Abstract
Background
Pathogenic hantaviruses geographically distributed in the Old World cause hemorrhagic fever with renal syndrome (HFRS), whereas New World hantaviruses are the etiological agents of hantavirus cardiopulmonary syndrome (HCPS). Ribavirin, a drug associated with toxicities, is presently indicated for treatment of HFRS, while treatment of the more frequently lethal HCPS is limited to supportive care. Because of the need for safe and effective antivirals to treat severe hantaviral infections, we evaluated favipiravir (T-705) against Dobrava and Maporal viruses as representative Old World and New World hantaviruses, respectively. Dobrava virus causes HFRS in Europe. Maporal virus (MPRLV), recently isolated from western Venezuela, is very similar phylogenetically to Andes virus (ANDV), the principal cause of HCPS in Argentina. Presently, there is no evidence that MPRLV can cause disease in humans.
Methods and Results
Here, we show that infection of Vero E6 cells with MPRLV is dependent on β3 integrins, similar to that reported for pathogenic hantaviruses. Further, by analysis of genetic determinants associated with the G1 glycoprotein cytoplasmic tail, we show the close genetic proximity of MPRLV to other HCPS-causing hantaviruses in these regions predictive of pathogenicity. Using focus-forming unit assays and measuring viral RNA by quantitative RT-PCR, we demonstrate anti-hantavirus activity by favipiravir with inhibitory concentrations ranging from 65 - 93 μM and selectivity indices > 50.
Conclusions
Our data suggest that MPRLV may serve as a safer alternative to modeling infection caused by the highly lethal ANDV and that hantaviruses are sensitive to the effects of favipiravir in cell culture.
1. Introduction
Hantaviruses (Bunyaviridae, Hantavirus) are enveloped RNA viruses with tripartite negative- or ambisense genomes that can cause severe and often fatal forms of disease in humans. Pathogenic hantaviruses geographically distributed in the Old World can cause hemorrhagic fever with renal syndrome (HFRS), whereas New World hantaviruses are the etiological agents of hantavirus cardiopulmonary syndrome (HCPS) [1]. The latter was initially described through an outbreak that occurred in the four corners region of southwestern United States in 1993 [2]. There are at least a dozen hantaviruses known to cause HCPS in North, Central, and South America [3-17]. The agents that have received the most attention are Sin Nombre and Andes hantaviruses which have caused significant disease outbreaks in North and South America, respectively [18]. The high mortality (> 40%) associated with this emerging infectious disease, the potential for intentional release of hantaviruses, and the lack of effective antiviral drugs or vaccines available for therapeutic use and prevention stress the importance of developing effective antivirals and treatment modalities.
For some time, the lack of a practical animal model of disease has hindered the development of therapeutics for the treatment and prevention of HCPS. In efforts to study the pathogenesis of HCPS, a hamster disease model based on Andes virus (ANDV) infection was initially established [18]. More recently, a hamster model of infection using a recent isolate, Maporal virus (MPRLV) was described [19]. The virus was isolated in 1997 from central Venezuela and is phylogenetically similar to hantaviruses known to cause HCPS in southern South America [20]. Milazzo and colleagues demonstrated that the MPRLV infection and disease in hamsters parallels that of the ANDV model which closely resembles human disease [19]. MPRLV-infected animals showed signs of cardiac depression and diffuse pulmonary edema with rapid onset comparable to findings in clinical cases of HPS.
Currently there is no evidence that MPRLV can infect humans and cause disease. Thus, unlike the ANDV hamster infection model that requires maximum containment facilities (BSL-4) not readily available to most researchers, studies with MPRLV have been performed under less stringent (BSL-3+) containment. In light of the above, the MPRLV hamster infection model may be a more accessible and a suitable surrogate for the study of disease caused by HCPS-causing hantaviruses. Although further characterization of this model will be necessary (especially at the level of immune response and cardiac involvement), initial work by Milazzo and coworkers suggest that it will be a valuable tool for the early stage evaluation of candidate therapeutics to counter HCPS [19].
