Abstract
Cancer-targeting drug delivery can be based on the rational design of a therapeutic platform. This approach is typically achieved by the functionalization of a nanoparticle with two distinct types of molecules, a targeting ligand specific for a cancer cell, and a cytotoxic molecule to kill the cell. The present study aims to evaluate the validity of an alternative simplified approach in the design of cancer-targeting nanotherapeutics: conjugating a single type of molecule with dual activities to nanoparticles, instead of coupling a pair of orthogonal molecules. Herein we investigate whether this strategy can be validated by its application to methotrexate, a dual-acting small molecule that shows cytotoxicity because of its potent inhibitory activity against dihydrofolate reductase and that binds folic acid receptor, a tumor biomarker frequently upregulated on the cancer cell surface. This article describes a series of dendrimer conjugates derived from a generation 5 polyamidoamine (G5 PAMAM) presenting a multivalent array of methotrexate and also demonstrates their dual biological activities by surface plasmon resonance spectroscopy, a cell-free enzyme assay, and cell-based experiments with KB cancer cells.
Keywords: Folate receptor, methotrexate, poly(amidoamine) dendrimer, targeted drug delivery, multivalent binding, dual activity
1. Introduction
Nanotechnology is uniquely suited for providing multifunctional platforms for targeted delivery in several life-threatening diseases, including cancers and inflammatory diseases.[1–6] This study aims to investigate a novel and simplified delivery strategy based on functionalization of a nanoplatform with a dual-acting small molecule, in lieu of two single-acting molecules, that serves as both a targeting ligand and an anticancer therapeutic. In this communication, we demonstrate the validity and effectiveness of this simplified strategy by designing multivalent NPs presenting methotrexate on the periphery of a generation 5 polyamidoamine (G5 PAMAM) dendrimer.
Applications of multifunctional NPs in anticancer therapeutic delivery have been well demonstrated by use of targeting ligands specific for a cell surface molecule overexpressed in cancer cells, such as folic acid receptor (FAR),[7–9] riboflavin receptor,[10,11] αvβ3 integrin,[12–14] prostate-specific membrane antigen,[15] Her2,[16] transferrin receptor,[17] and epidermal growth factor receptor.[16,18,19] The biological process for the effective uptake of such NPs requires multiple-ligand molecules attached to the NP surface. It begins with specific adhesion of a NP to the targeted cell surface in a mechanism that is characterized by multivalent interactions occurring collectively at the interface of multiple receptor-ligand pairs.[12,20] Such a multivalent mechanism is considered as highly important during the receptor-mediated endocytosis because it constitutes the basis for tight NP-cell adhesion and conformal contacts created during the formation of coated pits.[21–24] Therefore, in a rational design for targeted NPs, each NP is covalently conjugated with multiple copies of a targeting ligand on its periphery in order to achieve the multivalent effects, and each is further functionalized to carry therapeutic or imaging molecules as the payloads for cellular delivery.[1,25–27]
Despite the rational basis of the NP design and successful proof of concept studies demonstrated already, several challenging issues face the development of cancer-targeting therapeutic NPs. They are attributable simply to the complexity of the NP structure and the lack of methods to control the distribution of the particle size, ligand density, and drug loads.[27–31] Currently, there are only a few specialized methods demonstrated for the precise engineering and ligand functionalization of NPs such as PAMAM dendrimer,[28,30,32] polymer,[33] or gold[34,35]. To ease the complexity of the fabrication of targeted NP therapeutics, we have explored new design strategies. Here we evaluate the feasibility of using a dual-acting small molecule that can: i) function as a ligand for a cancer-specific receptor, and ii) induce cytotoxicity following cellular internalization. Because this approach is based on using a single type of small molecule for both targeting and functional activity, the precision by which these functionalized nanoparticles can be synthesized is higher than what can be obtained using the conventional two small molecule approach.
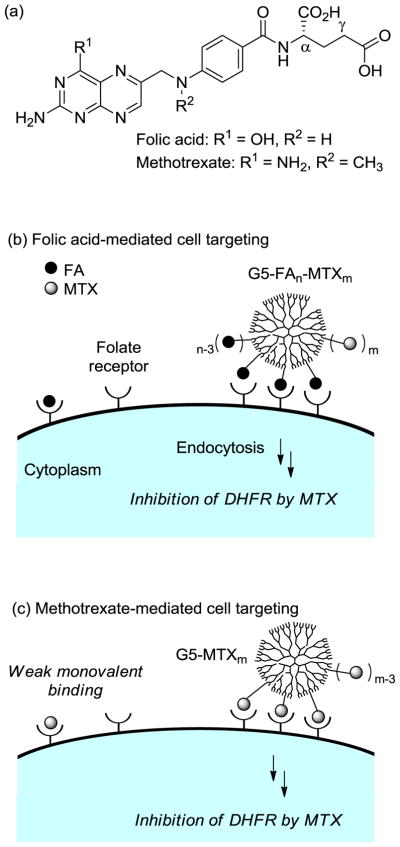
In our search for candidate molecules that could both target and function as a therapeutic, we were interested in methotrexate (MTX, Figure 1), which has been used as an anticancer drug.[5,6] This therapeutic molecule functions primarily by inhibiting the metabolic enzymes human dihydrofolate reductase (DHFR), an enzyme localized in the cytoplasm (Ki = 1.2 nM).[36] While the cellular uptake of MTX is mediated by reduced folate carrier proteins,[37] MTX is also able to bind FAR because of its high structural homology to FA, though at a lower affinity constant (KD = ~20–100 nM vs. KD (FA) = ~1 nM to kidney FAR).[38–40]
Figure 1.
(a) Structure of folic acid (FA), and methotrexate (MTX); (b) Schematic for the folic acid receptor (FAR)-mediated cancer cell targeting by a dendrimer nanoconjugate (G5-FAn-MTXm) presenting FA as a targeting ligand and carrying MTX as a cytotoxic drug; (c) Schematic for the proposed folic acid receptor (FAR)-mediated cancer cell targeting by a dendrimer nanoconjugate (G5-MTXn) presenting methotrexate (MTX) as a dual acting molecule for targeting and the cytotoxic activity. The figure is not drawn to scale.
Despite the lower FAR affinity of MTX, we hypothesize that MTX could still function as a targeting ligand if multiple copies per nanoparticle were utilized to form a multivalent nanoparticle.[21–24] Recently, we designed a class of synthetic multivalent nanoparticles, each composed of multiple FA ligands conjugated to a G5 PAMAM dendrimer scaffold, and demonstrated by surface plasmon resonance (SPR) spectroscopy that nanoparticles functionalized with more than one FA bound more tightly to a bovine folate binding protein (FBP)-coated surface than free FA.[20,41] Based on this observation, we aimed to determine whether a multivalent MTX-based dendrimer is still effective for FAR targeting. A recent theoretical analysis suggests that multivalent cooperativity can be kinetically limited if the binding of an individual receptor-ligand pair is too tight.[42] It further suggests that the targeting specificity for a particular cell type can be enhanced by making the affinity of each individual receptor-ligand pair weaker. In this study, we describe the synthesis of multivalent MTX conjugates and provide evidence, based on SPR and in vitro studies, that these conjugates possess the dual activity necessary for serving as an effective cancer therapeutic.
2. Results and Discussion
2.1. Surface plasmon resonance (SPR) spectroscopy of multivalent MTX conjugates
We studied the effect of multivalency on the interaction between FBP present on the surface and a dendrimer functionalized with MTX molecules by using SPR spectroscopy. As a bioanalytical method well established for studying binding kinetics of analytes to the biological surface on a real-time basis, SPR spectroscopy has been utilized for the studies of multivalent ligand-receptor interactions, including the system of the G5 PAMAM dendrimer conjugated with FA.[20,43–47]
2.1.1. Synthesis of dendrimer-based multivalent MTX conjugates
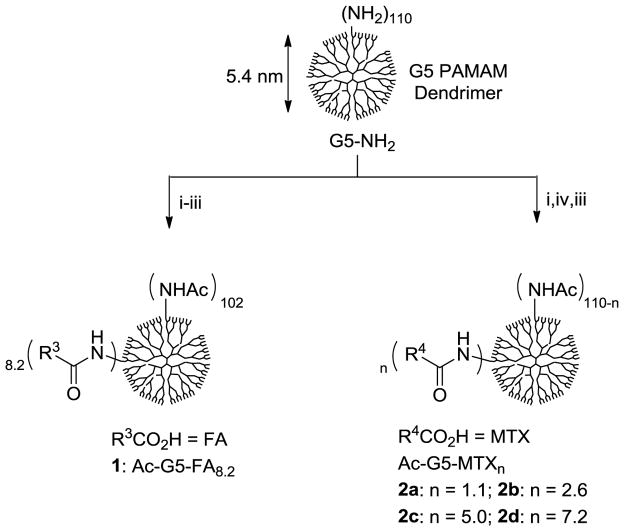
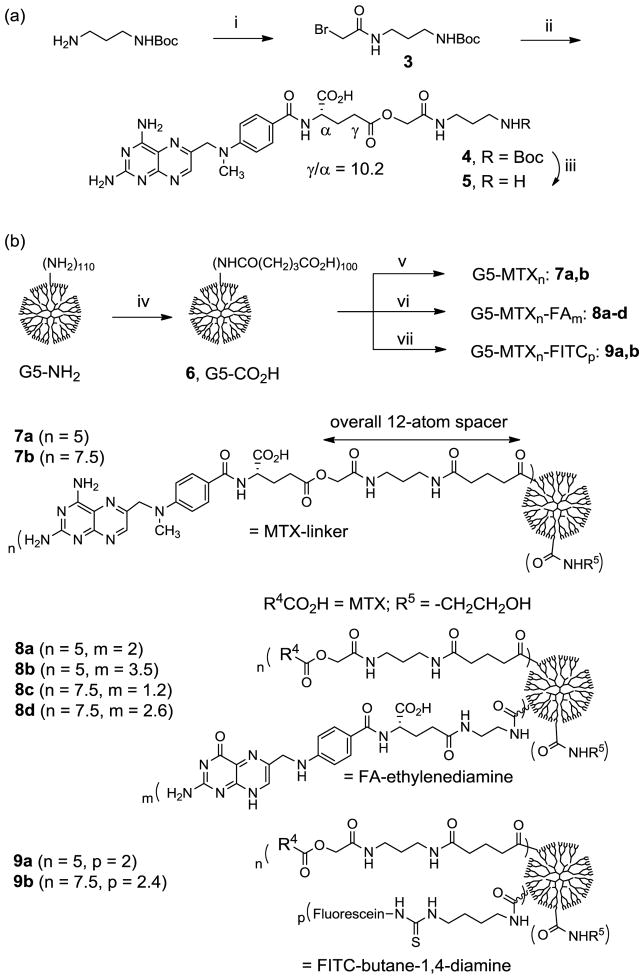
In designing multivalent MTX conjugates we followed the approach previously developed for the SPR studies of dendrimer-FA conjugates[20] (1, Scheme 1) because of the similarity in the structure and chemical reactivity of FA and MTX. Scheme 1 shows the synthetic method and structures for multivalent MTX conjugates 2a-d studied here. Each of the conjugates contains a variable number of MTX molecules presented on the neutral surface of G5 PAMAM dendrimer. Synthesis of these conjugates was performed by using an EDC-based coupling method where an amide bond is formed between the carboxylic acid of a MTX L-glutamate and a primary amine present on the periphery of a partially acetylated dendrimer molecule.[20] Following the purification using extensive dialysis (MWCO 10 kDa) first against phosphate-buffered saline (PBS) solution and then deionized water, each of these conjugates was characterized in combination with MALDI-TOF mass spectrometry, 1H NMR spectroscopy, and UV/Vis spectrometry. These analyses allowed us to calculate the number of ligand molecules attached per dendrimer, though on a mean basis: 1 (Ac-G5-FA8.2), and 2a-d (Ac-G5-MTXn; n = 1.1, 2.6, 5.0, 7.1).
