tup1-recruiting factors Nrg1, α2, and Rox1. These observations indicate that Tup1 can interact with activation domains in vivo and suggest that interactions with Tup1-recruiting repressor–activator proteins are stronger than those of generic activation domains.
Discussion
Tup1 repression occurs primarily by masking activation domains and blocking activator-mediated recruitment of coactivators
Considerable work from multiple laboratories has led to various models for the mechanism of repression by Tup1 (Smith and Johnson 2000; Malave and Dent 2006). These include (1) modulating Mediator function, (2) recruitment of histone deacetylases, (3) and various effects on chromatin structure. A major problem with these models is that mutations that inactivate putative Tup1 targets, individually or in combination, have very small effects on transcription of natural genes, especially in comparison with the dramatic effects observed upon loss of Tup1 (Lee et al. 2000; Green and Johnson 2004; Zhang and Reese 2004a).
Here, we used a kinetic approach to follow the process of Tup1-dependent derepression and rerepression in vivo. The major advantage of this approach is that molecular events that occur rapidly upon loss of Tup1 are very likely to reflect direct effects of Cyc8–Tup1, a key issue that cannot be addressed by standard deletion alleles that confer very slow growth and other serious physiological defects. Using this approach, we demonstrate that Cyc8–Tup1 inhibits transcription primarily by blocking the transcriptional activation domain of the recruiting protein, not by functioning as a bona fide corepressor (although a corepressor function appears to play a minor role) (see below).
Upon depletion of Tup1, the coactivators Swi/Snf, SAGA, and (slightly later) Mediator are rapidly recruited to Tup1-repressed promoters. Concomitant with coactivator recruitment, histone acetylation increases and nucleosomes are evicted. While histone acetylation and histone eviction are largely independent of each other, full Mediator recruitment depends on these chromatin changes. Conversely, upon reassociation of Tup1 (via galactose induction following Tup1 depletion), SAGA (as inferred from loss of H3 acetylation), Mediator, and (slightly later) Swi/Snf rapidly dissociate from target promoters. Importantly, these events occur long before redeposition of nucleosomes to promoters, indicating that Tup1 is able to block transcription before any repressive chromatin structure is established; such repression in the absence of nucleosomes has been previously demonstrated (Bryant et al. 2008). Previous studies (Redd et al. 1996; Papamichos-Chronakis et al. 2002, 2004; Proft and Struhl 2002; Zhang and Reese 2005) and our analysis of Gcn4 binding in the “mechanistic strain” indicate that Tup1 does not inhibit activator binding. As recruitment of all three coactivators is mediated by direct and independent interactions with activation domains of DNA-bound activator proteins, the kinetic profile of molecular events at target promoters indicates that Tup1 directly blocks activator-mediated recruitment of these coactivators.
The simplest model by which Cyc8–Tup1 directly inhibits activator-mediated recruitment of coactivators involves an interaction with the activation domain that masks its ability to interact with coactivators. Strong support for such a masking model comes from the observation that Tup1 can interact with Gcn4 and the VP16 activation domain in vivo. Presumably, this interaction is mediated by the Tup1 repression domain (Tzamarias and Struhl 1994), although other regions of Tup1 might contribute, and it is formally possible that a heretofore unknown protein may also be involved. We disfavor the formal possibility that Cyc8–Tup1, by virtue of being a large complex, occludes interactions between coactivators and DNA, thereby reducing the stable association of these coactivators at Tup1 target genes. Aside from being ad hoc, this formal possibility is inconsistent with previous observations that Tup1 can co-occupy target sites with Swi/Snf and SAGA (Proft and Struhl 2002) and that it can increase recruitment of these coactivators to target sites upon stress (Papamichos-Chronakis et al. 2002; Proft and Struhl 2002; Fragiadakis et al. 2004; Kim et al. 2005; Desimone and Laney 2010). In addition, repression models that invoke specific chromatin changes directly mediated by Cyc8–Tup1 do not explain the near-immediate kinetic link between the reciprocal occupancies by Cyc8–Tup1 and three distinct coactivator complexes. Thus, we suggest that transcriptional inhibition by the Cyc8–Tup1 complex occurs primarily by masking the activation domains of diverse proteins via a direct interaction.
