Abstract
The ATP-grasp enzymes consist of a superfamily of 21 proteins that contain an atypical ATP-binding site, called the ATP-grasp fold. The ATP-grasp fold is comprised of two α + β domains that “grasp” a molecule of ATP between them and members of the family typically have an overall structural design containing 3 common conserved focal domains. The founding members of the family consist of biotin carboxylase, D-ala-D-ala ligase and glutathione synthetase, all of which catalyze the ATP-assisted reaction of a carboxylic acid with a nucleophile via the formation of an acylphosphate intermediate. While most members of the superfamily follow this mechanistic pathway, studies have demonstrated that two enzymes catalyze only the phosphoryl transfer step and thus are kinases instead of ligases. Members of the ATP-grasp superfamily are found in several metabolic pathways including de novo purine biosynthesis, gluconeogenesis and fatty acid synthesis. Given the critical nature of these enzymes, researchers have actively sought the development of potent inhibitors of several members of the superfamily as antibacterial and anti-obseity agents. In this review, we will discuss the structure, function, mechanism and inhibition of the ATP-grasp enzymes.
Keywords: ATP-grasp fold, biotin carboxylase, carbamoyl phosphate synthetase, mechanism, inhibitors, structure
1. Introduction
ATP, in addition to its role in the synthesis of nucleic acids, is frequently used by enzymes either as a substrate in phosphoryl transfer (i.e. kinases) reactions or as a driver of energetically unfavorable reactions (i.e ligases). The broad class of ATP-utilizing enzymes is commonly encountered in numerous biological processes within the cell.[1-4] The distinction between the various superfamilies of these enzymes can be made based upon their proposed reaction mechanism, the enzyme's core structure, and the ATP-binding motif.[4] This review article focuses on a specific superfamily of enzymes possessing an atypical nucleotide-binding fold called the palmate, or more commonly, the “ATP-grasp” fold. [5, 6] This distinct superfamily of enzymes is involved in an eclectic array of central biological systems such as de novo purine biosynthesis, fatty acid synthesis, gluconeogenesis, and cell metabolism.[1, 3, 4, 7] Over the past 15 years, the ATP-grasp superfamily of enzymes has expanded well beyond the first three structurally and mechanistically characterized proteins: biotin carboxylase (BC), D-Alanine-D-Alanine ligase (DDLigase), and glutathione synthetase (GSHase).[5, 8-13] The last comprehensive article on the ATP-grasp superfamily was written in 1997 and Galperin and co-workers reported 15 enzymes belonging to this superfamily.[5] Recent research has shown that there are 6 new enzymes to be included in the superfamily (Table 1) and sequence analysis suggests the possibility of additional members. In this review we will discuss the general function of these enzymes, the mechanistic studies available on several members of the superfamily, their common structural features, and several classes of the ATP-grasp enzyme inhibitors.
Table. 1. ATP-grasp superfamily enzymes.
| Enzyme | Biological Function | Taxonomy | Nucleophilic Substrate | Carboxylate Substrate |
|---|---|---|---|---|
| Acetyl-CoA carboxylase (biotin carboxylase domain) (ACC) | Fatty acid biosynthesis | Bacteria, archaea, eukaryota | Biotin-enzyme | HCO3- |
| D-Alanine-D-Alanine ligase (DDLigase) | Peptidoglycan biosynthesis | Bacteria | D-Alanine | D-Alanine |
| Glutathione synthetase (GSHase) | Glutathione biosynthesis | Bacteria, eukaryota | Glycine | γ-Glutamyl-cysteine |
| Pyruvate carboxylase (PC) | Gluconeogenesis | Bacteria, eukaryota | Biotin-enzyme | HCO3- |
| Glycinamide ribonucleotide synthetase (PurD) | Purine biosynthesis | Bacteria, eukaryota | Phosphoribosylamine | Glycine |
| Formylglycinamide ribonucleotide synthetase (PurT) | Purine biosynthesis | Bacteria | Glycinamide ribonucleotide | HCOO− |
| N5-Carboxyaminoimidazole ribonucleotide synthetase (PurK) | Purine biosynthesis | Bacteria, yeast, and fungi | Aminoimidazole ribonucleotide | HCO3- |
| Flavin 5-aminoimidazole-4-carboxamide ribonucleotide synthetase (PurP) | Purine biosynthesis | Archaea | Aminoimidazole-4-carboxamide ribonucleotide | HCOO− |
| Ribosomal protein S6 modification protein (RimK) | Ribosome biogenesis | Bacteria | Glutamate | Ribosomal protein S6 |
| Propionyl-CoA carboxylase (PCC) | Amino acid catabolism | Bacteria, eukaryota | Biotin-enzyme | HCO3- |
| Urea amidolyase | Urea hydrolysis | Bacteria, eukaryota | Biotin-enzyme | HCO3- |
| Carbamoyl phosphate synthetase (CPS) | Arginine biosynthesis, pyrimidine biosynthesis | Bacteria, archaea, eukaryota | NH3 N/A |
HCO3- NH2COO- |
| Pyruvate phosphate dikinase (PPDK) | Gluconeogenesis, photosynthesis | Bacteria, eukaryota | Pyruvate | N/Aa |
| Carnosine synthase | Dipeptide synthesis | Bacteria, archaea, eukaryote, viruses | L-Histidine | β-Alanine |
| Inositol 1,3,4-triphosphate 5/6-kinase (IP56K) | Polyphosphate synthesis | Eukaryota | Inositol 1,3,4-trisphosphate | N/Aa |
| Synapsin Ia (SynC) | Neuronal function | Eukaryota | Unknown | Unknown |
| Tubulin-tyrosine ligase (TTL) | Microtubules assembly | Bacteria, eukaryota | Tyrosine | α-Tubulin |
| Succinyl-CoA synthetase β-chain (SCS) | Citric acid cycle | Bacteria, archaea, eukaryota | Coenzyme A | Succinate |
| Malate-CoA ligase β-chain | Growth on C-1 compounds | Bacteria, Viruses | Coenzyme A | Malate. succinate |
| ATP-citrate lyase | Lipid biosynthesis | Eukaryota | Coenzyme A | Citrate |
| Lysine biosynthesis enzyme (LysX) | Lysine biosynthesis pathway | Bacteria | α-Aminoadipate | Unknown |
kinases are involved in phosphoryl transfer rather than conversion of a carboxylate substrate into a reactive intermediate.