Scheme 1.
A representative synthetic scheme for G5 PAMAM dendrimers presenting either folic acid (FA) or methotrexate (MTX) molecule in multiple copies. (i) Ac2O, MeOH, rt; (ii) folic acid, EDC, DMSO, DMF, rt, 24 h; (iii) Ac2O, DMSO, rt; (iv) variable amounts of methotrexate, EDC, DMF, DMSO, rt, 24 h.
2.1.2. Surface plasmon resonance (SPR) spectroscopy
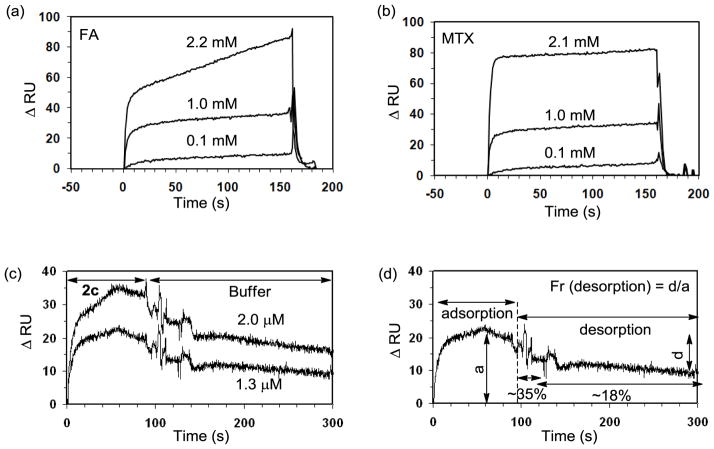
For the SPR experiments we utilized a CM5 sensor chip to present bovine FBP immobilized on the surface. Following an EDC-based amide coupling method as described previously,[20] the FBP-presenting chip was prepared at the surface protein density of 9.5 ng/mm2 (≈ 2 FBP per 10 nm2). It was first used for the binding studies of the monovalent control ligands (FA, MTX) and a set of their SPR sensorgrams are shown in Figure 2a,b. Each of the sensorgrams for the monovalent ligands was analyzed to extract its kinetic parameters (on rate = kon, off rate = koff) by using the BIAevaluation software, version 3 (Biacore). Given the monovalent mode of association for free FA or MTX, a 1:1 binding mode was utilized for a curve fitting based on the Langmuir model,[20] which led to determination of the kinetic parameters (koff, kon) and as a result, the equilibrium dissociation constant (KD = koff/kon; Table 1). The results suggest that MTX binds to bovine FBP with a high micromolar dissociation constant (KD = 2.4 × 10−5 M), an affinity ~2-fold lower than that of FA (KD = 1.1 (±1.0) × 10−5 M in this study; 5 (±3) × 10−6 M[20]), primarily as a result of its slightly higher off rate.
Figure 2.
Surface plasmon resonance (SPR) experiments measuring the binding (association, and dissociation) kinetics of FA, MTX, and a MTX-presenting dendrimer 2c to bovine folate binding protein (FBP) immobilized onto a CM5 sensor chip. (a, b) SPR sensorgrams for monovalent ligands FA, and MTX, respectively, each injected at three different concentrations. (c) SPR sensorgrams for Ac-G5-(MTX)5.0 2c, injected at 2.0 μM (an upper curve), and 1.3 μM (a lower curve). (d) An illustration for fraction of dendrimer 2c desorbed at the dissociation phase (%decrease in ΔRU), each corresponding to the mean value calculated from the two SPR sensorgrams (c).
Table 1.
Rate constants and equilibrium dissociation constants (KD) of folic acid (FA), methotrexate (MTX), Ac-G5-(FA)8 (1), and Ac-G5-(MTX)5 (2c) to the folate binding protein measured by surface plasmon resonance spectroscopy.
| Ligands | FA | MTX | 1 | 2c |
|---|---|---|---|---|
| KD (M)a | 1.1(±0.50)×10−5 | 2.4(±0.11)×10−5 | 2.3(±0.35)×10−9 | 2.6(±1.0)×10−8 |
| kon (M−1s−1) | 1.1(±1.0)×103 | 7.0(±5.6)×102 | 2.9 (±2.2)×105 | 3.4(±2.5)×104 |
| koff (s−1) | 1.2(±0.2)×10−2 | 1.7(±0.6)×10−2 | 6.6 (±0.76)×10−4 | 8.7(±2.5)×10−4 |
| βb | 1 | 1 | 4820 (588c) | 923 (185c) |
Each dissociation constant (KD = koff/kon) represents a mean value calculated by averaging the data obtained from at least three sets of measurement per injection concentration;
β = multivalent binding enhancement = [KDmono ÷ KDmulti];
valency (n)-corrected value = [β ÷ n] where n is equal to either 8.2 (1) or 5.0 (2c).
Binding studies of dendrimer-based multivalent MTX conjugates 2a-d were then performed using the same FBP-presenting CM5 chip. SPR sensorgrams for the conjugates to the FBP surface are illustrated in Figure 2c by the conjugate 2c where each set of the sensorgrams shows the association and dissociation curve. PAMAM dendrimer negative controls, conjugated without either MTX or FA, were also prepared for binding and include a fully acetylated neutral dendrimer (Ac-G5) and a glutaric acid-terminated dendrimer (G5-glutaric acid). Neither of these dendrimers showed any meaningful level of specific adsorption to the FBP surface when measured under comparable conditions, indicating the specificity of the FBP binding (Figure S8). As an evidence indicative of tight adsorption, the SPR profiles of 2c are characterized markedly by slow dissociation (Figure 2c,d), a kinetic feature suggesting the multivalent binding as consistently observed in numerous other multivalent systems.[20,43,44] Such a binding feature was also observed similarly as the binding of Ac-G5-FA8.2, a multivalent dendrimer comparator presenting folate ligand as a positive control (Table 1; Figure S8).[20] The dissociation curves for 2c suggest that it dissociates apparently in multiple phases, initially at a rapid off rate (= Δ(ΔRU) ÷ Δt) and subsequently at slower off rates. For example, ~35% fraction of desorption was observed per initial 20s, followed by ~18% of fractional desorption per following 20–200s until the end of data collection time (200s), when the dissociation appears to still be incomplete (Figure 2d).
2.1.3. Correlation between dendrimer population and fractional desorption
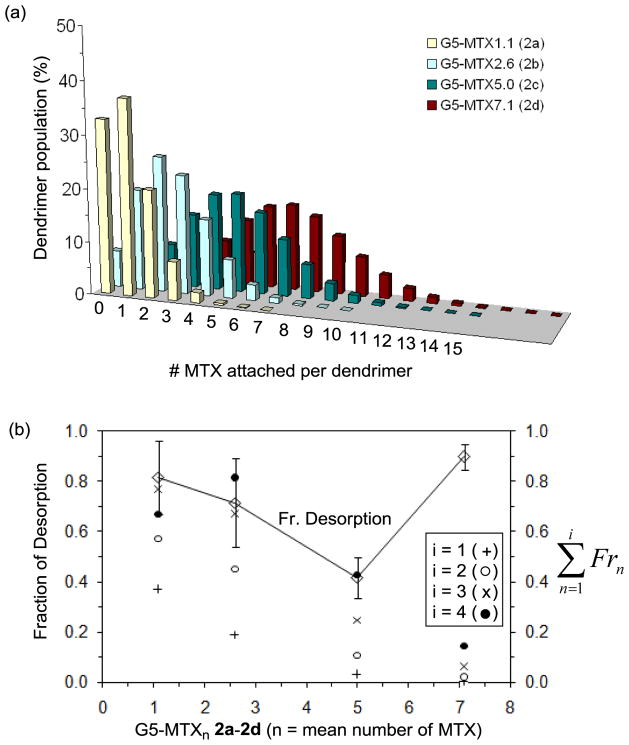
We hypothesize that such mixed dissociations observed from 2c could reflect the distribution of its subpopulations on the assumption that some subpopulations bind weakly and others bind more tightly and dissociate more slowly. The possibility of such mixed dissociations might be accounted for by the preexisting heterogeneity in the dendrimer population of 2c, in particular in regard to the distribution in ligand valency. While 2c Ac-G5-MTX5 is assigned to have five MTX ligands attached per dendrimer as calculated on an average basis, our recent studies[28,32] indicate that it is composed of diverse multivalent dendrimer species that can be grouped by their ligand density according to Poisson distribution (Figure 3a). Of those mixed dendrimers having variable ligand valency, we believe that the lower-valent species are likely to bind less tightly than the higher-valent species if predicted on the basis of the large number of related studies.[20–23] We address this hypothesis in Figure 3b, where the fractional desorption of G5-MTXn 2a – 2d is plotted as a function of the mean valency of the MTX for each conjugate. The plot shows a positive correlation for the three conjugates except for 2d, which showed otherwise deviation, perhaps due to its partial solubility, and suggests that the observed degree of desorption is greater for those conjugates having a lower MTX valency. Figure 3b also shows fractions ( ) for major low-valent species (n = 1 to 4) that are distributed in each of the conjugates, as calculated on the basis of their Poisson distribution (Figure 3a). It suggests that a fraction of dendrimer desorption could be accounted for further by the fraction of low-valent species—except for 2d. Specifically, the sum of fractions from monovalent to tri- or tetravalent species ( ; i = 3, or 4) is best correlated with the fraction of desorption within an experimental error range. A recent work by Waddell, et al.[41] hypothesizes that all desorption observed in the analogous G5-FA system is due to the dendrimer with a single ligand attached. This G5-FA work conforms to the i = 1 data ( Fr1) in Fig. 3b. Although the trend is similar in our work, greater agreement has been found for i = 3, where the desorption is more correlated with the fractions of dendrimers conjugated with up to 3 ligands.