The activation domain-masking model is consistent with, and helps explain, previous observations
Repression by Cyc8–Tup1 is associated with many changes in chromatin structure, leading to frequent suggestions that this corepressor functions through a direct effect on chromatin structure. Despite these suggestions, there are no experiments demonstrating a direct effect of Cyc8–Tup1 on chromatin structure. On the contrary, many (and perhaps all) effects of Cyc8–Tup1 on chromatin structure are more easily explained as an indirect consequence of its ability to mask activation domains and hence inhibit activator-mediated recruitment of coactivators that remodel nucleosomes (Swi/Snf and RSC) and acetylate histones (SAGA). For example, although many Tup1-regulated genes show Hda1-dependent deacetylation on H3 and H2B acetylation (Robyr et al. 2002), this effect can be explained by reduced recruitment of SAGA, which has the same acetylation specificity for H3 and H2B. Acetylation levels in vivo are strongly affected by a genome-wide dynamic equilibrium between nontargeted histone acetylases and deacetylases (Katan-Khaykovich and Struhl 2002). As low levels of SAGA are associated with Tup1-regulated promoters even under repressive conditions, loss of Hda1 results in increased acetylation at the site of Tup1 and SAGA recruitment. Last, by virtue of inhibiting coactivator recruitment, Tup1 also blocks transcription and hence the many transcription-coupled effects on chromatin structure, such as histone methylation and nucleosome occupancy and/or positioning.
The activator-blocking model also explains why mutations in putative Tup1 targets, by themselves or in combination, show very limited derepression that is far below that achieved in tup1Δ or cyc8Δ mutant strains (Lee et al. 2000; Zhang and Reese 2004a). The previous interpretation of these observations was that Tup1 repressed transcription by multiple mechanisms and hence had multiple targets of action. Even if true, this interpretation would require an additional unknown mechanism of repression to account for the full effect of Cyc8–Tup1. In contrast, our results indicate that Tup1 repression occurs primarily by a single mechanism, physically masking the transcriptional activation domain, thereby leading to inhibition of the multiple functions mediated by such activation domains. As such, mutations in one or more components that affect minor repression mechanisms would have minimal effects on gene expression. In accord with this model, Tup1 does not repress transcription when the normal activation process is bypassed by artificially recruiting Mediator to a promoter (Papamichos-Chronakis et al. 2000).
Cyc8–Tup1 is a general repressor of stress-inducible, repressor–activator proteins
Given that Tup1 blocks activator function, a key question is which activators and activation domains are targets of Cyc8–Tup1. There are several examples in which the repressor protein that recruits Tup1 under nonstressed conditions is converted to an activator upon stress (Papamichos-Chronakis et al. 2002; Proft and Struhl 2002; Mennella et al. 2003; Fragiadakis et al. 2004; Zhang and Reese 2005; Hickman and Winston 2007). Our results strongly argue that, in most and perhaps all cases, the activator inhibited by Tup1 is also the repressor that recruits Cyc8–Tup1 under nonstressed conditions. Specifically, upon loss of Tup1, Swi/Snf and Mediator are recruited to target sites that are at locations indistinguishable (<25 bp) from those where Tup1 was bound prior to depletion. Thus, repression by Cyc8–Tup1 appears to involve masking the activation domains of a diverse set of activator proteins that respond to specific environmental signals or to cell type.
There are several possible models by which Cyc8–Tup1 masks activation domains of a diverse set of DNA-binding activator proteins. Each Tup1-regulated, DNA-binding protein might possess a specific Cyc8–Tup1 interaction surface that overlaps a generic activation domain (or causes a conformational change that masks the activation domain), as is the case for Crt1 (Zhang and Reese 2005). Alternatively, Cyc8–Tup1 might have a general propensity to mask activation domains, but the interaction with a given activation domain could be augmented by the specific sequence within or near the activation surface. Our finding that Tup1 association with Gcn4 and the VP16 activation domain appears weaker than the association with classical Tup1-recruiting proteins is consistent with this model. It is unlikely that Cyc8–Tup1 masks all activation domains in a common manner, because different surfaces of Cyc8–Tup1 are required for interaction with specific DNA-binding proteins (Tzamarias and Struhl 1995).