2. Structure
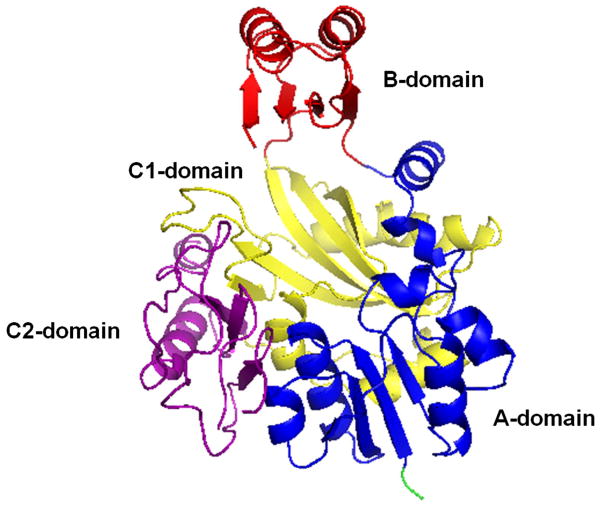
The ATP-grasp superfamily of enzymes is delineated by the unique structure of their ATP-binding site (ATP-grasp fold) which was first elucidated for GSHase, BC, succinyl-CoA synthetase (SCS), and DDLigase.[4, 8-10, 12, 13] The fold is comprised of two α + β domains that “grasp” a molecule of ATP between them. Proteins of the ATP-grasp family have an overall structural design containing 3 common conserved focal domains.[14, 15] These domains are commonly termed the A, B, and C domains although alternate terminology has been amply cited in the literature. The A, B, and C domains are additionally referred to as the N-terminal domain (which corresponds to the A-domain), the central domain (which corresponds to the B-domain), and the C-terminal domain (which corresponds to the C-domain) (Figure 1).[14, 15] For this paper, we will refer to the three domains as A, B, and C.
Figure 1.
General representation of the ATP-grasp fold based on the 3-D structure of the E. coli PurD enzyme (PDB ID: IGSO) [15]
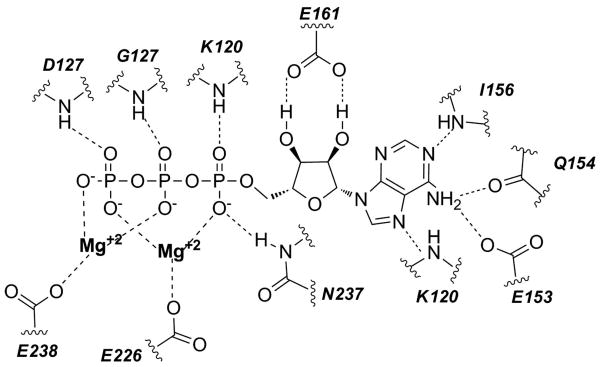
The C-domain can be further divided into C1 and C2 sub-domains, as seen with members of the de novo purine biosynthethetic pathway (glycinamide ribonucleotide synthetase (PurD), N5-carboxyaminoimidazole ribonucleotide synthetase (PurK), formylglycinamide ribonucleotide synthetase (PurT), and flavin 5-aminoimidazole-4-carboxamide ribonucleotide synthetase (PurP). The division of the C-domain varies in complexity among the different enzymes.[16, 17] Domains A and C assemble together and form a large central core that houses the essential substrate binding sites. Between the B domain and the large A/C domain, is the active site which contains a small cleft where all ATP-grasp proteins bind ATP.[18, 19] The polyphosphate moiety of the bound ATP typically interacts with one to three Mg2+ ions with the exception of synapsin Ia (SynC) which binds one Ca2+ ion.[6, 15, 20] The Mg2+ ions are surrounded by a triad of amino acids (as depicted in the Figure 2), typically containing two acidic amino acids and an additional Asn (N) or Asp (D) residue.[19]
Figure 2.
Schematic representation of the ATP binding interactions in E. coli PurK (PDB ID: 3ETH).
The B-domain provides a flexible lid over the active site and it extends away from the main body.[6, 11, 17, 18] The primary function of the B-domain is to provide the ATP binding site via the conserved phosphate-binding P-loop. Generally, the B-domain is flexible in the absence of ATP and it undergoes a transition to an ordered configuration upon nucleotide binding.[11, 17, 18] An exception to this is seen with the lysine biosynthesis enzyme, LysX, where the B-domain remains unaltered upon nucleotide binding.[39] Members of the ATP-grasp superfamily have high levels of structural similarity within their ATP-grasp domain, notably in their A and B domains (Figure 3). These domains contain a variety of structural motifs such as β-sheets, α-helices, 310-helices, loops, and turns. [11, 15, 17, 19]
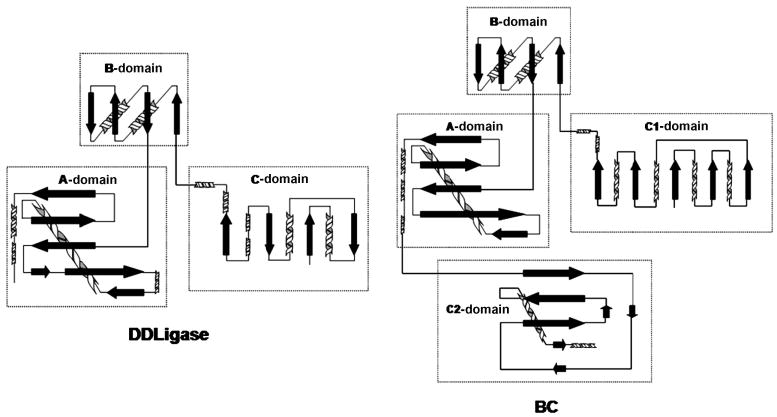
Figure 3.