The use of integrins as receptors that mediate cellular entry of hantaviruses has been extensively studied [21]. Considerable evidence indicates that pathogenic hantaviruses that cause HCPS and HFRS gain entry via β3 integrins whereas nonpathogenic hantaviruses utilize β1 integrins [22, 23]. β3 integrins participate in the control of vascular permeability [24, 25]. Thus, interaction of pathogenic hantaviruses with β3 integrins may disrupt normal function and contribute to the alteration of vascular biology that often leads to severe disease.
Within the hantavirus genome, the M segment is translated into a polyprotein that is cleaved into N-terminal G1 and C-terminal G2 glycoproteins [26, 27]. The G1 glycoprotein contains a long cytoplasmic tail that contains a conserved immunoreceptor tyrosine-based activation motif (ITAM) present in all HCPS-causing hantaviruses, but not HFRS or nonpathogenic hantaviruses [28]. Also in the same region of the conserved ITAM motifs, hantavirus G1 cytoplasmic tails can contain a C-terminal hydrophobic domain that directs proteasomal degradation. This so called ‘degron’ sequence directs ubiquitination and degradation of the cytoplasmic tail, which is linked to hantavirus virulence [27, 29, 30]. The hantavirus degron has been shown to consist of tyrosines that reside within the ITAM (Y619 and Y632), as well as an additional eight residues [28].
Although ribavirin has been shown to have some effectiveness in treating HFRS, it lacks specificity and has toxicity [31]. Recently, the antiviral favipiravir (T-705, 6-fluoro-3-hydroxy-2-pyrazinecarboxamide) has been reported to be active against a number of bunyaviruses both in vitro and in vivo in mouse and hamster models [32, 33]. Notably, favipirair has also been shown to be effective in treating severe influenza virus, arenavirus, and flavivirus infections in animal models [34-39]. Because favipiravir is active against other members of the Bunyaviridae family, we hypothesized that it would also inhibit hantavirus replication. Thus, in the present study, we investigated MPRLV β integrin utilization and genetic determinants of pathogenicity, and evaluated hantavirus sensitivity to the effects of favipiravir in cell culture.
2. Methods
2.1. Cells and Viruses
The monkey kidney cell line Vero E6 was purchased from American Type Culture Collection (ATCC, Manassas, VA). Cells were maintained in minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS), and gentamicin (Sigma, St. Louis, MO). All media and serum were from HyClone Laboratories (Logan, UT) unless otherwise stated. Prospect Hill virus (PHV), strain MP40 TVP6042, Dobrava virus (DOBV), strain Sotkamo, and MPRLV, strain HV9021050, were provided by Dr. Robert Tesh (World Reference Center for Emerging Viruses and Arboviruses, University of Texas Medical Branch, Galveston, TX). All three viruses were propagated in Vero E6 cells and their identity confirmed by quantitative RTPCR (data not shown). Infections were performed in Dulbecco's modified essential medium (DMEM) with 2% FBS and gentamicin. Biosafety level 3 (BSL-3) facilities were used for studies with DOBV and MPRLV, while BSL-2 containment was used for work with PHV.
2.2. Ligands, antibodies and staining reagents
Sin Nombre virus (SNV) hyperimmune mouse serum, kindly provided by Dr. Robert Tesh, was used as the primary anti-hantavirus antibody for focus-forming unit detection. The secondary antibody used was a goat anti-mouse horseradish peroxidase conjugated antibody from Kirkegaard & Perry Laboratories (KPL, Gaithersburg, MD). Both were used at a 1:500 dilution. Polyclonal function-blocking antibodies directed at β1 and β3 integrins for receptor usage determination studies were purchased from Millipore (Chemicon, Temecula, CA) and titrated for use at 1 μg/ml. Fibronectin and vitronectin were purchased from Sigma and titrated for use at concentrations of 20 and 1 μg/ml, respectively. Acidin and biotinylated horseradish peroxidase complex (ABC) and 3,3’-diaminobenzidine (DAB) staining kits were purchased from Vector Laboratories (Burlingame, CA).