With the exception of cell type-specific genes that are regulated by cell type-specific expression of α2, genes regulated by Tup1 are strongly induced upon environmental stress. As a specific stress condition activates only the subset of Cyc8–Tup1 target genes that are bound by a specific DNA-binding protein, pathway-specific regulation ultimately is due to changes in the function of the DNA-binding protein that recruits Cyc8–Tup1. In principle, modification of the DNA-binding protein (e.g., by phosphorylation) could abolish the interaction with Cyc8–Tup1 (similar to what occurs upon anchor-away) and/or alter the conformation of the activation domain such that it can recruit coactivators even when associated with Cyc8–Tup1. In many cases, Cyc8–Tup1 remains associated at the relevant target promoters under stress conditions and stimulates Swi/Snf and SAGA recruitment (Papamichos-Chronakis et al. 2002; Proft and Struhl 2002; Mennella et al. 2003). Thus, our results suggest that Cyc8–Tup1 inhibits a class of stress-activated, DNA-binding transcriptional activator proteins and that, in most or all cases, stress alters the conformation of the activation domain such that its function is no longer inhibited by Cyc8–Tup1.
It remains unclear why Cyc8–Tup1 blocks the function of diverse repressor–activator proteins, whereas it does not appear to repress the classical activators such as Gcn4 and Gal4. Consistent with our observations on Gcn4 and the VP16 activation domain, it is possible that Cyc8–Tup1 is only modestly associated with the classical activators under physiological conditions. An alternative, but not mutually exclusive, possibility is that the activating potential of these classical activators may be much stronger than those of Tup1-recruiting repressor–activator proteins. The affinity of these activators for coactivators may be much higher than the affinity for Tup1, such that Tup1 regulation is relatively insignificant.
The corepressor function of Cyc8–Tup1 has a modest effect on gene regulation
By definition, a corepressor inhibits transcription that is mediated by factors distinct from the recruiting protein; i.e., inhibiting activation is mechanistically distinct from repression. As such, the major inhibitory function of Cyc8–Tup1, masking the activation domains of the recruiting proteins, does not represent a corepressor function. Cyc8–Tup1 was defined as a corepressor based on experiments involving artificial, and otherwise active, promoters containing an ectopic site that recruits Cyc8–Tup1. In many cases, Cyc8–Tup1 was artificially recruited to these artificial promoters by fusing either Cyc8 or Tup1 to a heterologous DNA-binding domain. However, it appears that there are few, if any, natural promoters in which the Tup1-recruiting “repressor” is a different protein from the Tup1-regulated activator. Furthermore, the repression at artificial promoters is much weaker as compared with natural promoters. Thus, it is unclear whether the corepressor function being measured in these assays is physiological.
The DNA-binding proteins that recruit Cyc8–Tup1 were initially described as repressors because strains lacking these proteins typically show increased transcription of target genes. The phenotypic difference between removing the recruiting protein and blocking its activation function suggests that Cyc8–Tup1 does not function solely by blocking the activation domain of the recruiting protein. It is worth noting that there is no direct evidence that Cyc8–Tup1 is required to mediate repression, as opposed to activator inhibition, at natural genes. Such evidence would require mutations that selectively abrogate the corepressor function, and these are likely to be difficult to obtain. In addition, for at least one Cyc8–Tup1-recruiting repressor, Sko1, repression can occur by blocking the Aca1 and Aca2 coactivators from binding the same site (Garcia-Gimeno and Struhl 2000; Rep et al. 2001). Nevertheless, based on the corepressor function identified and characterized on artificial promoters, we suspect that Cyc8–Tup1 is required for at least some, and perhaps many or all, DNA-binding repressors to repress transcription.
There are several models, not mutually exclusive, for how Cyc8–Tup1 can function as a corepressor at natural target genes. First, Cyc8–Tup1 could stabilize the binding of the repressor to its target site, thereby blocking the binding of other activator proteins that recognize the same or overlapping site. Second, by analogy to the major mechanism of activator inhibition, Cyc8–Tup1 could mask activation domains of other proteins bound to nearby sites, and this mechanism may explain the corepressor function at artificial promoters. However, the coincidence of Tup1 and coactivator association throughout the genome suggests that transcriptional activation mediated by other proteins is weak. Third, as proposed previously, Tup1-mediated corepression of natural genes could be mediated by a direct effect on chromatin structure (e.g., binding histone tails and/or inhibiting nucleosome remodeling per se) or on the general transcriptional function of Mediator. The available evidence argues against recruitment of histone deacetylase complexes. Whatever specific mechanisms are involved, the corepressor function of Cyc8–Tup1 is considerably less important than the activator inhibition function in regulating target genes.