Topological comparison of DDLigase and BC.[15]
Despite structural similarities, the ATP-grasp superfamily displays an overall low sequence identity (e.g., ∼10-20% in the aligned regions between BC, DDLigase, GSHase and SynC). [2, 6, 21] It has been reported that there are multiple unique residues in the ATP-binding region (90% identical in the aligned regions of BC, GSHase, DDLigase, and PurD) present in almost all of the enzymes in this superfamily (Figure 4).[15] Specifically, thirteen fingerprint residues have been identified as characteristic of the ATP-grasp fold.[19] The high degree of topological similarity between the A and B domains and the characteristic fingerprint residues are shared amongst all ATP grasp family members and serve to define the ATP-grasp motif. [11, 15, 17, 19]
Figure 4.
Alignment and key fingerprint residues in the ATP-binding site. Resides in bold represent highly conserved or similar amino acids involved in the binding of ATP.[5]
4. Function
The ATP-grasp enzymes are situated within many diverse biological systems. The family of biotin carboxylases and the de novo purine biosynthetic pathway embody approximately 40% of all know ATP-grasp enzymes. De novo purine biosynthesis catalyzes the conversion of phosphoribosyl pyrophosphate to inosine monophosphate (IMP), the common intermediate for the synthesis of either GMP or AMP. These nucleotides are the building blocks for DNA and RNA synthesis and have various additional functions in the cell.[22] Within the pathway, four enzymes belong to the ATP-grasp superfamily: PurD, PurT, PurK, and PurP. Although members of the de novo purine biosynthesis catalyze different reactions, their structural and mechanistic similarity within the pathway suggests that there is an evolutionary relationship between these enzymes.[3, 14, 15, 23, 24]
The biotin carboxylase family is a specific group of nine enzymes that produces carboxybiotin for use in various prokaryotic and eukaryotic pathways. Four biotin carboxylase enzymes belong to the ATP-grasp superfamily of enzymes: pyruvate carboxylase (PC), urea amidolase, propionyl-CoA carboxylase (PCC), and acetyl-CoA carboxylase (ACC).[2, 7, 25-27] These enzymes all have diverse biological functions despite their analogous origin and classification. Urea amidolase catalyzes the ATP-dependent carboxylation of urea to yield allophanate. Urea amidolase is frequently fused to allophanate hydrolase to generate a bifunctional enzyme that is capable of converting urea to ammonia and carbon dioxide.[26, 28-30] PCC is a mitochondrial enzyme that catalyses the carboxylation of propionyl-CoA to form methylmalonyl-CoA (MMC) and is vital in the catabolism of cholesterol, fatty acids with an odd number of carbon chains, and several amino acids.[2, 25, 31] ACC is an essential enzyme in fatty acid synthesis that produces malonyl-CoA.[2] PC converts pyruvate into oxalacetate, a key intermediate in the Krebs cycle. PC is an important enzyme due to its pivotal role in regulating gluconeogenesis, lipogenesis, biosynthesis of neurotransmitters, and glucose-induced insulin secretion.[2, 7, 32]
Tubulin tyrosine ligase (TTL) is the only ATP-grasp enzyme involved in tubulin modifications. Tubulin, the precursor to microtubules, is extensively modified by a variety of post-translational modifications. One class of modification is the reversible addition of tyrosine onto the C-terminus of the protein. Tyrosine removal is catalyzed by tyrosine carboxypeptidase while addition of the amino acid is accomplished by the enzyme TTL. TTL activates the C-terminal glutamic acid for attack by tyrosine.[33, 34]
Another biologically important enzyme that belongs to the ATP-grasp superfamily is the modification enzyme of ribosomal protein S6 (RimK). This enzyme is involved in ribosome biogenesis.[35] RimK catalyzes addition of glutamic acid residues to the carboxyl terminus of ribosomal protein S6.[36, 37] It has been postulated that two additional ligases, RimKLA and RimKLB, should be accepted into the ATP-grasp superfamily of enzymes.[38] These newly reported enzymes synthesize N-acetylaspartylglutamate (NAAG), the most abundant dipeptide in vertebrate central nervous system and an agonist for glutamate receptors.[39] RimKLA and RimKLB utilize ATP, N-acetylaspartate (carboxyate donor), and L-glutamate (amine donor) to form NAAG. RimKLB has an additional catalytic function in the formation of β-citrylglutamate (a structural analog of NAAG) from citrate. RimKLA and RimKLB share 30% and 25% sequence identity with E. coli RimK and E. coli glutathione synthetase respectfully.[38]
The most recently reported member of the ATP-grasp superfamily is LysX. This enzyme is involved in the synthesis of lysine through a modified α-aminoadipate pathway comparable to that of fungi and yeast. The mechanism of LysX has not been fully elucidated, although it has been suggested that LysX utilizes ATP for catalytic modification of α-aminoadipate to N2-acetyl-α-aminoadipate.[40]
Currently, there are two members of the ATP-grasp superfamily that catalyze phosphoryl transfer, namely pyruvate phosphate dikinase (PPDK) and inositol 1,3,4-trisphosphate 5/6-kinase (IP56K). PPDK is a multi-domain enzyme involved in gluconeogenesis and photosynthesis in bacteria and eukarya.[5, 41]. IP56K is involved in the polyphosphate synthesis and plays an important role in the IP6 production pathway.[19, 42]
5. Mechanism
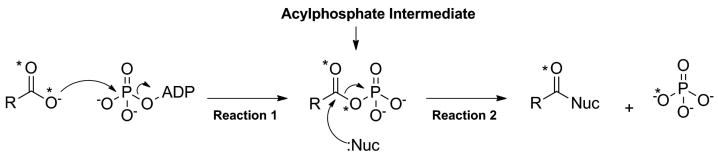
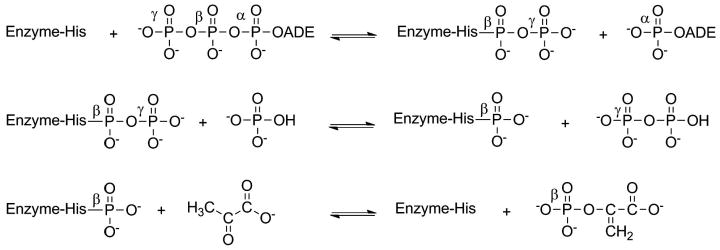
The mechanism of the ATP-grasp superfamily of enzymes, with the exception of the kinases, can be broken down into two partial reactions (Figure 5).[32, 34, 43, 44] In the first reaction, the substrate carboxylic acid reacts with ATP to yield a reactive acylphosphate intermediate.[4, 5, 11, 15, 21, 24, 45, 46] This step serves to convert the carboxylic acid into an electrophile. Evidence for the formation of this intermediate has been determined from positional isotope exchange studies which have shown that a labeled oxygen atom from the carboxylic acid is transferred to the inorganic phosphate product (Figure 5).[47-53] The existence of the acylphosphate intermediate is also supported by diazomethane trapping studies on carbamoyl phosphate synthetase which trapped the reactive intermediate, carboxyphosphate.[54] Formation of an acylphosphate intermediate has been implicated by the discovery of highly potent mechanistic inhibitors of DDLigase which mimic the reaction of a nucleophile with this phosphorylated intermediate (see the section 6 for additional details).[55, 56] Lastly, evidence for the formation of an acylphosphate intermediate is also suggested by studies showing substrate carboxylic acid stimulated ATPase activity for the ATP-grasp enzymes.[48, 57-59]
Figure 5.