Figure 3.
(a) Poisson distribution of dendrimer-methotrexate conjugates G5-MTXn 2a–2d, each having the MTX mean of 1.1, 2.6, 5.0, or 7.1, respectively. (b) A plot of fractional desorption of G5-MTXn 2a – 2d as a function of mean MTX number (valency). The fractional desorption was calculated from the SPR binding studies of the dendrimer conjugates. The sum of the fractions ( ) for multivalent species (G5-MTXn; n = 0 to 18) distributed in each of the dendrimer MTX conjugates is also plotted separately in the secondary y-axis. The value for Fr is defined as the fraction of n-valent species relative to all the species that comprise each conjugate. As an illustration, each value for (where i = 2) in 2a–2d refers to the sum of the two fractions for a monovalent (Fr 1) and divalent (Fr 2) species, each calculated by Poisson distribution of the conjugates above.
2.1.4. Determination of equilibrium dissociation constant (KD)
In an approach to better understand the complicated dissociation profiles for 2a-d, we estimate their dissociation constants by using global fittings (BIAevaluation software) that include Langmuir and bivalent analyte models. Each of these models is frequently employed for affinity analysis based on either a monovalent or bivalent binding mode and is also used for studies to determine reasonable estimates of the avidity constants for multivalent ligands,[43,46,48–50] including those that are nanoparticle-based.[20,47,51] Application of these analyses for the sensorgrams of Ac-G5-(MTX)5 2a-d allowed us to extract the estimates for their kinetic rate constants (kon, koff) and, consequently, for the steady state dissociation constants (KD) for the folate binding protein immobilized on the surface, as summarized in Table 1 and Figure 4 (see also Table S3). As an illustration, the dissociation constant estimated for 2c (KD = 2.6×10−8 M) suggests that its binding avidity is enhanced by a factor of ~900 (multivalent binding enhancement = β = [KDmono ÷ KDmulti]) relative to free MTX (KD = 2.4×10−5 M). Except for 2a (Ac-G5-MTXn: n = 1.1), which bound weakly, like a monovalent ligand, the other conjugates 2b and 2d showed the binding kinetics characterized by slow off rates (Table S3) with their dissociation constants (KD) lowered by two to three orders of magnitude relative to that of MTX (Figure 4). We believe that this observation is consistent with the hypothetical mechanism for multivalent association, where complete dissociation by a multivalent ligand occurs much more slowly because all of the ligands tethered to a single multivalent particle have to dissociate simultaneously from multiple receptor sites (observed off rate = koff, obsd ≈ kdn where kd refers to the mean rate of dissociation for a single receptor-ligand pair and n refers to the multivalency in bound states). [23,43,44] In summary, the SPR study demonstrates that the dendrimer-based multivalent MTX conjugate binds much more tightly to the multivalent FAR surface than free MTX. This SPR study provides evidence that FAR could be targeted by the MTX-based multivalent platform.
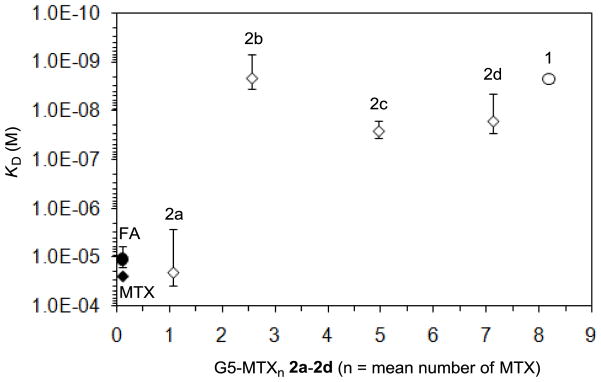
Figure 4.
A summary of equilibrium dissociation constants (KD, M) for dendrimer-based multivalent ligands (1, 2a-d) plotted against ligand valency (n). Dissociation constants for control ligands including folic acid (FA) and methotrexate (MTX) are placed arbitrarily around n ≈ 0 as the reference values indicative of monovalent association.
2.2. Design and synthesis of PAMAM dendrimers conjugated with MTX through a longer linker framework
After performing SPR binding studies at the receptor level, we proceeded to the biological assay at the cellular level for the dendrimers conjugated with MTX 2a-d to determine if these conjugates are biologically active in inhibiting cell growth. In a cell-based XTT assay using KB cancer cells, a sub-line of cervical carcinoma that expresses a high level of folate receptor,[52] all of these MTX conjugates were weakly cytotoxic at low micromolar concentrations (<10% inhibition of cell growth, data not shown). We hypothesized that the weak cytotoxicity might be attributable in part to the direct amide linkage used in 2a-d because this linker could be too stable for rapid release of the MTX, following the cellular uptake of the conjugates. In order to optimize the biological activity by the MTX conjugates at the cellular level, we designed a new MTX linker construct 5, made through two major modifications: i) the functional group at the linkage point that affects its chemical stability; ii) the tether length (Scheme 2). In this linker construct, we introduced a less stable ester linkage placed at the γ-carboxylate oxygen of the MTX L-glutamate and, in combination, a longer linker that consists of a 6-atom spacer terminated with a primary amine, the functional group introduced for conjugation with glutaric acid-terminated PAMAM dendrimer[53] later. The ester linkage is more likely hydrolyzed inside the cell by the actions of intracellular esterases and/or in acidic endosomes (pH ≈ 5 – 6.5) where FAR-associated conjugates are thought to be localized following cellular entry.[54,55] Therefore, under intracellular conditions, the ester linkage may be cleaved for drug release in a rate much faster than the amide linkage used in dendrimer conjugates 2. The longer spacer for MTX tethering, which is estimated to be 14.8 Å in distance from the dendrimer surface when calculated at a fully extended conformation (ChemBio3D Ultra 12.0), can provide the tethered MTX with more conformational flexibility of the linker and freedom of movement. Such a longer tether might play a critical role in the interaction at the enzyme-dendrimer interface when the tethered MTX molecule inhibits the catalytic activity of DHFR (cf. the crystal structure of hDHFR[56] in complex with a methotrexate molecule at its active site in Figure 5).[16,57]
Scheme 2.
(a) Synthesis of a new methotrexate-linker construct. Reagents and conditions: i) bromoacetyl chloride, DIPEA, CHCl3, 0°C to rt, 24 h, 64%; ii) methotrexate hydrate, Cs2CO3, K2CO3, NaI, DMF, rt, 48 h, 22%; iii) TFA, CH2Cl2, rt, 15 min; (b) Synthesis of generation 5 (G5) PAMAM dendrimer-based nanoconjugates 7a,b, 8a-d, and 9a,b with variable amounts of methotrexate attached singly or in combination with folic acid molecules. iv) EDC, NHS, DMF, rt, 36 h; v) 5, Et3N, DMF, rt, 24h; then ethanolamine; vi) 5, FA-CONH(CH2)2NH2, Et3N, DMF, rt, 24h; ethanolamine; vii) 5, fluorescein-NHC(=S)NH(CH2)4NH2, Et3N, DMF, rt, 24h; ethanolamine.
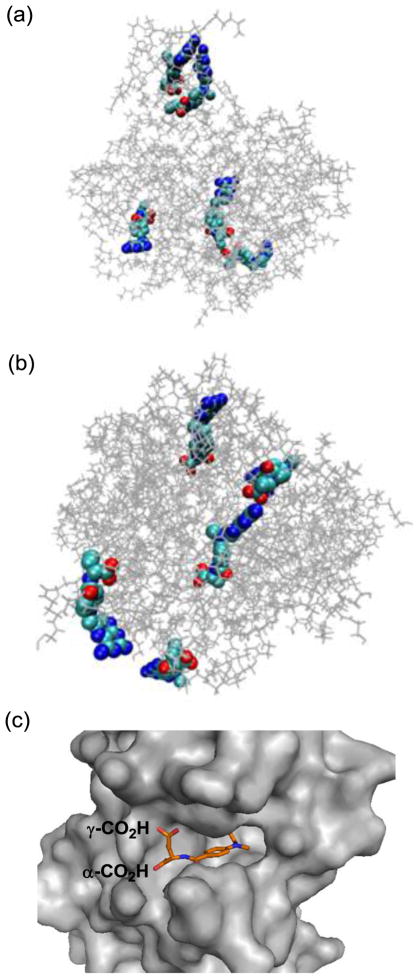
Figure 5.
(a, b) Molecular models generated by Molecular Dynamics simulation for the representative members of generation 5 PAMAM dendrimer conjugated with methotrexate, either directly via an amide bond (2c, Scheme 1) or through a medium length of spacer (7a, Scheme 2). Distribution of MTX molecules on each of the dendrimers was selected arbitrarily for this simulation purpose, and thus should be considered as one of the potential distributions. Final configurations for 2c (a) and 7a (b) were acquired after 1-ns MD simulations. The figures of the structures were generated with the software VMD (visual molecular dynamics) where the PAMAM dendrimer and MTX were shown as lines and van der Waals surface, respectively (Colors: grey: dendrimer, light blue: carbon, blue: nitrogen, red: oxygen); (c) A crystal structure of human dihydrofolate reductase (hDHFR) in complex with a methotrexate molecule at its active site (PDB code 1u72), where the L-glutamate carboxylic acids from the drug molecule are anchored near the entrance to the enzyme catalytic pocket, while a pteridine head group (hidden) is bound deep into the pocket.
2.2.1. Molecular Dynamics (MD) simulation of dendrimer-MTX conjugates
In an effort to corroborate this hypothesis for the structural features of the PAMAM dendrimers and the role played by the linker, we constructed molecular models for the two representative members of MTX-conjugated PAMAM dendrimers, each conjugated either directly via an amide linkage (2c, Scheme 1) or through the longer spacer (7a, the dendrimer conjugated with the longer linker construct 5, Scheme 2). Figure 5 shows these two models generated by Molecular Dynamics (MD) simulations based on an implicit solvent model and using the distribution of MTX molecules that were selected arbitrarily for the simulation purpose.[58,59] The radius of gyration estimated for each of the dendrimers, 2c and 7a, is 20.6 ± 0.2 and 22.0 ± 0.2 (Å), respectively. The model for 7a suggests that the longer linker is flexible in the conformation and can provide a variable distance for the spacer from the dendrimer surface. Accordingly, it can provide each of the attached MTX molecules with a greater level of exposure to the solvent.