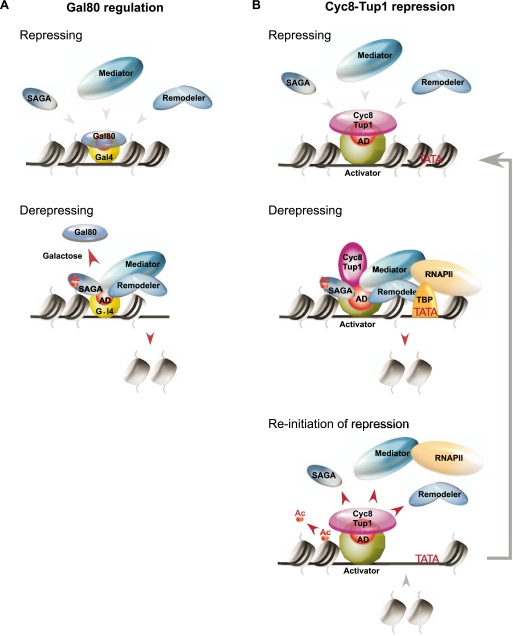
The biological role of Cyc8–Tup1, and a comparison with Gal80
Mechanistically, the primarily repression mechanism of Cyc8–Tup1 is analogous to that of Gal80, which directly masks the Gal4 activation domain (Fig. 7A; Kumar et al. 2008; Thoden et al. 2008). However, Cyc8–Tup1 and Gal80 differ in several respects. Cyc8–Tup1 associates with a variety of DNA-binding proteins that repress hundreds of genes in diverse stress pathways and cell type determination, whereas Gal80 specifically interacts with Gal4. In addition, the DNA-binding proteins that recruit Cyc8–Tup1 have been defined as repressors (loss of function results in increased expression under nonstressed conditions), whereas Gal4 does not appear to have any repressor function (loss of Gal4 does not affect expression in glucose medium). Upon the appropriate stress (galactose), Gal80 dissociates from Gal4, thereby exposing the Gal4 activation domain for function (Jiang et al. 2009), whereas Cyc8–Tup1 often remains associated and actually contributes to the activation process.
Figure 7.
Biological role of Cyc8–Tup1 and a comparison with Gal80. (A) Regulation of Gal4 activation by Gal80. Under repressing conditions (e.g., growth in the absence of galactose), the activation domain of Gal4 is blocked by a physical interaction with Gal80. Upon growth in glucose-limiting medium containing galactose, Gal80 dissociates from Gal4, exposing its activation domain for recruiting coactivators. (B) Cyc8–Tup1 is recruited to target promoters by a class of “repressor–activator” proteins. Under normal (repressing) conditions, Cyc8–Tup1 blocks the activating potential of these proteins by interacting with and masking their activation domains (ADs). Upon derepressing conditions, the responsive repressor–activator is modified, leading to a conformational change in the interaction with Cyc8–Tup1 such that the activation domain can now recruit the Swi/Snf, SAGA, and Mediator coactivators. Cyc8–Tup1 remains at the promoter and contributes to the activation process. During reinitiation of repression, Cyc8–Tup1 re-establishes a repressive state through removing bound coactivators at promoters. The acetylation on histone H3 is removed by global nontargeted activity of deacetylases, and nucleosomes are subsequently redeposited to promoters.
Our results suggest that Cyc8–Tup1 is a general coregulator of a class of stress-regulated, DNA-binding, repressor–activator proteins that are constitutively bound to their target sites. Under nonstressed conditions, Cyc8–Tup1 associates with these bound proteins and blocks their ability to activate transcription. In addition, we suggest that the corepressor function of Cyc8–Tup1 further reduces the level of transcription below that of a nonactivated gene. Upon stress, the appropriate DNA-binding proteins are modified (by phosphorylation in the known cases) (Proft and Struhl 2002; Papamichos-Chronakis et al. 2004), leading to a conformational change in the interaction with Cyc8–Tup1 such that the activation domain can now recruit the Swi/Snf, SAGA, and Mediator coactivators (Fig. 7B). For natural promoters regulated by multiple Cyc8–Tup1-recruiting repressors of different physiological pathways, our model suggests that repression mediated by individual complexes can be relieved independently, and under relevant conditions, they can even act synergistically to achieve different degrees of induction. When stress is over, or upon initiation of repression, Cyc8–Tup1 re-establishes a repressive state by blocking activation (i.e., removes coactivators from promoters), and consequently, acetylation on histone H3 is removed by global nontargeted deacetylases, and, at a later stage, nucleosomes are redeposited to promoters. Thus, Cyc8–Tup1 is a general and versatile stress regulatory complex that permits rapid transitions between repressed and activated states.