2.3. Focus-forming unit (FFU) assay
Vero E6 cells (~80% confluent) in 96-well plates were pretreated with known β integrin ligands or function-blocking antibodies for 1 h at 37°C before viral infection. Ligands or antibodies were removed and 100 μl volumes containing ~1000 FFUs of MPRLV or ~4,000 FFUs of DOBV or PHV were adsorbed onto the pretreated cells at 37°C, 5% CO2 for 90 min. After adsorption, virus inocula were removed, and the cells were incubated in DMEM with 2% FBS and gentamicin an additional 24 h for MPRLV, 3 days for DOBV, and 5 days for PHV prior to staining. All cell cultures were confluent at the time of immunoperoxidase staining.
Immunoperoxidase staining of the nucleocapsid protein in infected cells has been previously described [23]. In brief, cell monolayers were fixed with 100% methanol at 4°C, for 10 min, permeabilized with 0.25% Triton X-100 in PBS for 10 min, and blocked with 4% goat serum in 1% BSA in PBS for 1 h. Monolayers were incubated with SNV hyperimmune mouse serum (1/500) for 1 h at 37°C, and subsequently with goat anti-mouse horseradish peroxidase conjugated antibody (1/500). Infected cell foci were quantified after staining with ABC and working DAB solution per the manufacturer's instructions.
2.4. Phylogenetic analysis
Amino acid sequences from the Genbank database were compared using Molecular Evolutionary Genetics Analysis (MEGA) software (Center for Evolutionary Medicine and Informatics, Tempe, AZ). Analysis of phylogenetic relationships were made using the neighbor-joining and bootstrap consensus methods [40] analyzing only the ITAM/degron 30 amino acid region [28, 29] of the M segment of the genome. The evolutionary distances were computed using the poisson correction method and are in the units of the number of amino acid substitutions per site [41]. All positions containing gaps and missing data were eliminated from the dataset (complete deletion option).
Hantavirus M segment Genbank sequence accession numbers were as follows: Andes (AAO86638.1), Bayou (AAA61690.1), Bermejo (AAB87911), Choclo (ABB90558.1), Dobrava (AAY27875.1), Hantaan (AAK27683.2), Laguna Negra (AAB87603), Maporal (AAR14889.1), New York-1 (AAC54560), Prospect Hill (CAA38922.1), Puumula (AB297666.2), Seoul (NP_942557.1), Sin Nombre (AAG03036.1), and Tula (NP_942586).
2.5. Drug efficacy studies
Favipiravir was provided by the Toyama Chemical Company (Toyama, Japan). Ribavirin was obtained from ICN Pharmaceuticals (Costa Mesa, CA). The antiviral activity of favipiravir and ribavirin was determined as follows. Vero E6 cells (~80% confluent) in 96-well plates were infected with hantavirus FFUs as specified above. MPRLV and DOBV inocula were adsorbed for one hour, and PHV was absorbed for six hours. After infection, virus was removed and cells were treated with eight serial half-log10 dilutions of the drugs. Plates were incubated (37°C, 5% CO2, DMEM with 2% FBS and gentamicin) for 24 h for MPRLV, 3 days for DOBV, and 5 days for PHV. To determine cell cytotoxicity, drugs were added to uninfected parallel cultures and cell viability was determined at 24 h, 3 days, or 5 days (consistent with the various hantavirus assay time requirements) by neutral red (NR) dye uptake, as previously described [32]. Toxicity was presented as the 50% cell cytotoxic concentration (CC50), defined as the concentration of the drug that resulted in a 50% decrease in cell viability compared to the cell controls. Drug efficacy was determined by quantification of infected cells and determining the 50% inhibitory concentration (IC50), defined as the concentration of the drug required to reduce viral FFUs by 50%. The selectivity (or therapeutic) index (SI) is defined as the ratio of the CC50 to the IC50.