2.2.2. Synthesis of dendrimer conjugated with MTX via longer linker
MTX linker 5 was then prepared using a two-step process based on the synthesis of 4 by the O-alkylation of the MTX L-glutamate with a bifunctional linker 3 (Scheme 2a; regioselective ratio for γ-ester to α-ester = 10.2 after purification). Subsequent deprotection of the N-Boc group with the TFA led to 5 isolated as a TFA salt. Synthesis of G5 PAMAM dendrimers conjugated with the new MTX linker construct (7a,b, 8a-d, and 9a,b) was performed by the covalent attachment of 5 to the G5-CO2H 6 (Mn = 42730 gmol−1, PDI = Mw/Mn ~1.046)[53] through an amide linkage (Scheme 2b). Members of the conjugates include 8a-d, which contain MTX as well as FA molecules co-attached as the targeting ligand. Other dendrimer conjugates include 9a,b, each containing FITC as a fluorescent imaging molecule. Each of the mentioned dendrimers was synthesized by a one-pot reaction process in which the carboxylic acids located at the periphery of the PAMAM dendrimer (~100 CO2H per dendrimer molecule) were preactivated to N-hydroxysuccinimide esters and coupled with 5, singly or in combination with FA-CONH(CH2)2NH2[60] and/or fluorescein isothiocyanate (FITC)-diaminobutane as an imaging molecule[61]. Each of the resultant dendrimer conjugates was purified by extensive dialysis using membrane tubing (MWCO 10 kDa) against phosphate-buffered saline (PBS) solution and then water and was characterized by analytical methods including MALDI-TOF mass spectrometry, 1H NMR spectroscopy, UV/Vis spectrometry, and analytical HPLC (purity >96%) developed in our laboratories. Analysis of these data allowed us to calculate the number of FA, MTX, and imaging molecules attached per dendrimer particle on a mean basis (Table S2). The molecular masses for these dendrimer conjugates are in the range of 45,000–48,000 (gmol−1): 7a = 45,700, 7b = 47,200 (PDI = 1.103 by gel permeation chromatography), 8a = 46,700, 8b = 47,200, 8c = 47,800, 8d = 48,100, 9a = 46700, 9b = 48,350.
2.3. Cell-based experiments in KB cancer cells
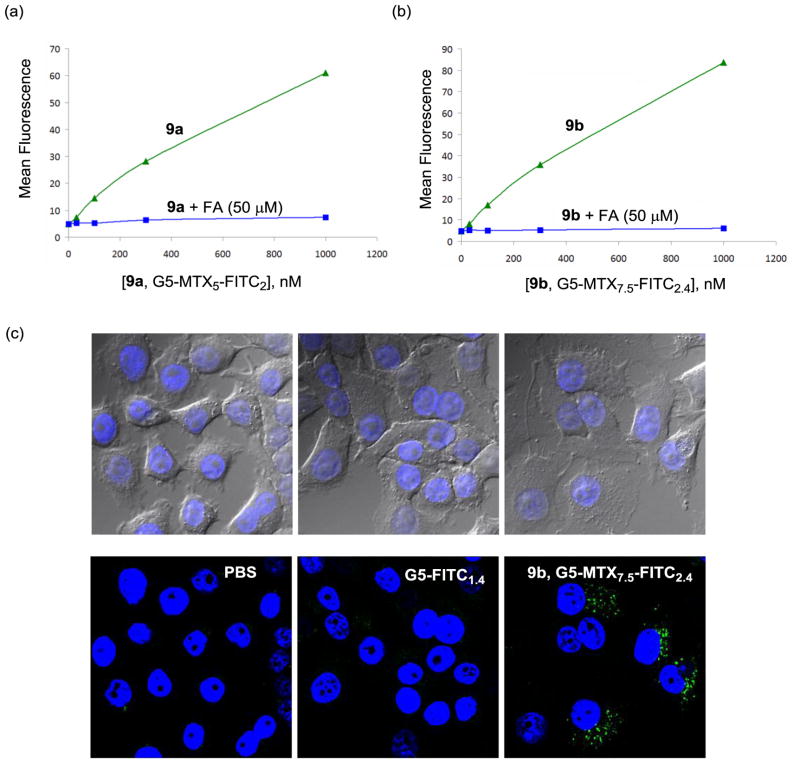
First, we performed cellular binding and uptake in KB cells that express a high level of folate receptor (FAR),[52] using fluorescent dendrimer conjugates 9a and b, each presenting MTX alone without FA attached. Figure 6 shows that each of these conjugates bound to the cells in a dose-dependent manner at concentrations up to 1000 nM. In addition, under this binding condition, considered identical or comparable, 9b (7.5 MTX per dendrimer) showed a slightly greater level of cellular association than 9a (5 MTX per dendrimer). The binding activity of each of the MTX conjugates could be blocked completely when each conjugate was co-incubated with free FA dosed at a higher concentration (50 μM). However, it is notable that the dendrimers conjugated with MTX alone did not show concentration-dependent saturation curves, a phenomenon which is often displayed by FA-presenting dendrimer conjugates under comparable assay conditions.[26,52,62] It is possible that the lack of such binding saturation might be attributable to a lower avidity binding in combination with a slower rate of cell uptake, compared to FA-mediated cell targeting. The uptake of the conjugate 9b in KB cells was then studied by confocal microscopy, as illustrated in Figure 6c in which the green fluorescence in the cytoplasm indicates that the dendrimer particles were internalized. The results from the cell binding studies suggest that FAR molecules are involved in the surface receptor targeted by these MTX conjugates.
Figure 6.
(a,b) Dose-dependent binding and uptake of PAMAM dendrimer conjugates 9a,b in KB cells. The cells were incubated with each of the conjugates at different concentrations for 2 h, rinsed and measured for its mean fluorescence in a flow cytometer. For competitive ligand displacement experiments, the cells were treated with the conjugates under the condition identical to the above except in the presence of free folic acid (50 μM); (c) Confocal microscopy of KB cells treated with 9b (G5-MTX7.5-FITC2.4; 100 nM). KB cells were incubated with the indicated conjugate for 18 h, fixed and treated with a nuclear staining agent, DAPI (4′,6-diamidino-2-phenylindole) prior to the measurement of DAPI (blue) and FITC (green) fluorescence using a confocal microscope. Control experiments were performed under an identical condition using G5-FITC1.4 (300 nM) as a non-targeting dendrimer conjugate, and PBS alone. An optical microscopic image for each confocal image is shown in the upper part.
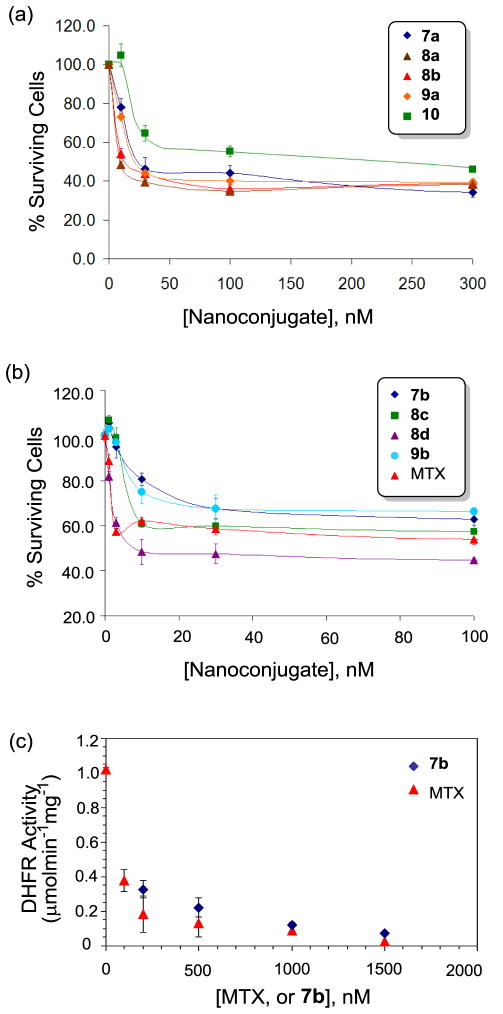
Second, we evaluated the cytotoxicity of the dendrimer-MTX conjugates 7a, 7b, 8a-d, 9a, and 9b, each conjugated with the new MTX linker, against FAR-overexpressing KB cells in an XTT assay.[29] As shown in Figure 7a, the dendrimer conjugates 7a and 9a, each presenting MTX molecules alone even without FA, potently inhibited cell growth in a dose-dependent manner in which a maximal inhibition (~50% inhibition of cell growth) was achieved, each at ~25 nM. In contrast, negative controls (such as a fully acetylated PAMAM dendrimer, and other dendrimers conjugated with FA and/or dye alone) did not show any cytotoxicity to KB cells under an identical assay condition (not shown)[10,52]. Each of the conjugates 7a, 9a was more cytotoxic than 10 (G5-FA5-MTX7.5), the internal standard having FA ligands co-attached directly through the amide bond.[1,52] This positive control is structurally analogous to 1 (Scheme 1) but contains fewer FA (5 copies per dendrimer) as well as MTX (7.5 per dendrimer) molecules which are attached in an ester linkage.[29,52] Figure 7a also shows cytotoxic activities for other analogous MTX conjugates, 8a and 8b, as the positive controls, each targeted by co-attached FA ligands. Co-presentation of FA in each of the comparators led to a slight increase in the inhibition activity, possibly as a result of the targeting effect contributed by FA ligands during the cellular uptake. The other series of dendrimer MTX conjugates—7b, 8c, 8d, and 9b—each having a higher number of MTX molecules (7.5) carried per dendrimer, also showed potent cytotoxicity against the FAR-overexpressing KB cells. These conjugates were generally more active than those comparable conjugates otherwise having a lower number of MTX molecules (5) (Figure 7b). While it is consistently observed that presentation of MTX alone in the conjugates 7b and 9b was sufficient to induce cytotoxicity, the MTX conjugates 8c and 8d that present FA as well were more active than those without co-attached FA. Also, one of the conjugates, 8d, was more potent than free MTX, an activity which is rarely observed among the standards of our previous results from other FA-targeted MTX conjugates.[29,52] In summary, the cell assay demonstrated that PAMAM dendrimers conjugated with MTX alone could cause potent cytotoxicity in KB cells with the IC50 values of 25 (7a, G5-MTX5 series) to 10 nM (7b, G5-MTX7.5 series) calculated on a conjugate basis. The co-attachment of FA as a targeting ligand led to a synergy in the biological activity and led to greater cytotoxicity. This suggests that a MTX molecule tethered to a dendrimer nanoparticle is both a ligand required for targeting a FAR-expressing cell and a drug molecule that kills the cancer cell. The level of cellular activity appeared to be tunable in response to the variation of the MTX valency and the co-presentation with FA ligands.
Figure 7.