Materials and methods
Yeast strains and plasmids
Yeast strains are listed in Supplemental Table S1. PCR products used in the generation of different tagged strains were amplified from plasmids (pFA6-FRB-HIS, pFA6-FRB-KAN, pFA6-KAN, pFA6-HPH, pFA6-3xHA-HPH, and pFA6-MYC) using primers listed in Supplemental Table S2 (Longtine et al. 1998). The anchor-away strains were generated as described previously (Fan et al. 2011).
Anchor-away experiments
Strains were first grown in synthetic complete (SC) medium overnight at 30°C. Subsequently, cells were diluted with fresh SC medium and grew until OD600∼0.4. A fraction of the culture was cross-linked. To induce anchor-away, rapamycin was then added to the remaining culture (final concentration of 1 μg/mL), and cells were cross-linked at the indicated time. In all experiments, the depletion of Tup1-FRB was confirmed at the promoter of ENA1 that has high level of Tup1 binding, and thus a sensitive test for Tup1 depletion. For the experiments involving induction of Tup1 from the GAL1(p)TUP1 plasmid, strains were first grown in SC medium supplemented with raffinose as the sole carbon source until OD600∼0.4. A fraction of the culture was cross-linked. Rapamycin was then added to the remaining culture, and cells were grown for an additional hour, after which, another fraction of the culture was cross-linked. For the induction of Tup1, galactose was added to a final concentration of 2%, and cells were cross-linked at the indicated amount of time.
ChIP
Chromatin was prepared as described previously (Fan et al. 2008). Two microliters of 8WG16 (Covance), 2.4 μg of anti-H3 (Abcam ab1791), 3 μL of anti-acetyl-H3 (Upstate Biotechnologies), 2 μL of anti-FRB (Alexis), 2 μg of anti-Gcn5 (Santa Cruz Biotechnology), 2 μg of anti-HA (Santa Cruz Biotechnology), 2 μg of anti-MYC, or 2 μg of anti-Gcn4 was used in the respective ChIPs. Quantitative PCR in real time was performed using gene-specific primers listed in Supplemental Table S2.
Sample preparation for multiplex ChIP-seq and data analyses
Sequencing libraries were constructed according to Illumina's protocol, starting with ∼1 ng of immune-precipitated DNA fragments. For multiplex sequencing, each library was generated separately with custom-made adaptors that contain a unique 6-bp bar code at the ligating end, the sequences of which are available on request. Twelve to 24 bar-coded libraries were mixed and sequenced together in the same reaction. Raw sequence reads (NCBI accession no. SRA044839.1) were separated according to their bar codes, and sequences were mapped to the S. cerevisiae genome sequences using Bowtie with the [−m] option specified as 1. Binding peaks and their summits were identified using the model-based analysis for ChIP-seq (MACS) algorithm (Zhang et al. 2008). Binding peaks were then assigned to the closest translational start codon (ATG) of annotated ORF. Tup1-repressed promoters were classified by two criteria: the presence of Tup1 binding and increased Pol II occupancy, as measured by Pol II ChIP-seq, after Tup1 depletion (Supplemental Table S3). For the binding profiles of Tup1-FRB, Gal11-MYC, Snf2-HA, and Gcn4, 1-kb regions spanning the Tup1 peak summits were divided into 10 windows, and the number of reads within each window was summed and normalized to the total number of mapped reads. The peak summits of Tup1, Gal11, and Snf2 used in the pairwise comparisons were defined by the highest point within each ChIP-seq peak.
Acknowledgments
We thank Xiaochun Fan, Zarmik Moqtaderi, Joseph Geisberg, Nathan Lamarre-Vincent, and Yi Jin for technical advice and insightful discussions, and Zarmik Moqtaderi and Nathan Lamarre-Vincent for generously sharing their bioinformatic programs. This work was supported by a postdoctoral fellowship to K.H.W. from the Croucher Foundation (Hong Kong) and a research grant to K.S. from the National Institutes of Health (GM30186).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.179275.111.