Depiction of the general reaction catalyzed by the ligase members of the ATP-grasp superfamily of enzymes. The asterisks represent labeled oxygen atoms of the carboxylic acid and demonstrate the fate of these atoms during the course of the reaction. Notably, one oxygen atom is incorporated into phosphate suggesting the presence of an acylphosphate intermediate.
The second half reaction is the reaction of the acylphosphate intermediate. The fate of this intermediate depends upon the carboxylic acid substrate used in the first step. For most carboxylic acids, it has been postulated that there is a direct nucleophilic attack on the carbonyl carbon of the acylphosphate intermediate to yield a tetrahedral intermediate.[23] This tetrahedral intermediate is mimicked by the phosphinophosphate inhibitors of DDLigase.[55, 56] Decomposition of the tetrahedral intermediate results in the final product and inorganic phosphate. For reactions that utilize bicarbonate as the carboxylic acid, the fate of carboxyphosphate is still uncertain. Carboxyphosphate has been reported to potentially serve as the electrophile which is directly attacked by the nucleophilic substrate; a reaction analogous to that for other carboxylic acid substrates.[23] Alternatively, it has been suggested that carboxyphosphate decarboxylates to yield the reactive carboxylating agent CO2 and inorganic phosphate. [32, 43, 44] While structure-based molecular models have supported this mechanism, direct experimental evidence has, so far, failed to detect the production of CO2 by these enzymes. [1, 23, 60]
The second half reaction is thought to require an active site base for removal of the proton from either the thiol- or amine-containing substrate.[23] For several ATP-grasp enzymes, a specific active site base has been postulated based upon structural as well as mutagenesis studies. However, the identity of the active site base utilized for biotin carboxylation is still an unresolved issue. Numerous site-directed mutagenesis studies examining a variety of active site bases have so far failed to definitively identify the base.[1] In the absence of an identified base, it has been postulated that inorganic phosphate generated from decomposition of carboxyphosphate may act as the base to remove the uredio proton from biotin to generate the more nucleophilic enol form of biotin. [1, 44]
Currently, there are two kinase members of the ATP-grasp superfamily that catalyze phosphoryl transfer. These enzymes possess the ATP-grasp fold, but are mechanistically distinct from the “classical” ATP-grasp enzymes typified by biotin carboxylase. PPDK is thought to function by transfer of pyrophosphate to a histidine residue followed by attack of the enzyme-bound pyrophosphate by inorganic phosphate (Figure 6).[53] This yields pyrophosphate containing the γ-phosphate group and a phosphorylated enzyme where the phosphate group originated from the beta-phosphate group. In the final step, pyruvate is phosphorylated to yield phosphoenolpyruvate.[61-63]
Figure 6.
Conversion of ATP, Pi, and pyruvate to AMP, PPi, and phoshoenolpyruvate by PPDK.[62]
The catalytic mechanism for the second ATP-grasp kinase, inositol 1,3,4-trisphosphate 5/6-kinase (IP56K), has been postulated based upon analysis of the crystal structure. It has been suggested that the enzyme catalyzes an attack of the hydroxyl group at either the 5 or 6 position of inositol 1,3,4 trisphosphate on the γ-phosphate of ATP.[19]
6. Inhibitors of ATP-grasp Enzymes
6.1. Types of Inhibitors
Given the medicinal importance of the pathways that utilize ATP-grasp enzymes, it is not surprising that researchers have sought to identify inhibitors of these enzymes for the treatment of a variety of diseases.[64] In addition to the medicinal benefits of these inhibitors, researchers have also used enzyme inhibitors as probes to study the enzyme mechanisms.[41, 56] There are several distinct classes of inhibitors of ATP-grasp enzymes. The first class includes the mechanism-based or transition-state analogs that function by mimicking the transition state of the enzyme. The second class encompasses inhibitors which target the ATP-binding pocket of the grasp enzymes while the third class contains reversible agents that either compete with one or both substrates or bind to a non-active site pocket. A discussion of each inhibitor class is presented below.
6.2. Mechanism-based and Transition-state Analogs
Compounds that are stable transition-state analogs are generally highly potent inhibitors and also serve as effective probes of the mechanism of the reaction. These transition-state inhibitors are also useful in the study of other grasp enzymes that share analogous chemistry, exhibit structural similarities, or are evolutionarily linked with the original enzyme. The utility of this approach is demonstrated by work performed on DDLigase and GSHase.[9, 41, 55, 56]
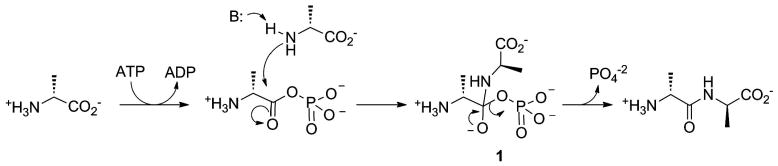
DDLigase is an attractive target for the development of novel antibiotics due to the essential role it plays in bacterial cell wall biosynthesis. [9, 65] The proposed mechanism of DDLigase is believed to function in a manner analogous to that previously discussed above (Figure 5). One molecule of D-alanine attacks ATP to generate an acylphosphate intermediate which is then attacked by a second molecule of D-alanine to yield a tetrahedral intermediate, compound 1 (Figure 7). Collapse of this intermediate with expulsion of phosphate produces the dipeptide. Researchers from Merck and Harvard rationalized that a phosphinate analog of the tetrahedral intermediate (1) would be a potent inhibitor of the enzyme.[9, 66, 67]
Figure 7.