(a,b) Cytotoxicity of G5 PAMAM conjugates 7a,b, 8a-d, 9a,b, and 10 in FAR-over expressing KB cells. KB cells were incubated with each of the dendrimer conjugates for 2 d, and allowed to grow further for 2 d. The level of cytotoxicity was quantified by a colorimetric assay based on XTT (sodium 3-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis(4-methoxy-6-nitro)benzene sulfonic acid hydrate); (c) Inhibition of human dihydrofolate reductase (hDHFR) by 7b (G5-MTX7.5) conducted in a cell-free enzyme assay. The unit for DHFR activity is defined as μmol×min−1×mg−1, and concentrations for 7b on the X-axis are given on the basis of MTX rather than dendrimer conjugate.
2.4. Dihydrofolate reductase (DHFR) inhibition assay
Our present studies based on SPR and cell-based binding experiments provided important pieces of evidence suggesting that methotrexate molecules attached to the dendrimer carrier could target FAR for cancer-specific delivery. However, details that define the mechanisms of drug action after cellular entry are less understood including the release kinetics of MTX inside the cell. Previously we showed in a cell-free enzyme assay using human DHFR that this enzyme could be inhibited by free MTX as well as by a MTX-conjugated PAMAM dendrimer 10 (Ac-G5-FA-MTX) in the assay condition where MTX molecules are still covalently attached.[63] In a similar effort to understand the enzyme inhibitory activity by the current set of dendrimer-MTX conjugates, we chose 7b and investigated whether it is able to inhibit the human DHFR. In a standard enzyme assay condition as described elsewhere,[63] 7b potently inhibited the enzyme activity, though less potently than free MTX (Figure 7c). This result is in close agreement with our previous observation from the conjugate 10.[63] We examined the stability of 7b against the hydrolytic release of MTX under the assay condition (PBS, pH 7.4, ambient temperature, incubation time = 5 min.). The stability test showed that MTX was not released from the conjugate to any meaningful extent (<2% over 48 hours). This indicates that the dendrimer conjugate inhibits DHFR activity even if MTX molecules are still tethered. The inhibition activity documented in 7b is supported by numerous structure-activity relationship studies performed with various types of MTX derivatives. For example, the activity of human DHFR is potently inhibited by MTX γ-glutamate esters[64,65] and its poly-γ-glutamates,[66] the metabolites that are formed from free MTX in the cell. However we speculate that this in vitro access is quite different from the in vivo activity where MTX could be cleaved much faster by cytosolic esterases and/or by chemical mechanisms after being internalized to the acidic compartments of the cell.[54] In summary, the enzyme assay shows potent inhibition of the DHFR by the intact MTX conjugate as another possible mechanism of drug action in addition to the MTX release.
3. Conclusion
We investigated a simplified strategy for cancer-targeted delivery by using MTX as a dual-acting molecule, serving as both a chemotherapeutic and a targeting agent. These MTX-functionalized PAMAM dendrimers effectively kill and also target cancer cells. Our SPR study proves that the multivalent dendrimer-MTX conjugate binds FAR on the surface. The cell-based studies provide evidence that the MTX nanoconjugates are internalized through FAR because the MTX on the dendrimer cannot get through the reduced folate carrier because of its nanometer size. In conclusion, this approach provides novel bifunctional capabilities for cancer therapy, demonstrates the MTX-mediated cytotoxicity and targeting to cancer cells, and suggests broad applicability for other MTX-like pemetrexed and raltitrexed antifolate metabolites[67] as potential anticancer therapeutics.
4. Experimental Section
4.1 General
General synthetic methods, and the details of instrumental analysis for the characterization of small molecules and dendrimers by using 1H, and 13C NMR spectroscopy, MALDI TOF mass spectrometry, gel permeation chromatography (GPC), and analytical HPLC are provided in the Supplementary Material (pages S2–3).[10,29,53,68]
4.2. Synthesis of MTX-linker 5
To a solution of methotrexate hydrate (0.684 mg, 1.45 mmol) in anhydrous DMF (35 mL) was added cesium carbonate (708 mg, 2.17 mmol), and potassium carbonate (0.3 g, 2.17 mmol). While stirring this mixture under nitrogen atmosphere, N-(bromoacetyl)-N′-Boc-1,3-diaminopropane 3[69] (0.47 g, 1.59 mmol) and sodium iodide (0.217 g, 1.45 mmol) were added to it. The final mixture was stirred at room temperature for 48 h under nitrogen atmosphere. The mixture was concentrated in vacuo, yielding yellow oily material. It was diluted with 15 mL of 10% MeOH/CH2Cl2, and purified by flash column chromatography on silica gel (100 g) by eluting with 10% MeOH/CH2Cl2 to 2% AcOH/15% MeOH/CH2Cl2. Fractions with an Rf value of 0.27 (2% AcOH in 20% MeOH/CH2Cl2) were combined and evaporated to afford the desired product 4 as yellow solid (213 mg, 22%). An HPLC analysis of the purified product suggested that the regioselectivity of the O-alkylation is 10.2 (γ-ester to α-ester). 1H NMR (400 MHz, DMSO-d6): δ 8.53 (s, 1H), 8.45-8.44 (d, NH-Glu, 1H, J = 7.2 Hz), 7.82-7.80 (t, 1H, J = 5.6 Hz), 7.70-7.68 (d, 2H, J = 8.8 Hz), 6.78-6.76 (d, 2H, J = 8.8 Hz), 6.73-6.70 (m, 1H), 4.74 (s, 2H), 4.42 (s, 2H), 4.36-4.35 (m, 1H), 3.17 (s, 3H), 3.02-2.88 (m, 2H), 2.86-2.83 (m, 1H), 1.94-1.85 (m, 1H), 1.48-1.44 (quin, 2H, J = 6.8 Hz), 1.30 (s, 9H) ppm. 13C NMR (100 MHz, DMSO-d6): δ 174.38, 172.23, 167.36, 166.90, 163.15, 151.58, 149.54, 129.53, 122.0, 120.88, 111.54, 77.94, 62.94, 55.28, 52.69, 36.63, 30.59, 29.90, 28.70, 26.11, 14.36 ppm. MS (ESI): m/z (relative intensity, %) = 669.3 (100) [M+H]+, 569.3 (56) [M-Boc+H]+. HRMS (ESI) calcd for C30H41N10O8 669.3109, found 669.3123. The N-Boc group in 4 was deprotected by treating with TFA. Typically 4 (10 mg, 15 μmol) was mixed with 50% TFA/CH2Cl2 (1 mL) and stirred for 15 min at room temperature. Evaporation of the solution in vacuo afforded 5 as pale yellow oily residue. It was used immediately for next step. 1H NMR (400 MHz, CD3OD): δ 8.60 (s, 1H), 7.74-7.72 (d, 2H, J = 7.2 Hz), 6.84-6.81 (d, 2H, J = 7.2 Hz), 4.89 (s, 2H), 4.66-4.62 (d, 1H, J = 15.2 Hz), 4.55-4.51 (d, 1H, J = 15.2 Hz), 4.53-4.49 (t, 1H, J = 5.6 Hz), 3.34-3.31 (t, 2H, J = 5.6 Hz), 3.24 (s, 3H), 2.93-2.89 (t, 2H, J = 5.6 Hz), 2.49-2.46 (t, 2H, J = 5.6 Hz), 2.26-2.23 (m, 1H), 2.16-2.12 (m, 1H), 1.88-1.81 (quin, 2H, J = 5.6 Hz). 13C NMR (100 MHz, CD3OD): δ 175.20, 171.76, 169.33, 163.51, 156.37, 152.25, 151.75, 148.93, 145.51, 128.99, 121.98, 120.72, 116.86, 115.76, 113.50, 111.34, 62.62, 55.19, 52.94, 36.76, 35.36, 29.85, 29.80, 27.17, 25.49, 15.19 ppm. MS (ESI): m/z (relative intensity, %) = 569.3 (100) [M+H]+, 1137.5 (3) [2M+H]+. HRMS (ESI) calcd for C25H33N10O6 569.2585, found 569.2573.
4.3. A representative synthesis for G5-MTXn 7b (n = 7.5)
To a suspension of G5-(CO2H)100 6[53] (50 mg, 1.24 μmol) in anhydrous DMF (15 mL) was added 4-dimethylaminopyridine (15.2 mg, 125 μmol), N-hydroxysuccinimde (14.4 mg, 125 μmol), and N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC, 23.8 mg, 124 μmol). After stirring for 36 h at room temperature, methotrexate-linker 5 (18.7 μmol, 15 eq to G5 dendrimer) dissolved in 1 mL of DMF and triethylamine (6.3 mg, 50 eq to G5 dendrimer) were added to the reaction mixture, and the mixture continued to be stirred for 24 h at room temperature. At that time, the mixture was treated with ethanolamine (15.2 mg, 249 μmol) and the final mixture was stirred for an additional period of 6 h. At the conclusion of the reaction, this mixture was concentrated in vacuo to ~3 mL in volume and diluted with 20 mL of phosphate-buffered saline (PBS, pH 7.2). The solution was loaded into a membrane dialysis tubing (MWCO 10 kDa), and dialyzed against PBS (4L×2), and deionized water (4L×2) over 3 days. The duration of dialysis performed here was not optimized and could be reduced to fewer days by replacing each dialysis solution more frequently until the purity of the dialyzed solution becomes acceptable by analytical HPLC. The aqueous solution was collected and lyophilized to afford G5-MTX7.5 as pale yellow solid (37 mg) and in 98% purity based on analytical HPLC analysis. MALDI TOF mass spectrometry: m/z = 47200 gmol−1. GPC: Mw = 47280 gmol−1, PDI = 1.103. UV/vis (PBS, pH 7.2): 365 (λmax), 305, 258 nm. 1H NMR (400 MHz, DMSO-d6): δ 8.4 (s), 8.05-7.9 (broad s), 7.9-7.75 (broad d), 7.7-7.6 (m), 6.75 (broad), 4.75 (s), 4.4 (s), 4.35 (m), 4.2-3.9 (broad), 3.6 (broad s), 3.2-2.8 (m), 2.7-2.0 (m), 1.65 (m), 1.5-1.4 (m) ppm. The resultant nanoconjugate contains 7.5 methotrexate covalently attached to the surface of PAMAM G5 dendrimer per mean basis as calculated by dividing the difference in the molecular weights between two directly comparable conjugates: .
Other analogous conjugate G5-MTXn 7a (n = 5) was prepared in a similar manner otherwise using a lower amount of 5 (15 μmol, 12 eq to 6) while adjusting the amounts of other reagents accordingly. MALDI TOF mass spectrometry: m/z = 45700 gmol−1. The number (n) of methotrexate covalently attached to the dendrimer surface is estimated to be 5, and on a mean basis.