References
- Bryant GO, Prabhu V, Floer M, Wang X, Spagna D, Schreiber D, Ptashne M 2008. Activator control of nucleosome occupancy in activation and repression of transcription. PLoS Biol 6: 2928–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D 2006. Tethering RITS to a nascent transcript initiates RNAi-and heterochromatin-dependent gene silencing. Cell 125: 873–886 [DOI] [PubMed] [Google Scholar]
- Cooper JP, Roth SY, Simpson RT 1994. The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev 8: 1400–1410 [DOI] [PubMed] [Google Scholar]
- Davie JK, Edmondson DG, Coco CB, Dent SY 2003. Tup1–Ssn6 interacts with multiple class I histone deacetylases in vivo. J Biol Chem 278: 50158–50162 [DOI] [PubMed] [Google Scholar]
- Desimone AM, Laney JD 2010. Corepressor-directed preacetylation of histone H3 in promoter chromatin primes rapid transcriptional switching of cell-type-specific genes in yeast. Mol Cell Biol 30: 3342–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen RJ, Varshavsky A 2005. Heat-inducible degron and the making of conditional mutants. Methods Enzymol 399: 799–822 [DOI] [PubMed] [Google Scholar]
- Ducker CE, Simpson RT 2000. The organized chromatin domain of the repressed yeast a cell-specific gene STE6 contains two molecules of the corepressor Tup1p per nucleosome. EMBO J 19: 400–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Smith MM, Roth SY 1996. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev 10: 1247–1259 [DOI] [PubMed] [Google Scholar]
- Fan X, Lamarre-Vincent N, Wang Q, Struhl K 2008. Extensive chromatin fragmentation improves enrichment of protein binding sites in chromatin immunoprecipitation experiments. Nucleic Acids Res 36: e125 doi: 10.1093/nar/gkn535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Geisberg JV, Wong KH, Jin Y 2011. Conditional depletion of nuclear proteins by the anchor away system. Curr Protoc Mol Biol 93: 13.10B.1–13.10B.8 doi: 10.1002/0471142727.mb1310bs93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AB, Pennings S 2001. Antagonistic remodelling by Swi–Snf and Tup1–Ssn6 of an extensive chromatin region forms the background for FLO1 gene regulation. EMBO J 20: 5219–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AB, Pennings S 2007. Tup1–Ssn6 and Swi–Snf remodelling activities influence long-range chromatin organization upstream of the yeast SUC2 gene. Nucleic Acids Res 35: 5520–5531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragiadakis GS, Tzamarias D, Alexandraki D 2004. Nhp6 facilitates Aft1 binding and Ssn6 recruitment, both essential for FRE2 transcriptional activation. EMBO J 23: 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gimeno MA, Struhl K 2000. Aca1 and Aca2, ATF/CREB activators in Saccharomyces cerevisiae, are important for carbon-source utilization but not the response to stress. Mol Cell Biol 20: 4340–4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin IM, Kladde MP, Simpson RT 2000. Tup1p represses Mcm1p transcriptional activation and chromatin remodeling of an a-cell-specific gene. EMBO J 19: 5875–5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, Struhl K 2000. TATA-binding protein mutants that increase transcription from enhancerless and repressed promoters in vivo. Mol Cell Biol 20: 1478–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gligoris T, Thireos G, Tzamarias D 2007. The Tup1 corepressor directs Htz1 deposition at a specific promoter nucleosome marking the GAL1 gene for rapid activation. Mol Cell Biol 27: 4198–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SR, Johnson AD 2004. Promoter-dependent roles for the Srb10 cyclin-dependent kinase and the Hda1 deacetylase in Tup1-mediated repression in Saccharomyces cerevisiae. Mol Biol Cell 15: 4191–4202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SR, Johnson AD 2005. Genome-wide analysis of the functions of a conserved surface on the corepressor Tup1. Mol Biol Cell 16: 2605–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Elgin SC 2007. Transcription and RNA interference in the formation of heterochromatin. Nature 447: 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S 2007. Heterochromatin revisited. Natl Rev 8: 35–46 [DOI] [PubMed] [Google Scholar]
- Gromoller A, Lehming N 2000. Srb7p is a physical and physiological target of Tup1p. EMBO J 19: 6845–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SJ, Lee JS, Kang JS, Kim YJ 2001. Med9/Cse2 and Gal11 modules are required for transcriptional repression of distinct group of genes. J Biol Chem 276: 37020–37026 [DOI] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK 2008. The anchor-away technique: Rapid, conditional establishment of yeast mutant phenotypes. Mol Cell 31: 925–932 [DOI] [PubMed] [Google Scholar]