Proposed mechanism of DDLigase
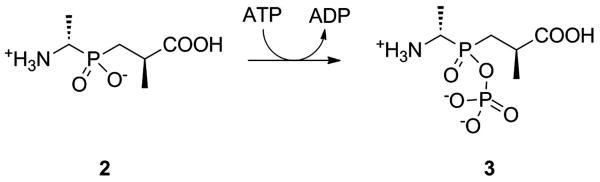
These investigators found that in presence of ATP, compound 2 (Figure 8) bound to the active site, in a slow tight manner, and acted to inhibit the enzyme.[9, 65] The slow, tight-binding nature of the kinetics indicated that compound 2 was converted to a more potent compound, subsequent to binding in the active site. NMR and crystallographic studies verified that 2 was phosphorylated by ATP to generate a phosphinophosphate (Figure 8, compound 3).[9, 66, 67] Compound 2 is thus a mechanism-based inhibitor which dissociates from the enzyme on the time scale of days.[9, 66, 67]
Fig. 8.
Phosphinate inhibitor of DDLigase
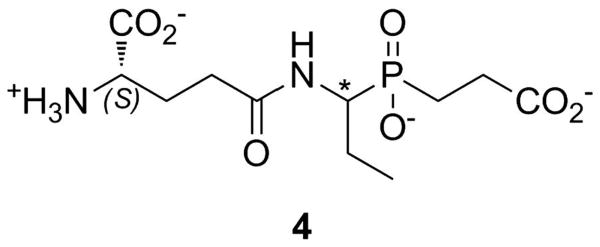
A similar mechanism-based inhibitor approach has also been reported for GSHase. Compound 4 (Figure 9) is a slow, tight-binding inhibitor of the synthetase with overall inhibition constant in the 20-50 nM range (depending upon stereochemistry). As seen for the DDLigase inhibitors, ATP was essential for the development of potent inhibition by phosphorylating the inhibitor to yield the potent transition state mimic. The phosphorylation of compound 4 was confirmed by X-ray crystallography.[41]
Figure 9.
Glutathione synthetase inhibitors. The asterisk indicates a chiral center that was varied to examine the role of stereochemistry on inhibition.
6.3. ATP-competitive Inhibitors
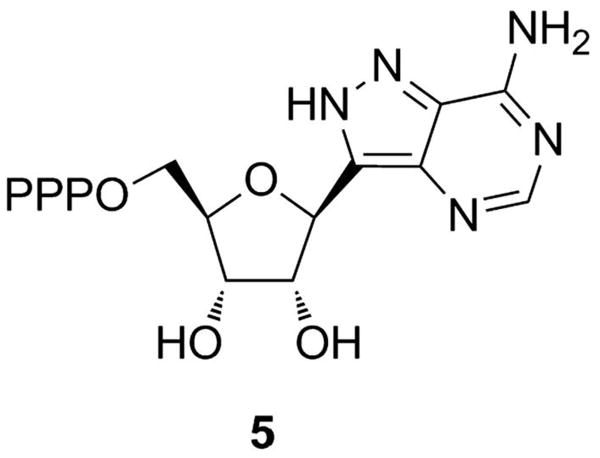
Compounds which bind to the ATP-binding site of the ATP-grasp enzymes have been explored as potential inhibitors and many of these compounds have been identified as inhibitors of other ATP utilizing enzymes. For example, nucleotide analogs, such as formycin A triphosphate (compound 5, Figure 10) or non-hydrolyzable analogs of ATP, have been shown to be modest inhibitors of BC.[64, 68] These molecules did not show an appreciable degree of selectivity between the ATP-grasp enzymes versus other ATP-utilizing enzymes. Thus, the use of these agents as potential drug targets is limited.
Figure 10.
Structure of formycin A triphosphate
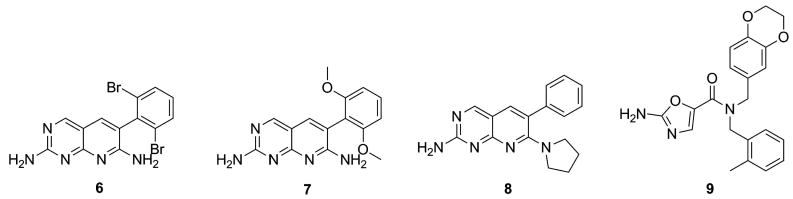
The design and synthesis of enzyme inhibitors that function as ATP-competitive molecules with high selectivity profiles was initially thought unfeasible in the realm of drug discovery. The primary obstacle associated with ATP-competitive inhibition was the development of compounds with sufficient potency to compete with large amounts of ATP in cells. Despite the issues with utilizing existing ATP-binding agents to target ATP-grasp enzymes, researchers utilized kinase inhibitors as starting points in drug discovery. Researchers from Pfizer investigated the use of protein kinase inhibitors by screening a large proprietary library of compounds for inhibition of bacterial growth.[64, 69] Several pyridopyrimidines 6-8 (Figure 11) were identified from the screening and additional studies revealed that these agents were potent inhibitors of BC. [64, 69] Mechanistic studies of compounds 6-8 revealed that these agents targeted the ATP-binding site of BC with IC50 values ranging from 5-150 nM.[64, 69, 70] Crystallographic analysis verified this result and revealed the binding orientation of these agents in the ATP-grasp fold.[69] Finally, the investigators showed that these agents were remarkably selective for BC versus a family of protein kinases. This result indicated that the ATP-grasp binding site has distinct features that allowed it to be specifically targeted relative to other ATP-binding sites.[64, 69, 70] In silico and fragment based screening methods were implemented to identify additional inhibitors of BC. This study lead to the discovery of compound 9 (Figure 11), with a reported IC50 of 7 nM and displayed a high selectivity profile for BC relative to protein kinases.[70]
Figure 11.