4.4. A representative synthesis for G5-MTXn-FAm 8c (n = 7.5, m = 1.2)
G5-(CO2H)100 6 (50 mg, 1.24 μmol) was activated to its NHS ester and reacted with 5 (18.7 μmol) as described above for 7b. Prior to quenching with ethanolamine, FA-CONH(CH2)2NH2[60] (0.9 mg, 1.9 μmol) suspended in 1 mL of DMF (or dissolved in DMSO by brief sonication) was added to the reaction mixture, and the mixture was stirred for 24 h at ambient temperature, and followed by treatment with ethanolamine (15.2 mg, 249 μmol). The final mixture was stirred for additional 6 h at room temperature, and the mixture was concentrated in vacuo to ~3 mL in volume. After dilution with 20 mL of phosphate-buffered saline (PBS, pH 7.2), the solution was loaded into a membrane dialysis tubing (MWCO 10 kDa), and dialyzed against PBS (4L×2), and deionized water (4L×2) over 3 days. The aqueous solution was collected and lyophilized to afford G5-MTX7.5-FA1.2 as pale yellow solid (35 mg) and in 98% purity based on analytical HPLC. MALDI TOF mass spectrometry: m/z = 47800 gmol−1. 1H NMR (400 MHz, DMSO-d6): δ 8.55 (broad), 8.5 (s), 8.1-7.9 (broad), 7.9-7.75 (broad d), 7.7-7.6 (m), 6.75 (broad), 6.6-6.5 (broad), 4.7 (s), 4.4 (s), 4.35-4.1 (m), 3.9-3.4 (broad), 3.2-2.8 (m), 2.7-2.0 (m), 1.65 (m), 1.5-1.4 (m) ppm. The nanoconjugate contains 1.2 folic acid covalently attached to PAMAM G5 dendrimer on a mean basis as calculated by dividing the molecular weight difference between two directly comparable conjugates: . The number (m) for attached FA could be also estimated from the 1H NMR spectrum where the relative ratio between MTX and FA was calculated by integration analysis (m ≈ 1.18), and is in good agreement with the number estimated from the previous mass data.
G5-MTXn-FAm 8d (n = 7.5, m = 2.6) was prepared in a manner similar to 8c otherwise using a higher amount of folic acid-ethylenediamine amide (1.8 mg, 3.7 μmol). MALDI TOF mass spectrometry: m/z = 48100 gmol−1. Average number of folic acid covalently attached to the surface of PAMAM G5-glutaric acid is 2.6 (MALDI-based), and 2.0 (1H NMR-based) on a mean basis.
Two other conjugates of this class G5-MTXn-FAm 8a (n = 5, m = 2) and 8b (n = 5, m = 3.5) were also prepared following the above procedure for 8c except that different amounts of 5 (15 μmol, 12 eq to G5-CO2H) and FA-CONH(CH2)2NH2 (m = 2: 2.5 μmol; m = 3.7: 5.0 μmol) were used in each of the conjugation reactions.[60]
4.5. A representative synthesis for G5-MTXn-FITCp 9b (n = 7.5, p = 2.4)
G5-(CO2H)100 (50 mg, 1.24 μmol) was activated to NHS ester and reacted with methotrexate-linker 5 (18.7 μmol) as described above for 7b. Prior to quenching with ethanolamine, fluorescein-butane-1,4-diamine[61] (2.4 mg, 5.0 μmol) suspended in 1 mL of DMF was added to the mixture and stirred for 24 h at ambient temperature. The mixture was then added with ethanolamine (15.2 mg, 249 μmol) and stirred for additional 6 h. At the conclusion of the reaction, the mixture was concentrated in vacuo to ~3 mL in volume and diluted with 20 mL of phosphate-buffered saline (PBS, pH 7.4). The solution was loaded into a membrane dialysis tubing (MWCO 10 kDa), and dialyzed against PBS (4L×2), and deionized water (4L×2) over 3 days. The aqueous solution was collected and lyophilized to afford G5-MTX7.5-FITC2.4 as pale orange solid (39 mg) and in 97% purity based on analytical HPLC. MALDI TOF mass spectrometry: m/z = 48000 gmol−1. UV/Vis (PBS, pH 7.2): 498 (λmax), 364, 305, 258 nm. 1H NMR (400 MHz, DMSO-d6): δ 8.5 (s), 8.3-8.2 (broad), 8.1-7.9 (broad), 7.9-7.75 (broad d), 7.7-7.6 (m), 7.1-7.0 (m), 6.8-6.65 (broad), 6.6 (s), 6.5-6.4 (m), 4.7 (s), 4.4 (s), 4.3-4.1 (m), 4.1-4.1 (broad), 4.0-3.9 (broad), (broad), 3.75-3.4 (m), 3.2-2.8 (m), 2.7-1.8 (m), 1.65 (m), 1.5-1.4 (m), 1.3 (m) ppm. This conjugate contains 2.4 fluorescein covalently attached to PAMAM dendrimer on a mean basis: . The number (p) for attached FITC was also estimated from the relative ratio between MTX and FITC based on the analysis of 1H NMR spectrum: p (FITC) ≈ 2.7 on a mean basis.
G5-MTXn-FIp 9a (n = 5, p = 2) was prepared in a manner similar to 9b otherwise using a lower amount of 5 (15 μmol, 12 eq to G5-CO2H). MALDI TOF mass spectrometry: m/z = 46700 gmol−1. The number (p) of fluorescein covalently attached to the surface of PAMAM G5-glutaric acid is estimated to be 2.3 (MALDI-based), and 2.4 (1H NMR-based) on a mean basis.
4.6. Surface plasmon resonance (SPR) spectroscopy
SPR experiments were performed in Biacore® X (Pharmacia Biosensor AB, Uppsala, Sweden). Folate binding protein (FBP, bovine milk) was immobilized on the surface of a CM5 sensor chip through protein conjugation chemistry to a carboxymethylated dextran-coated layer on gold following a standard amide coupling protocol.[20] The immobilization process of FBP on channel 2 resulted in 9500 response unit (RU) equivalent to 9.5 ng/mm2. SPR signals for FBP binding were obtained by injection of each ligand dissolved in HBS-EP buffer at a flow rate of 30 μL/min (FA, MTX), or 50 μL/min (dendrimer conjugates 1, 2). After each measurement, the surface of the chip was regenerated by injection of 10 μL of 10 mM glycine-HCl (pH 2.5). SPR sensorgrams needed to evaluate binding parameters for each ligand were obtained from the SPR signals from channel 2 subtracted by those from channel 1 (ΔRU = RU2 − RU1). Kinetic binding parameters, the rate of association (kon), and the rate of dissociation (koff), were extracted by fitting each binding curve separately using the Langmuir kinetic model as described[20] in Biacore evaluation software and were analyzed (dR/dt vs. R) to ensure the absence of the effect associated with mass transport as discussed elsewhere.[70] Dissociation constants (KD = koff/kon) for each ligand were calculated by averaging the data obtained from at least three sets of measurement per injection concentration which had chi square (χ2) values lower than 5.
4.7. Molecular Dynamics (MD) simulations
MD simulations on G5 PAMAM dendrimers (2c and 7a) were carried out using the CHARMM program based on an implicit solvent model with a 1 fs time step at 300 K.[58] The dendrimer parameters were obtained from the CHARMM parameters for generic proteins.[59] The initial structures for the dendrimers were produced by a recursive script in CHARMM. After steepest descent and adopted basis Newton-Raphson minimizations for 10000 steps, the structures were further annealed at high temperature and cooled down to room temperature to obtain lower energy configurations. The systems were equilibrated for 200 ps and then molecular dynamics were carried out for 600 ps. The total potential energy function (Utotal) for MD simulations is described as,
where ε is the minimum energy of the Lennard-Jones potential, σ the distance yielding a minimum Lennard-Jones potential, q the partial charge on the atom, D the dielectric constant, r the distance between i and j and i, j are nonbonded atom pairs. Electrostatic interactions were modeled by a distance-dependent dielectric function of the type E(r) = 4r without a long-range nonbonded cut-off in the simulations.[71]
4.8. Flow cytometry
KB Cells were incubated in the presence of the different concentrations of dendrimer methotrexate conjugates 9a-b for 2 h and the mean FL1 fluorescence of 10,000 cells were taken by flow cytometry.[29,52] Competitive binding experiments with FA were performed by pre-incubating with 50 uM of free FA for 15 min prior to adding the conjugates.
4.9. Confocal fluorescence microscopy
KB Cells were incubated in the presence of 9a (300 nM) for 2 h and the cells were fixed and stained for DAPI. The DAPI (blue) and FITC (green) fluorescence were measured using a confocal microscope.[29,52]
4.10. In vitro cytotoxicity assay
Cytotoxicity of free MTX, and PAMAM dendrimer nanoconjugates (7a-b, 8a-d, 9a-b, 10) was measured using XTT assay in KB cells, a sub-line of the cervical carcinoma HeLa cells (ATCC, Manassas, VA, USA) as described elsewhere.[29,52] The cells were grown as a monolayer cell culture at 37 °C and under 5% CO2 in folic acid-deficient RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS). The 10% FBS provided folic acid concentration equivalent to that present in the human serum (~20 nM). For the cytotoxicity experiments, the cells were seeded in 96-well microtiter plates (3000 cells/well) in serum-containing medium. Two days after plating, the cells were treated with different concentrations of MTX or conjugates in tissue culture medium for 4 days. A colorimetric XTT (sodium 3-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis (4-methoxy-6-nitro) benzene sulfonic acid hydrate) assay (Roche Molecular Biochemicals, Indianapolis, IN) was performed following the vendor’s protocol. After incubation with the XTT labeling mixture, the microtiter plates were read on an ELISA reader (Synergy HT, BioTek) at 492 nm with the reference wavelength at 690 nm.
4.11. Dihydrofolate reductase (DHFR) assay
MTX and 7b were tested for the inhibitory activity against human dihydrofolate reductase (Sigma assay kit) according to the assay protocol as provided.[63] The assay was performed at room temperature by following the decrease of NADPH and dihydrofolate concentrations through absorbance measurements at 340 nm at different concentrations of the inhibitor as indicated in Figure 5c. The ΔOD (min−1) required for calculating specific enzyme activity (Units/mg P = μmolmin−1mg−1) in each of the inhibition reactions was obtained through a linear fitting in the initial phase of reaction kinetics (0–2.5 min).
Supplementary Material
Acknowledgments
This work was supported by the National Cancer Institute, National Institutes of Health under award 1 R01 CA119409 (JRB). We thank Dr. Pascale Leroueil for the proofreading of this manuscript.