- Hickman MJ, Winston F 2007. Heme levels switch the function of Hap1 of Saccharomyces cerevisiae between transcriptional activator and transcriptional repressor. Mol Cell Biol 27: 7414–7424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DE, Hope IA, Macke JP, Struhl K 1986. Saturation mutagenesis of the yeast HIS3 regulatory site: Requirements for transcriptional induction and for binding by GCN4 activator protein. Science 234: 451–457 [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Brown SA, Clark CD, Winston F 1992. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev 6: 2288–2298 [DOI] [PubMed] [Google Scholar]
- Ho S, Biggar SR, Spencer DM, Schreiber SL, Crabtree GR 1996. Dimeric ligands define a role for transcriptional activation domains in reinitiation. Nature 382: 822–826 [DOI] [PubMed] [Google Scholar]
- Huang L, Zhang W, Roth SY 1997. Amino termini of histones H3 and H4 are required for a1-α2 repression in yeast. Mol Cell Biol 17: 6555–6562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Frey BR, Evans ML, Friel JC, Hopper JE 2009. Gene activation by dissociation of an inhibitor from a transcriptional activation domain. Mol Cell Biol 29: 5604–5610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan-Khaykovich Y, Struhl K 2002. Dynamics of global histone acetylation and deacetylation in vivo: Rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev 16: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher CA, Redd MJ, Schultz J, Carlson M, Johnson AD 1992. Ssn6–Tup1 is a general repressor of transcription in yeast. Cell 68: 709–719 [DOI] [PubMed] [Google Scholar]
- Kim SJ, Swanson MJ, Qiu H, Govind CK, Hinnebusch AG 2005. Activator Gcn4p and Cyc8p/Tup1p are interdependent for promoter occupancy at ARG1 in vivo. Mol Cell Biol 25: 11171–11183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Inai T, Mizunuma M, Okada I, Shitamukai A, Hirata D, Miyakawa T 2008. Identification of Tup1 and Cyc8 mutations defective in the responses to osmotic stress. Biochem Biophys Res Commun 368: 50–55 [DOI] [PubMed] [Google Scholar]
- Komachi K, Redd MJ, Johnson AD 1994. The WD repeats of Tup1 interact with the homeo domain protein α2. Genes Dev 8: 2857–2867 [DOI] [PubMed] [Google Scholar]
- Kuchin S, Carlson M 1998. Functional relationships of Srb10–Srb11 kinase, carboxy-terminal domain kinase CTDK-1, and transcriptional corepressor Ssn6–Tup1. Mol Cell Biol 18: 1163–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PR, Yu Y, Sternglanz R, Johnston SA, Joshua-Tor L 2008. NADP regulates the GAL induction system. Science 319: 1090–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Struhl K 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399: 609–612 [DOI] [PubMed] [Google Scholar]
- Lee M, Chatterjee S, Struhl K 2000. Genetic analysis of the role of Pol II holoenzyme components in repression by the Cyc8–Tup1 corepressor. Genetics 155: 1535–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Reese JC 2001. Ssn6–Tup1 regulates RNR3 by positioning nucleosomes and affecting the chromatin structure of the upstream repression sequence. J Biol Chem 276: 33788–33797 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Malave TM, Dent SY 2006. Transcriptional repression by Tup1–Ssn6. Biochem Cell Biol 84: 437–443 [DOI] [PubMed] [Google Scholar]
- Matallana E, Franco L, Perez-Ortin JE 1992. Chromatin structure of the yeast SUC2 promoter in regulatory mutants. Mol Gen Genet 231: 395–400 [DOI] [PubMed] [Google Scholar]
- Mennella TA, Klinkenberg LG, Zitomer RS 2003. Recruitment of Tup1–Ssn6 by yeast hypoxic genes and chromatin-independent exclusion of TATA binding protein. Eukaryot Cell 2: 1288–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z, Bai Y, Poon D, Weil PA, Struhl K 1996. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature 382: 188–191 [DOI] [PubMed] [Google Scholar]
- Morohashi N, Nakajima K, Kurihara D, Mukai Y, Mitchell AP, Shimizu M 2007. A nucleosome positioned by α2/Mcm1 prevents Hap1 activator binding in vivo. Biochem Biophys Res Commun 364: 583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N, Shi Y 2010. Reversal of histone methylation: Biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem 79: 155–179 [DOI] [PubMed] [Google Scholar]