Quantitative (q)RT-PCR was also used to determine drug efficacy against MPRLV by measuring relative viral RNA concentrations after incubation of infected cells with the drugs. Triplicate wells of a 24-well plate containing ~80% confluent Vero E6 cultures were infected with 250 μl containing ~300 FFU of MPRLV for six hours, at which time the virus inocula were removed and the cells treated with serial dilutions of ribavirin or favipiravir. After three days in culture, RNA was collected using an RNeasy total RNA purification kit following the manufacturer's recommendations (Qiagen, Valencia, CA). The MGB Eclipse probe system from Epoch Biosciences (Bothell, WA) was used in combination with the SuperScript III Platinum One-Step Quantitative RT-PCR system from Invitrogen (Carlsbad, CA), as recommended by the manufacturers for the qRT-PCR reactions. The MPRLV primers and probe combination were designed using the MGB Eclipse Online Design software and targeted the nucleocapsid coding sequence. The forward primer was 5’-GGA CAT TTC CAT AAC GCA GTG-3’ and the reverse primer was 5’-TGG CAG CTC AGA AAC TGG CTT CAA A-3’. The probe target sequence was (MGB EDQ)-5’-GTC ATC AGG TTC AAG C-3’ (FAM). Viral RNA levels were assayed in duplex with, and normalized to, β2-microglobulin expression. All reactions were performed using the DNA Engine Opticon 2 Real-Tim PCR detection system (Bio-Rad, South San Francisco, CA). The IC50 was calculated as the dose of drug reducing the relative MPRLV RNA levels by 50% compared to infected untreated controls. At the lower concentrations of compounds tested, cytotoxicity was not observed in parallel cultures that were not infected with MPRLV (data not shown).
3. Results
3.1. β3 integrin chain-specific antibodies and vitronectin block MPRLV infectivity
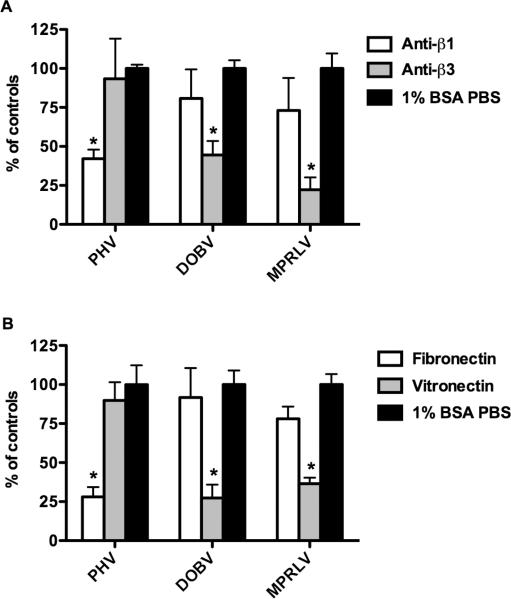
β3 integrin usage is a characteristic feature shared by pathogenic hantaviruses. To assess MPRLV integrin usage, Vero E6 cell monolayers were treated with antibodies or integrin antagonists to block hantavirus infectivity. Following incubation, infected cells were quantified by FFU assay and experimental treatment groups were compared to untreated virus-infected controls. PHV infection was significantly inhibited by anti-β1-specific integrin antibodies, but not anti-β3 integrin antibodies (Figure 1A). Conversely, pretreatment of Vero E6 cells with polyclonal antibodies to β3 integrins significantly decreased MPRLV and DOBV infectivity.
Figure 1. MPRLV infection is mediated by β3 integrins.
Duplicate wells of Vero E6 cells were pretreated with A) integrin-specific antibodies (1 μg/ml) or B) vitronectin and fibronectin (1 and 20 μg/ml, respectively) for 1 hour prior to viral infection. After infection and incubation for 1-5 days (depending on virus) cell monolayers were fixed and stained as described in the methods. Data are presented as percent FFU of infected 1% BSA PBS-treated controls. Data are representative of three independent experiments. *P < 0.05 compared to respective controls by two-way analysis of variance with Bonferroni posttest.
We next assayed the ability of specific integrin ligands to inhibit MPRLV infectivity. Vitronectin and fibronectin are ECM proteins with high affinity for αVβ3 and α5β1 integrins, respectively. Consistent with the antibody studies, pretreatment of Vero E6 cells with vitronectin significantly reduced MPRLV and DOBV infectivity. In contrast, PHV infection was most dramatically reduced with fibronectin (Figure 1B). Collectively, these data suggest that MPRLV host cell infection is facilitated principally by β3 integrins, similar to the known pathogen, DOBV.