Abbreviation List
- FA
folic acid
- FAR
folic acid receptor
- MTX
methotrexate
- PAMAM
polyamidoamine
- SPR
surface plasmon resonance
- DHFR
dihydrofolate reductase
Footnotes
Supplementary Material Available: Two tables (SPR kinetic parameters for 2; characterization data of 6, 7, 8, 9), SPR sensorgrams (1, 2), and spectral data (1H NMR spectra, mass or MALDI-TOF spectra, and analytical HPLC chromatograms) for 4, 5, and 7-9 are provided. These materials can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kukowska-Latallo JF, Candido KA, Cao Z, Nigavekar SS, Majoros IJ, Thomas TP, Balogh LP, Khan MK, Baker JR., Jr Nanoparticle Targeting of Anticancer Drug Improves Therapeutic Response in Animal Model of Human Epithelial Cancer. Cancer Res. 2005;65:5317–5324. doi: 10.1158/0008-5472.CAN-04-3921. [DOI] [PubMed] [Google Scholar]
- 2.Low PS, Henne WA, Doorneweerd DD. Discovery and Development of Folic-Acid-Based Receptor Targeting for Imaging and Therapy of Cancer and Inflammatory Diseases. Acc Chem Res. 2008;41:120–129. doi: 10.1021/ar7000815. [DOI] [PubMed] [Google Scholar]
- 3.Myc A, Majoros IJ, Thomas TP, Baker JR., Jr Dendrimer-Based Targeted Delivery of an Apoptotic Sensor in Cancer Cells. Biomacromol. 2007;8:13–18. doi: 10.1021/bm060815l. [DOI] [PubMed] [Google Scholar]
- 4.Pan X, Lee RJ. Tumour-selective drug delivery via folate receptor-targeted liposomes. Expert Opin Drug Delivery. 2004;1:7–17. doi: 10.1517/17425247.1.1.7. [DOI] [PubMed] [Google Scholar]
- 5.Majoros IJ, Williams CR, Baker J., Jr Current Dendrimer Applications in Cancer Diagnosis and Therapy. Curr Top Med Chem. 2008;8:1165–1179. doi: 10.2174/156802608785849049. [DOI] [PubMed] [Google Scholar]
- 6.Majoros IJ, Williams CR, Becker A, Baker JR., Jr Methotrexate delivery via folate targeted dendrimer-based nanotherapeutic platform. WIREs: Nanomed Nanobiotech. 2009;1:502–510. doi: 10.1002/wnan.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilgenbrink AR, Low PS. Folate receptor-mediated drug targeting: From therapeutics to diagnostics. J Pharm Sci. 2005;94:2135–2146. doi: 10.1002/jps.20457. [DOI] [PubMed] [Google Scholar]
- 8.Lee RJ, Low PS. Folate-mediated tumor cell targeting of liposome-entrapped doxorubicin in vitro. Biochim Biophys Acta (BBA) - Biomembranes. 1995;1233:134–144. doi: 10.1016/0005-2736(94)00235-h. [DOI] [PubMed] [Google Scholar]
- 9.Low PS, Kularatne SA. Folate-targeted therapeutic and imaging agents for cancer. Curr Opin Chem Biol. 2009;13:1–7. doi: 10.1016/j.cbpa.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 10.Thomas TP, Choi SK, Li MH, Kotlyar A, Baker JR., Jr Design of Riboflavin-presenting PAMAM Dendrimers as a New Nanoplatform for Cancer-targeted Delivery. Bioorg Med Chem Lett. 2010;20:5191–5194. doi: 10.1016/j.bmcl.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plantinga A, Witte A, Li M-H, Harmon A, Choi SK, Banaszak Holl MM, Orr BG, Baker JR, Jr, Sinniah K. Bioanalytical Screening of Riboflavin Antagonists for Targeted Drug Delivery—A Thermodynamic and Kinetic Study. ACS Med Chem Lett. 2011;2:363–367. doi: 10.1021/ml100296z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montet X, Funovics M, Montet-Abou K, Weissleder R, Josephson L. Multivalent Effects of RGD Peptides Obtained by Nanoparticle Display. J Med Chem. 2006;49:6087–6093. doi: 10.1021/jm060515m. [DOI] [PubMed] [Google Scholar]
- 13.Shukla R, Thomas TP, Peters J, Kotlyar A, Myc A, Baker JR., Jr Tumor angiogenic vasculature targeting with PAMAM dendrimer-RGD conjugates. Chem Commun. 2005:5739–5741. doi: 10.1039/b507350b. [DOI] [PubMed] [Google Scholar]
- 14.Temming K, Lacombe M, Schaapveld RQJ, Orfi L, Kéri G, Poelstra K, Molema G, Kok RJ. Rational Design of RGD-Albumin Conjugates for Targeted Delivery of the VEGF-R Kinase Inhibitor PTK787 to Angiogenic Endothelium. ChemMedChem. 2006;1:1200–1203. doi: 10.1002/cmdc.200600201. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Foss CA, Byun Y, Nimmagadda S, Pullambhatla M, Fox JJ, Castanares M, Lupold SE, Babich JW, Mease RC, Pomper MG. Radiohalogenated Prostate-Specific Membrane Antigen (PSMA)-Based Ureas as Imaging Agents for Prostate Cancer. J Med Chem. 2008;51:7933–7943. doi: 10.1021/jm801055h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shukla R, Thomas TP, Desai AM, Kotlyar A, Park SJ, Baker JR., Jr HER2 specific delivery of methotrexate by dendrimer conjugated anti-HER2 mAb. Nanotechnology. 2008;19:295102. doi: 10.1088/0957-4484/19/29/295102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qian ZM, Li H, Sun H, Ho K. Targeted Drug Delivery via the Transferrin Receptor-Mediated Endocytosis Pathway. Pharmacol Rev. 2002;54:561–587. doi: 10.1124/pr.54.4.561. [DOI] [PubMed] [Google Scholar]
- 18.Thomas TP, Shukla R, Kotlyar A, Liang B, Ye JY, Norris TB, Baker JR., Jr Dendrimer-Epidermal Growth Factor Conjugate Displays Superagonist Activity. Biomacromolecules. 2008;9:603–609. doi: 10.1021/bm701185p. [DOI] [PubMed] [Google Scholar]
- 19.Raha S, Paunesku T, Woloschak G. Peptide-mediated cancer targeting of nanoconjugates, WIREs: Nanomed. Nanobiotech. 2010:n/a–n/a. doi: 10.1002/wnan.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong S, Leroueil PR, Majoros IJ, Orr BG, Baker JR, Jr, Banaszak Holl MM. The Binding Avidity of a Nanoparticle-Based Multivalent Targeted Drug Delivery Platform. Chem Biol. 2007;14:107–115. doi: 10.1016/j.chembiol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 21.Lee YC, Lee RT. Carbohydrate-Protein Interactions: Basis of Glycobiology. Acc Chem Res. 1995;28:321–327. [Google Scholar]
- 22.Kiessling LL, Gestwicki JE, Strong LE. Synthetic multivalent ligands in the exploration of cell-surface interactions. Curr Opin Chem Biol. 2000;4:696–703. doi: 10.1016/s1367-5931(00)00153-8. [DOI] [PubMed] [Google Scholar]
- 23.Mammen M, Choi SK, Whitesides GM. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew Chem, Int Ed. 1998;37:2755. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 24.Roy R. Syntheses and some applications of chemically defined multivalent glycoconjugates. Curr Opin Struct Biol. 1996;6:692–702. doi: 10.1016/s0959-440x(96)80037-6. [DOI] [PubMed] [Google Scholar]
- 25.Majoros I, Baker J Jr, editors. Dendrimer-Based Nanomedicine. Pan Stanford; Hackensack, NJ: 2008. [Google Scholar]
- 26.Choi Y, Thomas T, Kotlyar A, Islam MT, Baker JR., Jr Synthesis and Functional Evaluation of DNA-Assembled Polyamidoamine Dendrimer Clusters for Cancer Cell-Specific Targeting. Chem Biol. 2005;12:35–43. doi: 10.1016/j.chembiol.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 27.Lu Y, Low PS. Folate-mediated delivery of macromolecular anticancer therapeutic agents. Adv Drug Del Rev. 2002;54:675–693. doi: 10.1016/s0169-409x(02)00042-x. [DOI] [PubMed] [Google Scholar]
- 28.Mullen DG, Desai AM, Waddell JN, Cheng X-m, Kelly CV, McNerny DQ, Majoros InJ, Baker JR, Jr, Sander LM, Orr BG, Banaszak Holl MM. The Implications of Stochastic Synthesis for the Conjugation of Functional Groups to Nanoparticles. Bioconj Chem. 2008;19:1748–1752. doi: 10.1021/bc8002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majoros IJ, Thomas TP, Mehta CB, Baker JR., Jr Poly(amidoamine) Dendrimer-Based Multifunctional Engineered Nanodevice for Cancer Therapy. J Med Chem. 2005;48:5892–5899. doi: 10.1021/jm0401863. [DOI] [PubMed] [Google Scholar]
- 30.Mullen D, Borgmeier E, Desai A, van Dongen M, Barash M, Cheng Xm, Baker JR, Jr, Banaszak Holl M. Isolation and Characterization of Dendrimer with Precise Numbers of Functional Groups. Chem Eur J. 2010;16:10675–10678. doi: 10.1002/chem.201001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hakem IF, Leech AM, Johnson JD, Donahue SJ, Walker JP, Bockstaller MR. Understanding Ligand Distributions in Modified Particle and Particlelike Systems. J Am Chem Soc. 2010;132:16593–16598. doi: 10.1021/ja107139c. [DOI] [PubMed] [Google Scholar]
- 32.Mullen DG, Fang M, Desai A, Baker JR, Jr, Orr BG, Banaszak Holl MM. A Quantitative Assessment of Nanoparticle-Ligand Distributions: Implications for Targeted Drug and Imaging Delivery in Dendrimer Conjugates. ACS Nano. 2010;4:657–670. doi: 10.1021/nn900999c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu F, Zhang L, Teply BA, Mann N, Wang A, Radovic-Moreno AF, Langer R, Farokhzad OC. Precise engineering of targeted nanoparticles by using self-assembled biointegrated block copolymers. Proc Natl Acad Sci USA. 2008;105:2586–2591. doi: 10.1073/pnas.0711714105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Y, Qian H, Drake Bethany A, Jin R. Atomically Precise Au25SR18 Nanoparticles as Catalysts for the Selective Hydrogenation of alpha,beta-Unsaturated Ketones and Aldehydes13. Angew Chem Intl Ed. 2010;49:1295–1298. doi: 10.1002/anie.200906249. [DOI] [PubMed] [Google Scholar]
- 35.Qian H, Jin R. Controlling Nanoparticles with Atomic Precision: The Case of Au144(SCH2CH2Ph)60. Nano Letters. 2009;9:4083–4087. doi: 10.1021/nl902300y. [DOI] [PubMed] [Google Scholar]