- Oliphant AR, Brandl CJ, Struhl K 1989. Defining sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: Analysis of the yeast GCN4 protein. Mol Cell Biol 9: 2944–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Conlan RS, Gounalaki N, Copf T, Tzamarias D 2000. Hrs1/Med3: A Cyc8–Tup1 corepressor target in the RNA polymerase II holoenzyme. J Biol Chem 275: 8397–8403 [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Petrakis T, Ktistaki E, Topalidou I, Tzamarias D 2002. Cti6, a PHD domain protein, bridges the Cyc8–Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol Cell 9: 1297–1305 [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Gligoris T, Tzamarias D 2004. The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8–Tup1 co-repressor. EMBO Rep 5: 368–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M, Struhl K 2002. Hog1 kinase converts the Sko1–Cyc8–Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell 9: 1307–1317 [DOI] [PubMed] [Google Scholar]
- Redd MJ, Stark MR, Johnson AD 1996. Accessibility of α2-repressed promoters to the activator Gal4. Mol Cell Biol 16: 2865–2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Proft M, Remize F, Tamas M, Serrano R, Thevelein JM, Hohmann S 2001. The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol Microbiol 40: 1067–1083 [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo JM, Mieczkowski PA, and Buck MJ 2011. Tup1 stabilizes promoter nucleosome positioning and occupancy at transcriptionally plastic genes. Nucl Acids Res 39: 8803–8819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, Grunstein M 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109: 437–446 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V 2007. Polycomb silencing mechanisms and the management of genomic programmes. Natl Rev 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE 2009. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Smith RL, Johnson AD 2000. Turning genes off by Ssn6–Tup1: A conserved system of transcriptional repression in eukaryotes. Trends Biochem Sci 25: 325–330 [DOI] [PubMed] [Google Scholar]
- Struhl K 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev 12: 599–606 [DOI] [PubMed] [Google Scholar]
- Thoden JB, Ryan LA, Reece RJ, Holden HM 2008. The interaction between an acidic transcriptional activator and its inhibitor. The molecular basis of the Gal4 recognition by Gal80. J Biol Chem 283: 30266–30272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzamarias D, Struhl K 1994. Functional dissection of the yeast Cyc8–Tup1 transcriptional corepressor complex. Nature 369: 758–761 [DOI] [PubMed] [Google Scholar]
- Tzamarias D, Struhl K 1995. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8–Tup1 co-repressor complex to differentially regulated promoters. Genes Dev 9: 821–831 [DOI] [PubMed] [Google Scholar]
- Varanasi US, Klis M, Mikesell PB, Trumbly RJ 1996. The Cyc8 (Ssn6)–Tup1 corepressor complex is composed of one Cyc8 and four Tup1 subunits. Mol Cell Biol 16: 6707–6714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C, et al. 2011. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol Cell 41: 480–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson AD, Edmondson DG, Bone JR, Mukai Y, Yu Y, Du W, Stillman DJ, Roth SY 2000. Ssn6–Tup1 interacts with class I histone deacetylases required for repression. Genes Dev 14: 2737–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Suka N, Carlson M, Grunstein M 2001. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol Cell 7: 117–126 [DOI] [PubMed] [Google Scholar]
- Zhang H, Emmons SW 2002. Caenorhabditis elegans unc-37/groucho interacts genetically with components of the transcriptional mediator complex. Genetics 160: 799–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Reese JC 2004a. Redundant mechanisms are used by Ssn6–Tup1 in repressing chromosomal gene transcription in Saccharomyces cerevisiae. J Biol Chem 279: 39240–39250 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Reese JC 2004b. Ssn6-Tup1 requires the ISW2 complex to position nucleosomes in Saccharomyces cerevisiae. EMBO J 23: 2246–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Reese JC 2005. Molecular genetic analysis of the yeast repressor Rfx1/Crt1 reveals a novel two-step regulatory mechanism. Mol Cell Biol 25: 7399–7411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Varanasi U, Trumbly RJ 2002. Functional dissection of the global repressor Tup1 in yeast: Dominant role of the C-terminal repression domain. Genetics 161: 957–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137 doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]