3.2. MPRLV shares ITAM and degron sequence homology with pathogenic hantaviruses
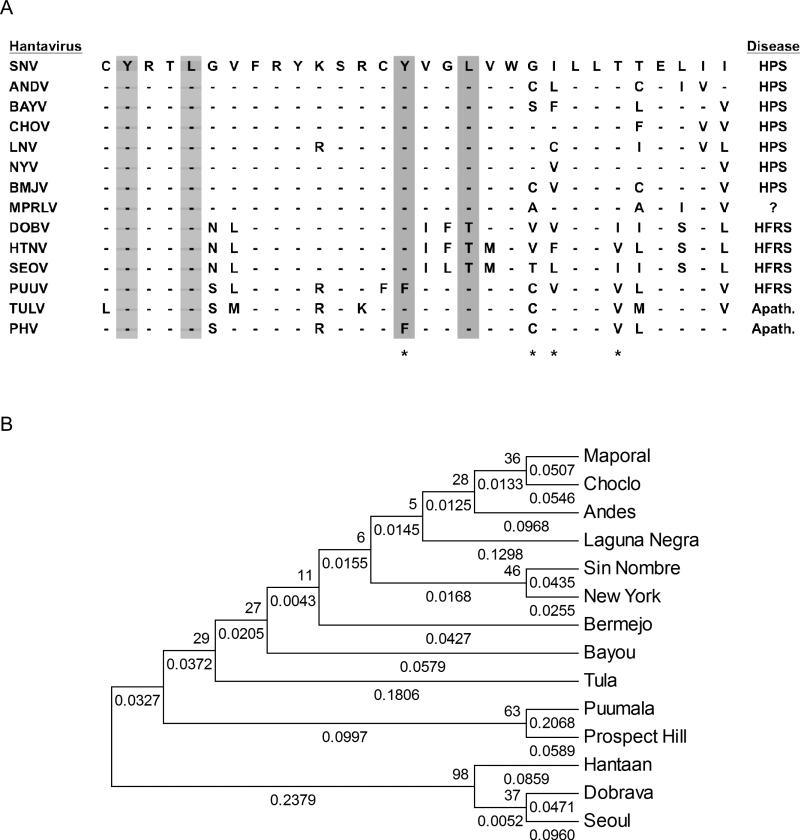
We performed a phylogenetic analysis to compare ITAM and degron protein sequences of thirteen hantaviruses with that of MPRLV as a means to obtain insights into potential pathogenicity of MPRLV. An alignment of hantavirus sequences obtained from Genbank that contain the overlapping ITAM and degron regions is shown in Figure 2A. This ClustalW alignment indicated that the G1 cytoplasmic tail of MPRLV contained the characteristic ITAM sequence found in all HCPS-causing hantaviruses. The comparison also indicated that MPRLV does not contain the conserved residues that stabilize the G1 tail in nonpathogenic hantaviruses, but does contain residues consistent with pathogenic hantaviruses (Figure 2A). Phylogentic analysis restricted to the 30 amino acid sequence encompassing the ITAM/degron region shows that MPRLV is more closely related to the HCPS-causing hantaviruses (Figure 2B). Notably, similarity of this region with ANDV was striking.
Figure 2. Amino acid comparison and phylogenetic relationships among the ITAM/degron sequences of fourteen different hantaviruses.
A) Alignment of the G1 protein cytoplasmic tail region containing the ITAM/degron residues. The asterisks indicate amino acids important in the proteosomal degradation of G1 tails. The gray columns represent the ITAM motif residues. B) Phylogentic analysis based on ITAM/degron sequence. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches. The tree is not drawn to scale, but indicates branch lengths (next to the branches) in the same units as those of the evolutionary distances used to infer the phylogenetic tree. BAYV, Bayou virus; BMJV, Bermejo virus; CHOV, Choclo virus, HTNV, Hantaan virus; LNV, Laguna Negra virus; NYV, New York-1 virus, PUUV, Puumula virus; SEOV, Seoul virus; TULV, Tula virus.