Inhibitors of biotin carboxylase
6.4. Reversible non-competitive inhibitors
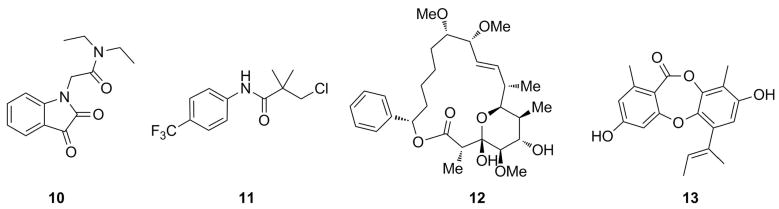
The third class of inhibitors includes compounds that are reversible inhibitors that bind to sites other than the ATP-binding site. A number of these inhibitors have been found to be non-competitive with the substrates. Compound 10 (Figure 12) was identified as a non-competitive inhibitor of PurK, an enzyme found in the microbial de novo purine biosynthesis pathway, but not in higher eukaryotes.[71] Research conducted by Pfizer identified compound 11 as a non-competitive allosteric inhibitor of DDLigase.[65] Researchers at Columbia showed that the natural product, soraphen A (compound 12) is a non-competitive inhibitor of BC and that the inhibitor binds 25 Å away from the active site.[72] Lastly, researchers have investigated inhibitors of PPDK as potential herbicides due to the role that this enzyme plays in the growth of C4 plants. Motti and co-workers found that the compound, unguinol (I) (compound 13) was a moderate non-competitive inhibitor (with respect to ATP) of this PPDK. The mechanism by which these compounds induce their respective enzymes to form a non-productive complex is not know but likely involved perturbations to critical residues in the active site.[73]
Figure 12.
Non-competitive inhibitors of ATP-grasp enzymes
7. Conclusions
It has been estimated that between 9-10% of all enzymes bind ATP. Not surprisingly, there are numerous structural folds that have been identified which serve to bind this nucleotide. In this review, we have focused on a specific superfamily of enzymes possessing an atypical nucleotide-binding fold called the “ATP-grasp” fold. This unique ATP-binding structure was first elucidated for GSHase, BC, and DDLigase but subsequent studies have added an additional 18 enyzmes to this superfamily. The mechanism of these enzymes, with the exception of the kinases, is believed to involve the reaction of the substrate carboxylic acid reacts with ATP to yield a reactive acylphosphate intermediate. The acylphosphate intermediate, in turn, is then reacted with the nucleophilic substrate to generate the final product. For the enzymes which utilize bicarbonate as the carboxylic acid substrate, the fate of the acylphosphate intermediate, carboxyphosphate, still remains uncertain. The enzymes of the superfamily can be found throughout metabolism and thus have medicinal importance. Researchers have developed potent transition-state analogs and perhaps most interestingly, have discovered that the ATP-binding site of the grasp enzymes is distinct from other ATP-binding enzymes. This provides the possibility that the ATP-binding site of the ATP-grasp enzymes may be a rational target for drug design. Finally, the discovery of non-competitive inhibitors for many of these enzymes suggests that there may be undiscovered allosteric mechanisms that serve to regulate the function of the enzyme. There is no doubt that the ATP-grasp proteins represent an important and interesting class of enzymes which will continue to challenge researchers for years to come.
Acknowledgments
The authors thank the National Institute of General Medical Sciences for funding (GM087467 to S.M.F).
References
- 1.Chou CY, Yu LPC, Tong L. J Biol Chem. 2009;284:11690–11697. doi: 10.1074/jbc.M805783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jitrapakdee S, Wallace JC. Curr Protein Pept Sci. 2003;4:217–229. doi: 10.2174/1389203033487199. [DOI] [PubMed] [Google Scholar]
- 3.Li H, Fast W, Benkovic SJ. Protein Sci. 2009;18:881–892. doi: 10.1002/pro.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murzin AG. Curr Opin Struct Biol. 1996;6:386–394. doi: 10.1016/s0959-440x(96)80059-5. [DOI] [PubMed] [Google Scholar]
- 5.Galperin MY, Koonin EV. Protein Sci. 1997;6:2639–2643. doi: 10.1002/pro.5560061218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esser L, Wang CR, Hosaka M, Smagula CS, Sudhof TC, Diesenhofer J. Embo J. 1998;17:977–984. doi: 10.1093/emboj/17.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jitrapakdee S, St Maurice M, Rayment I, Cleland WW, Wallace JC, Attwood PV. Biochem J. 2008;413:369–387. doi: 10.1042/BJ20080709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan C, Moews PC, Shi Y, Walsh CT, Knox JR. Proc Natl Acad Sci U S A. 1995;92:1172–1176. doi: 10.1073/pnas.92.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan C, Moews PC, Walsh CT, Knox JR. Science (Washington, D C) 1994;266:439–443. doi: 10.1126/science.7939684. [DOI] [PubMed] [Google Scholar]
- 10.Hibi T, Nishioka T, Kato H, Tanizawa K, Fukui T, Katsube Y, Oda Ji. Nat Struct Biol. 1996;3:16–18. doi: 10.1038/nsb0196-16. [DOI] [PubMed] [Google Scholar]
- 11.Sloane V, Blanchard CZ, Guillot F, Waldrop GL. J Biol Chem. 2001;276:24991–24996. doi: 10.1074/jbc.M101472200. [DOI] [PubMed] [Google Scholar]