- 36.Ercikan-Abali EA, Waltham MC, Dicker AP, Schweitzer BI, Gritsman H, Banerjee D, Bertino JR. Variants of human dihydrofolate reductase with substitutions at leucine-22: effect on catalytic and inhibitor binding properties. Mol Pharmacol. 1996;49:430–437. [PubMed] [Google Scholar]
- 37.Worm J, Kirkin AF, Dzhandzhugazyan KN, Guldberg P. Methylation-dependent Silencing of the Reduced Folate Carrier Gene in Inherently Methotrexate-resistant Human Breast Cancer Cells. J Biol Chem. 2001;276:39990–40000. doi: 10.1074/jbc.M103181200. [DOI] [PubMed] [Google Scholar]
- 38.Kamen BA, Capdevila A. Receptor-mediated folate accumulation is regulated by the cellular folate content. Proc Natl Acad Sci USA. 1986;83:5983–5987. doi: 10.1073/pnas.83.16.5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nandini-Kishore SG, Frazier WA. [3H]Methotrexate as a ligand for the folate receptor of Dictyostelium discoideum. Proc Natl Acad Sci USA. 1981;78:7299–7303. doi: 10.1073/pnas.78.12.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rijnboutt S, Jansen G, Posthuma G, Hynes JB, Schornagel JH, Strous GJ. Endocytosis of GPI-linked Membrane Folate Receptor-α. J Cell Biol. 1996;132:35–47. doi: 10.1083/jcb.132.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waddell JN, Mullen DG, Orr BG, Banaszak Holl MM, Sander LM. Origin of broad polydispersion in functionalized dendrimers and its effects on cancer-cell binding affinity. Phys Rev E. 2010;82:036108. doi: 10.1103/PhysRevE.82.036108. [DOI] [PubMed] [Google Scholar]
- 42.Licata NA, Tkachenko AV. Kinetic Limitations of Cooperativity-Based Drug Delivery Systems. Phys Rev Lett. 2008;100:158102. doi: 10.1103/PhysRevLett.100.158102. [DOI] [PubMed] [Google Scholar]
- 43.Arranz-Plaza E, Tracy AS, Siriwardena A, Pierce JM, Boons GJ. High-Avidity, Low-Affinity Multivalent Interactions and the Block to Polyspermy in Xenopus laevis. J Am Chem Soc. 2002;124:13035–13046. doi: 10.1021/ja020536f. [DOI] [PubMed] [Google Scholar]
- 44.Adler P, Wood SJ, Lee YC, Lee RT, Petri WA, Jr, Schnaar RL. High Affinity Binding of the Entamoeba histolytica Lectin to Polyvalent N-Acetylgalactosaminides. J Biol Chem. 1995;270:5164–5171. doi: 10.1074/jbc.270.10.5164. [DOI] [PubMed] [Google Scholar]
- 45.Gestwicki JE, Cairo CW, Mann DA, Owen RM, Kiessling LL. Selective Immobilization of Multivalent Ligands for Surface Plasmon Resonance and Fluorescence Microscopy. Anal Biochem. 2002;305:149–155. doi: 10.1006/abio.2002.5652. [DOI] [PubMed] [Google Scholar]
- 46.Rao J, Yan L, Xu B, Whitesides GM. Using Surface Plasmon Resonance to Study the Binding of Vancomycin and Its Dimer to Self-Assembled Monolayers Presenting d-Ala-d-Ala. J Am Chem Soc. 1999;121:2629–2630. [Google Scholar]
- 47.Tassa C, Duffner JL, Lewis TA, Weissleder R, Schreiber SL, Koehler AN, Shaw SY. Binding Affinity and Kinetic Analysis of Targeted Small Molecule-Modified Nanoparticles. Bioconj Chem. 2010;21:14–19. doi: 10.1021/bc900438a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakajima H, Kiyokawa N, Katagiri YU, Taguchi T, Suzuki T, Sekino T, Mimori K, Ebata T, Saito M, Nakao H, Takeda T, Fujimoto J. Kinetic Analysis of Binding between Shiga Toxin and Receptor Glycolipid Gb3Cer by Surface Plasmon Resonance. J Biol Chem. 2001;276:42915–42922. doi: 10.1074/jbc.M106015200. [DOI] [PubMed] [Google Scholar]
- 49.Hidari K, Shimada S, Suzuki Y, Suzuki T. Binding kinetics of influenza viruses to sialic acid-containing carbohydrates. Glycoconj J. 2007;24:583–590. doi: 10.1007/s10719-007-9055-y. [DOI] [PubMed] [Google Scholar]
- 50.Mann DA, Kanai M, Maly DJ, Kiessling LL. Probing Low Affinity and Multivalent Interactions with Surface Plasmon Resonance: Ligands for Concanavalin A. J Am Chem Soc. 1998;120:10575–10582. [Google Scholar]
- 51.Kensinger RD, Yowler BC, Benesi AJ, Schengrund CL. Synthesis of Novel, Multivalent Glycodendrimers as Ligands for HIV-1 gp120. Bioconj Chem. 2004;15:349–358. doi: 10.1021/bc034156a. [DOI] [PubMed] [Google Scholar]
- 52.Thomas TP, Majoros IJ, Kotlyar A, Kukowska-Latallo JF, Bielinska A, Myc A, Baker JR., Jr Targeting and Inhibition of Cell Growth by an Engineered Dendritic Nanodevice. J Med Chem. 2005;48:3729–3735. doi: 10.1021/jm040187v. [DOI] [PubMed] [Google Scholar]
- 53.Choi SK, Thomas T, Li M, Kotlyar A, Desai A, Baker JR., Jr Light-controlled release of caged doxorubicin from folate receptor-targeting PAMAM dendrimer nanoconjugate. Chem Commun. 2010;46:2632–2634. doi: 10.1039/b927215c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee RJ, Wang S, Low PS. Measurement of endosome pH following folate receptor-mediated endocytosis. Biochim Biophys Acta (BBA) - Mol Cell Res. 1996:237–242. doi: 10.1016/0167-4889(96)00041-9. [DOI] [PubMed] [Google Scholar]
- 55.Yang J, Chen H, Vlahov IR, Cheng JX, Low PS. Characterization of the pH of Folate Receptor-Containing Endosomes and the Rate of Hydrolysis of Internalized Acid-Labile Folate-Drug Conjugates. J Pharmacol Exp Ther. 2007;321:462–468. doi: 10.1124/jpet.106.117648. [DOI] [PubMed] [Google Scholar]
- 56.Cody V, Luft JR, Pangborn W. Understanding the role of Leu22 variants in methotrexate resistance: comparison of wild-type and Leu22Arg variant mouse and human dihydrofolate reductase ternary crystal complexes with methotrexate and NADPH. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 2):147–155. doi: 10.1107/S0907444904030422. [DOI] [PubMed] [Google Scholar]
- 57.Wu G, Barth RF, Yang W, Kawabata S, Zhang L, Green-Church K. Targeted delivery of methotrexate to epidermal growth factor receptor-positive brain tumors by means of cetuximab (IMC-C225) dendrimer bioconjugates. Mol Cancer Ther. 2006;5:52–59. doi: 10.1158/1535-7163.MCT-05-0325. [DOI] [PubMed] [Google Scholar]
- 58.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. Charmm: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4 :187–217. [Google Scholar]
- 59.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 60.Whiteley JM, Henderson GB, Russell A, Singh P, Zevely EM. The isolation of dihydrofolate reductases by affinity chromatography on folate-Sepharose. Anal Biochem. 1977;79:42–51. doi: 10.1016/0003-2697(77)90376-1. [DOI] [PubMed] [Google Scholar]
- 61.Gapski GR, Whiteley JM, Rader JI, Cramer PL, Hendersen GB, Neef V, Huennekens FM. Synthesis of a fluorescent derivative of amethopterin. J Med Chem. 2002;18:526–528. doi: 10.1021/jm00239a020. [DOI] [PubMed] [Google Scholar]
- 62.Quintana A, Raczka E, Piehler L, Lee I, Myc A, Majoros I, Patri AK, Thomas T, Mulé J, Baker JR., Jr Design and Function of a Dendrimer-Based Therapeutic Nanodevice Targeted to Tumor Cells Through the Folate Receptor. Pharm Res. 2002;19:1310–1316. doi: 10.1023/a:1020398624602. [DOI] [PubMed] [Google Scholar]
- 63.Thomas TP, Kukowska-Latallo JR. Biological application of PAMAM dendrimer nanodevices in vitro and in vivo. In: Majoros I, Baker JR Jr, editors. Dendrimer-Based Nanomedicine. Pan Stanford; Hackensack, NJ: 2008. pp. 175–207. [Google Scholar]
- 64.Rosowsky A, Forsch RA, Yu CS, Lazarus H, Beardsley GP. Methotrexate analogs. 21. Divergent influence of alkyl chain length on the dihydrofolate reductase affinity and cytotoxicity of methotrexate monoesters. J Med Chem. 1984;27:605–609. doi: 10.1021/jm00371a009. [DOI] [PubMed] [Google Scholar]
- 65.Piper JR, Montgomery JA, Sirotnak FM, Chello PL. Syntheses of α- and γ-substituted amides, peptides, and esters of methotrexate and their evaluation as inhibitors of folate metabolism. J Med Chem. 1982;25:182–187. doi: 10.1021/jm00344a018. [DOI] [PubMed] [Google Scholar]
- 66.Schilsky RL, Bailey BD, Chabner BA. Methotrexate polyglutamate synthesis by cultured human breast cancer cells. Proc Natl Acad Sci USA. 1980;77:2919–2922. doi: 10.1073/pnas.77.5.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heijden JWVD, Oerlemans R, Dijkmans BAC, Qi H, Laken CJVD, Lems WF, Jackman AL, Kraan MC, Tak PP, Ratnam M, Jansen G. Folate receptor beta as a potential delivery route for novel folate antagonists to macrophages in the synovial tissue of rheumatoid arthritis patients. Arthritis Rheum. 2009;60:12–21. doi: 10.1002/art.24219. [DOI] [PubMed] [Google Scholar]
- 68.Choi SK, Leroueil P, Li M-H, Desai A, Zong H, Van Der Spek AFL, Baker JR., Jr Specificity and Negative Cooperativity in Dendrimer–Oxime Drug Complexation. Macromol. 2011;44:4026–4029. [Google Scholar]
- 69.Kalesse M, Loos A. Transesterification of Phosphodiester by a Zinc-Containing Cyclen Derivative: Identification of the Active Species. Liebigs Ann. 1996;1996:935–939. [Google Scholar]
- 70.Glaser RW. Antigen-Antibody Binding and Mass Transport by Convection and Diffusion to a Surface: A Two-Dimensional Computer Model of Binding and Dissociation Kinetics. Analytical Biochemistry. 1993;213:152–161. doi: 10.1006/abio.1993.1399. [DOI] [PubMed] [Google Scholar]
- 71.Pickersgill RW. A rapid method of calculating charge-charge interaction energies in proteins. Protein Eng. 1988;2:247–248. doi: 10.1093/protein/2.3.247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.