3.3. Efficacy of favipiravir and ribavirin against MPRLV
The inhibitory activity of favipiravir and ribavirin against hantaviruses was determined by infectious FFU assay (Table 1). Ribavirin, known to be active against HFRS- and HCPS-causing hantaviruses in vitro [42-44], inhibited MPRLV infectivity in a dose-dependant manner, with an IC50 of 47 μM and therapeutic index of 22. As expected, ribavirin was also active against DOBV and PHV. Favipiravir, reported to be effective at inhibiting replication of related bunyaviruses [32], was also found to be similarly active against MPRLV with an IC50 of 65 μM and therapeutic index of 74. It was also effective against DOBV and PHV.
Table 1.
In vitro inhibitory effects of favipiravir and ribavirin against hantaviruses by FFU assaya.
| Virus | Favipiravir |
Ribavirin |
||||
|---|---|---|---|---|---|---|
| CC50± SDb | IC50 ± SDb | SIc | CC50± SDb | IC50 ± SDb | SIc | |
| PHV | 3819 ± 64 | 66 ± 26 | 58 | 1018 ± 866 | 23 ± 1.9 | 44 |
| DOBV | 4816 ± 662 | 93 ± 18 | 52 | 1215 ± 628 | 72 ± 2.4 | 17 |
| MPRLV | 4795 ± 1186 | 65 ± 17 | 74 | 1051 ± 135 | 47 ± 2.9 | 22 |
Data are the mean and standard deviations from 3-4 separate experiments in Vero E6 cells.
CC50 and IC50 values are in μM.
SI (selectivity index) = ratio of 50% cytotoxic concentration (CC50) to the 50% virus inhibitory concentration (IC50).
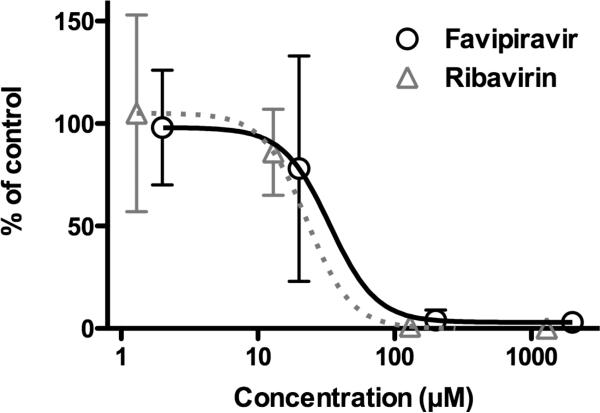
A qRT-PCR assay was also employed to evaluate the anti-MPRLV activities of favipiravir and ribavirin. As shown in Figure 3, favipiravir activity was verified by the reduction of MPRLV RNA (IC50 of 33 μM). Ribavirin also reduced MPRLV RNA levels in a dose-dependent manner with an EC50 of 22 μM (Figure 3). As observed with the FFU assay, the EC50 of ribavirin was slightly less compared to the EC50 of favipiravir by the qRT-PCR method. Notably, the qRTPCR assay generally resulted in lower EC50s μM ranges. Presumably, this difference resulted from differences in assay formats (96-well versus 24-well plates), the duration of the assays, and the lower multiplicity of infection.
Figure 3. Favipiravir reduces MPRLV RNA during infection.
Viral RNA was measured by qRT-PCR from total RNA extracted from infected Vero E6 cells treated with varying concentrations of favipiravir or ribavirin. Data are presented as percent of virus-infected, untreated controls and are representative of 2-3 experiments.
4. Discussion
Our findings suggest that MPRLV infectivity in Vero cells is principally mediated by β3 integrins. To date, only pathogenic hantaviruses have been demonstrated to ultilize β3 integrins for gaining entry into host cells, while the nonpathogenic hantaviruses require β1 integrins for cellular entry [22, 23]. αvβ3 integrins help maintain vascular integrity through interaction with extracellular matrix and participate in the regulation of vascular permeability [24, 25, 45, 46]. Alteration of αvβ3 integrin function through interaction with pathogenic hantaviruses may contribute vascular dysregulation associated with severe forms of disease. The preponderance of evidence suggests that β3 integrin usage by hantaviruses is highly predictive of their ability to cause disease in humans.