- 12.Waldrop GL, Rayment I, Holden HM. Biochemistry. 1994;33:10249–10256. doi: 10.1021/bi00200a004. [DOI] [PubMed] [Google Scholar]
- 13.Wolodko WT, Fraser ME, James MNG, Bridger WA. J Biol Chem. 1994;269:10883–10890. doi: 10.2210/pdb1scu/pdb. [DOI] [PubMed] [Google Scholar]
- 14.Thoden JB, Firestine S, Nixon A, Benkovic SJ, Holden HM. Biochemistry. 2000;39:8791–8802. doi: 10.1021/bi000926j. [DOI] [PubMed] [Google Scholar]
- 15.Wang W, Kappock TJ, Stubbe J, Ealick SE. Biochemistry. 1998;37:15647–15662. doi: 10.1021/bi981405n. [DOI] [PubMed] [Google Scholar]
- 16.Thoden JB, Kappock TJ, Stubbe J, Holden HM. Biochemistry. 1999;38:15480–15492. doi: 10.1021/bi991618s. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, White RH, Ealick SE. Biochemistry. 2008;47:205–217. doi: 10.1021/bi701406g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thoden JB, Blanchard CZ, Holden HM, Waldrop GL. J Biol Chem. 2000;275:16183–16190. doi: 10.1074/jbc.275.21.16183. [DOI] [PubMed] [Google Scholar]
- 19.Miller GJ, Wilson MP, Majerus PW, Hurley JH. Mol Cell. 2005;18:201–212. doi: 10.1016/j.molcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 20.Gitler D, Xu Y, Kao HT, Lin D, Lim S, Feng J, Greengard P, Augustine GJ. J Neurosci. 2004;24:3711–3720. doi: 10.1523/JNEUROSCI.5225-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polekhina G, Board PG, Gali RR, Rossjohn J, Parker MW. Embo J. 1999;18:3204–3213. doi: 10.1093/emboj/18.12.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welin M, Grossmann JG, Flodin S, Nyman T, Stenmark P, Tresaugues L, Kotenyova T, Johansson I, Nordlund P, Lehtioe L. Nucleic Acids Res. 2010;38:7308–7319. doi: 10.1093/nar/gkq595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thoden JB, Holden HM, Firestine SM. Biochemistry. 2008;47:13346–13353. doi: 10.1021/bi801734z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Morar M, Ealick SE. Cell Mol Life Sci. 2008;65:3699–3724. doi: 10.1007/s00018-008-8295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang H, Rao KS, Yee VC, Kraus JP. J Biol Chem. 2005;280:27719–27727. doi: 10.1074/jbc.M413281200. [DOI] [PubMed] [Google Scholar]
- 26.Kanamori T, Kanou N, Atomi H, Imanaka T. J Bacteriol. 2004;186:2532–2539. doi: 10.1128/JB.186.9.2532-2539.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novak BR, Moldovan D, Waldrop GL, de Queiroz MS. Proteins: Struct, Funct Bioinf. 2010;79:622–632. doi: 10.1002/prot.22910. [DOI] [PubMed] [Google Scholar]
- 28.Roon RJ, Hampshire J, Levenberg B. J Biol Chem. 1972;247:7539–7545. [PubMed] [Google Scholar]
- 29.Roon RJ, Levenberg B. Methods Enzymol. 1970;17:317–324. [Google Scholar]
- 30.Strope PK, Nickerson KW, Harris SD, Moriyama EN. BMC Evol Biol. 2011;11:80. doi: 10.1186/1471-2148-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang CS, Sadre-Bazzaz K, Shen Y, Deng B, Zhou ZH, Tong L. Nature (London, U K) 2010;466:1001–1005. doi: 10.1038/nature09302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lasso G, Yu LPC, Gil D, Xiang S, Tong L, Valle M. Structure (Cambridge, MA, U S) 2010;18:1300–1310. doi: 10.1016/j.str.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dal Piaz F, Vassallo A, Lepore L, Tosco A, Bader A, De Tommasi N. J Med Chem. 2009;52:3814–3828. doi: 10.1021/jm801637f. [DOI] [PubMed] [Google Scholar]
- 34.Erck C, Peris L, Andrieux A, Meissirel C, Gruber AD, Vernet M, Schweitzer A, Saoudi Y, Pointu H, Bosc C, Salin PA, Job D, Wehland J. Proc Natl Acad Sci U S A. 2005;102:7853–7858. doi: 10.1073/pnas.0409626102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kosuge T, Hoshino T. FEMS Microbiol Lett. 1997;157:73–79. doi: 10.1111/j.1574-6968.1997.tb12755.x. [DOI] [PubMed] [Google Scholar]
- 36.Kang WK, Icho T, Isono S, Kitakawa M, Isono K. MGG, Mol Gen Genet. 1989;217:281–288. doi: 10.1007/BF02464894. [DOI] [PubMed] [Google Scholar]
- 37.Nishida H, Nishiyama M, Kobashi N, Kosuge T, Hoshino T, Yamane H. Genome Res. 1999;9:1175–1183. doi: 10.1101/gr.9.12.1175. [DOI] [PubMed] [Google Scholar]
- 38.Collard F, Stroobant V, Lamosa P, Kapanda CN, Lambert DM, Muccioli GG, Poupaert JH, Opperdoes F, Van Schaftingen E. J Biol Chem. 2010;285:29826–29833. doi: 10.1074/jbc.M110.152629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neale JH. J Neurochem. 2011 [Google Scholar]
- 40.Sakai H, Vassylyeva MN, Matsuura T, Sekine Si, Gotoh K, Nishiyama M, Terada T, Shirouzu M, Kuramitsu S, Vassylyev DG, Yokoyama S. J Mol Biol. 2003;332:729–740. doi: 10.1016/s0022-2836(03)00946-x. [DOI] [PubMed] [Google Scholar]
- 41.Hiratake J. Chem Rec. 2005;5:209–228. doi: 10.1002/tcr.20045. [DOI] [PubMed] [Google Scholar]
- 42.Wilson MP, Hugge C, Bielinska M, Nicholas P, Majerus PW, Wilson DB. Proc Natl Acad Sci U S A. 2009;106:9831–9835. doi: 10.1073/pnas.0904172106. S9831/9831-S9831/9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Attwood PV. Int J Biochem Cell Biol. 1995;27:231–249. doi: 10.1016/1357-2725(94)00087-r. [DOI] [PubMed] [Google Scholar]
- 44.Attwood PV, Wallace JC. Acc Chem Res. 2002;35:113–120. doi: 10.1021/ar000049+. [DOI] [PubMed] [Google Scholar]
- 45.Eroglu B, Powers-Lee SG. Arch Biochem Biophys. 2002;407:1–9. doi: 10.1016/s0003-9861(02)00510-6. [DOI] [PubMed] [Google Scholar]
- 46.Li H, Xu H, Graham DE, White RH. Proc Natl Acad Sci U S A. 2003;100:9785–9790. doi: 10.1073/pnas.1733391100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Healy VL, Mullins LS, Li X, Hall SE, Raushel FM, Walsh CT. Chem Biol. 2000;7:505–514. doi: 10.1016/s1074-5521(00)00135-6. [DOI] [PubMed] [Google Scholar]
- 48.Marolewski AE, Mattia KM, Warren MS, Benkovic SJ. Biochemistry. 1997;36:6709–6716. doi: 10.1021/bi962961p. [DOI] [PubMed] [Google Scholar]
- 49.Mueller EJ, Meyer E, Rudolph J, Davisson VJ, Stubbe J. Biochemistry. 1994;33:2269–2278. doi: 10.1021/bi00174a038. [DOI] [PubMed] [Google Scholar]
- 50.Mullins LS, Lusty CJ, Raushel FM. J Biol Chem. 1991;266:8236–8240. [PubMed] [Google Scholar]
- 51.Mullins LS, Zawadzke LE, Walsh CT, Raushel FM. J Biol Chem. 1990;265:8993–8998. [PubMed] [Google Scholar]
- 52.Ogita T, Knowles JR. Biochemistry. 1988;27:8028–8033. doi: 10.1021/bi00421a009. [DOI] [PubMed] [Google Scholar]
- 53.Wang HC, Ciskanik L, Dunaway-Mariano D, Von der Saal W, Villafranca JJ. Biochemistry. 1988;27:625–633. doi: 10.1021/bi00402a020. [DOI] [PubMed] [Google Scholar]
- 54.Powers SG, Meister A. Proc Natl Acad Sci U S A. 1976;73:3020–3024. doi: 10.1073/pnas.73.9.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ellsworth BA, Tom NJ, Bartlett PA. Chem Biol. 1996;3:37–44. doi: 10.1016/s1074-5521(96)90082-4. [DOI] [PubMed] [Google Scholar]
- 56.McDermott AE, Creuzet F, Griffin RG, Zawadzke LE, Ye QZ, Walsh CT. Biochemistry. 1990;29:5767–5775. doi: 10.1021/bi00476a018. [DOI] [PubMed] [Google Scholar]
- 57.Climent I, Rubio V. Arch Biochem Biophys. 1986;251:465–470. doi: 10.1016/0003-9861(86)90353-x. [DOI] [PubMed] [Google Scholar]
- 58.Meyer E, Leonard NJ, Bhat B, Stubbe J, Smith JM. Biochemistry. 1992;31:5022–5032. doi: 10.1021/bi00136a016. [DOI] [PubMed] [Google Scholar]
- 59.Wimmer MJ, Rose IA, Powers SG, Meister A. J Biol Chem. 1979;254:1854–1859. [PubMed] [Google Scholar]
- 60.Gibson GE, Mullins LS, Raushel FM. Bioorg Chem. 1998;26:255–268. [Google Scholar]
- 61.Carroll LJ, Xu Y, Thrall SH, Martin BM, Dunaway-Mariano D. Biochemistry. 1994;33:1134–1142. doi: 10.1021/bi00171a012. [DOI] [PubMed] [Google Scholar]
- 62.Lin Y, Lusin JD, Ye D, Dunaway-Mariano D, Ames JB. Biochemistry. 2006;45:1702–1711. doi: 10.1021/bi051816l. [DOI] [PubMed] [Google Scholar]
- 63.Ye D, Wei M, McGuire M, Huang K, Kapadia G, Herzberg O, Martin BM, Dunaway-Mariano D. J Biol Chem. 2001;276:37630–37639. doi: 10.1074/jbc.M105631200. [DOI] [PubMed] [Google Scholar]
- 64.Skedelj V, Tomasic T, Masic LP, Zega A. J Med Chem. 2011;54:915–929. doi: 10.1021/jm101121s. [DOI] [PubMed] [Google Scholar]
- 65.Triola G, Wetzel S, Ellinger B, Koch MA, Huebel K, Rauh D, Waldmann H. Bioorg Med Chem. 2009;17:1079–1087. doi: 10.1016/j.bmc.2008.02.046. [DOI] [PubMed] [Google Scholar]
- 66.Chakravarty PK, Greenlee WJ, Parsons WH, Patchett AA, Combs P, Roth A, Busch RD, Mellin TN. J Med Chem. 1989;32:1886–1890. doi: 10.1021/jm00128a033. [DOI] [PubMed] [Google Scholar]
- 67.Duncan K, Walsh CT. Biochemistry. 1988;27:3709–3714. doi: 10.1021/bi00410a028. [DOI] [PubMed] [Google Scholar]
- 68.Zeczycki TN, St Maurice M, Attwood PV. Open Enzyme Inhib J. 2010;3:8–26. doi: 10.2174/1874940201003010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miller JR, Dunham S, Mochalkin I, Banotai C, Bowman M, Buist S, Dunkle B, Hanna D, Harwood J, Huband MD, Karnovsky A, Kuhn M, Limberakis C, Liu JY, Mehrens S, Mueller WT, Narasimhan L, Ogden A, Ohren J, Vara Prasad JVN, Shelly JA, Skerlos L, Sulavik M, Thomas VH, VanderRoest S, Wang L, Wang Z, Whitton A, Zhu T, Stover CK. Proc Natl Acad Sci U S A. 2009;106:1737–1742. doi: 10.1073/pnas.0811275106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mochalkin I, Miller JR, Narasimhan L, Thanabal V, Erdman P, Cox PB, Prasad JVNV, Lightle S, Huband MD, Stover CK. ACS Chem Biol. 2009;4:473–483. doi: 10.1021/cb9000102. [DOI] [PubMed] [Google Scholar]
- 71.Firestine SM, Paritala H, McDonnell JE, Thoden JB, Holden HM. Bioorg Med Chem. 2009;17:3317–3323. doi: 10.1016/j.bmc.2009.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen Y, Volrath SL, Weatherly SC, Elich TD, Tong L. Mol Cell. 2004;16:881–891. doi: 10.1016/j.molcel.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 73.Motti CA, Bourne DG, Burnell JN, Doyle JR, Haines DS, Liptrot CH, Llewellyn LE, Ludke S, Muirhead A, Tapiolas DM. Appl Environ Microbiol. 2007;73:1921–1927. doi: 10.1128/AEM.02479-06. [DOI] [PMC free article] [PubMed] [Google Scholar]