In our studies characterizing MPRLV, pretreatment of cells with function-blocking β3 integrin antibodies and vitronectin significantly reduced infectivity. Consistent with previous work by others [22], anti-β3 integrin and vitronectin also inhibited the HFRS-causing DOBV. Notably, complete inhibition could not be achieved, suggesting that other mechanisms of virus entry are likely. Recent studies have shown that in addition to the β integrins, hantaviruses also exploit decay-accelerating factor (DAF) to successfully infect cells [47, 48]. This may explain why, after incubation with ECM proteins or antibodies to the β integrins, viruses still were able to infect cells with ‘blocked’ receptors resulting in less than complete inhibition. Studies blocking both β integrin and DAF proteins would likely result in more complete blockage of hantaviral infection. Partial inhibition of pathogenic hantavirus replication following treatment of cells with β1 integrin-specific blocking antibodies or fibronectin may be attributed to steric hindrance of viral binding, as suggested in experiments with West Nile virus [49].
In addition to integrin utilization pattern as a predictor of pathogenicity, several motifs present in the cytoplasmic tail of the G1 glycoprotein of hantaviruses can also serve as distinguishing features. Our comparative analysis focused on the stretch of amino acids encompassing both the ITAM and degron sequences demonstrate extensive similarity between MPRLV and other HCPS-causing hantaviruses. ITAM motifs participate in intracellular signaling that directs cellular mechanisms. Viruses may be able to regulate ITAM signaling responses of host cells [29]. The presence of such motifs almost exclusively in pathogenic HCPS-causing hantaviruses may suggest an additional mechanism by which virulence and host cell function can be regulated by the virus. Notably, the apathogenic hantavirus, Tula virus (TULV), contains the ITAM sequence. However, its G1 tail is missing an adjacent cysteine residue conserved in all other hantaviruses that likely alters the conformation of the ITAM in the TULV G1 tail [28].
The degron sequence is a C-terminal hydrophobic domain of the G1 cytoplasmic tail that directs proteasomal degradation and is also linked to the virulence of hantaviruses. A recent study comparing differences between PHV and the New York-1 hantavirus (NYV) identified a series of four amino acid residues that are critical to the unstable state of the NYV G1 tail [27]. In MPRLV, like other pathogenic hantaviruses, the C-terminal hydrophobic region of the G1 cytoplasmic tail has interspersed hydrophilic residues. Replacement of hydrophobic amino acids in this region of the nonpathogenic PHV with polar residues results in its destabilization [27]. Experiments investigating tail-directed proteasomal degradation of the MPRLV and other pathogenic and nopathogenic hantavirus C1 cytoplasmic tails would further support the hypothesis that MPRLV may emerge as a human pathogen.
In our phylogenetic analysis restricted to the ITAM and degron sequences, Puumala virus (PUUV), a hantavirus that causes a mild form of HFRS [50, 51], segregates with the apathogenic PHV. Phylogenetic analysis not only serves to reveal relationships between hantaviruses that cause the same disease, but also a correlation with the phylogeny of their rodent hosts [52, 53]. To this end, the sharing of rodents hosts, such as that of PHV and PUUV (Arvicolinae), likely drives these associations. Notably, MPRLV had significant phylogentic similarities with pathogenic, and more specifically, HCPS-causing hantaviruses such as SNV and ANDV.
In the present study, ribavirin and favipiravir were evaluated against MPRLV and found to be active by FFU and qRT-PCR assays measuring infectious virus and viral RNA, respectively. DOBV was also found to be sensitive to the effects of favipiravir. We hypothesize that the mechanism by which favipiravir inhibits hantavirus replication is through inhibition of the viral polymerase, as has been suggested for influenza virus and arenaviruses [54, 55]. However, functional assays for hantavirus RNA-dependent RNA polymerases are not presently available to thoroughly investigate this proposed mode of action. The 65-93 μM inhibitory range observed was consistent with previously reported data for other members of the Bunyaviridae family, including the Punta Toro phlebovirus [32, 33]. More importantly, favipiravir has been demonstrated to be highly efficacious in the mouse and hamster Punta Toro virus infection models [32, 33]. It is hypothesized that the remarkable in vivo activity may be due to increased activity or abundance of the host cellular enzymes that covert favipiravir into the active triphosphate form of the drug.
Favipiravir is presently in clinical trials in Japan (Phase 3) and the United States (Phase 2) for the treatment of influenza virus infections, so the safety of the compound is being comprehensively addressed. Based on the well documented activity of favipiravir in mouse and hamster models of Punta Toro virus infection, and IC50s for this related bunyavirus ranging from 55-191 μM [32, 33], we would predict efficacy in hantavirus infection models. Collectively, our findings support future pre-clinical development of favipiravir in the hamster MPRLV or ANDV infection models [18, 19] or the mouse HNTV infection model [56, 57] as proof-of-concept for in vivo efficacy.
Acknowledgements
This work was supported by contract N01-AI-30048 from the Virology Branch, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Disclosure statement
YF is an employee of the Toyama Chemical Co., Ltd., the manufacturer of T-705. All other authors declare no competing interests.
References
- 1.Schmaljohn C, Hjelle B. Hantaviruses: a global disease problem. Emerging Infectious Diseases. 1997;3:95–104. doi: 10.3201/eid0302.970202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC Outbreak of acute illness--southwestern United States, 1993. MMWR. Morbidity and Mortality Weekly Report. 1993;42:421–424. [PubMed] [Google Scholar]
- 3.Gonzalez Della Valle M, Edelstein A, Miguel S, Martinez V, Cortez J, Cacace ML, Jurgelenas G, Estani SS, Padula P. Andes virus associated with hantavirus pulmonary syndrome in northern Argentina and determination of the precise site of infection. American Journal of Tropical Medicine and Hygiene. 2002;66:713–720. doi: 10.4269/ajtmh.2002.66.713. [DOI] [PubMed] [Google Scholar]
- 4.Hjelle B, Lee SW, Song W, Torrez-Martinez N, Song JW, Yanagihara R, Gavrilovskaya I, Mackow ER. Molecular linkage of hantavirus pulmonary syndrome to the white-footed mouse, Peromyscus leucopus: genetic characterization of the M genome of New York virus. Journal of Virology. 1995;69:8137–8141. doi: 10.1128/jvi.69.12.8137-8141.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson AM, Bowen MD, Ksiazek TG, Williams RJ, Bryan RT, Mills JN, Peters CJ, Nichol ST. Laguna Negra virus associated with HPS in western Paraguay and Bolivia. Virology. 1997;238:115–127. doi: 10.1006/viro.1997.8840. [DOI] [PubMed] [Google Scholar]
- 6.Johnson AM, de Souza LT, Ferreira IB, Pereira LE, Ksiazek TG, Rollin PE, Peters CJ, Nichol ST. Genetic investigation of novel hantaviruses causing fatal HPS in Brazil. Journal of Medical Virology. 1999;59:527–535. doi: 10.1002/(sici)1096-9071(199912)59:4<527::aid-jmv17>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 7.Khan AS, Gaviria M, Rollin PE, Hlady WG, Ksiazek TG, Armstrong LR, Greenman R, Ravkov E, Kolber M, Anapol H, Sfakianaki ED, Nichol ST, Peters CJ, Khabbaz RF. Hantavirus pulmonary syndrome in Florida: association with the newly identified Black Creek Canal virus. American Journal of Medicine. 1996;100:46–48. doi: 10.1016/s0002-9343(96)90010-8. [DOI] [PubMed] [Google Scholar]
- 8.Khan AS, Spiropoulou CF, Morzunov S, Zaki SR, Kohn MA, Nawas SR, McFarland L, Nichol ST. Fatal illness associated with a new hantavirus in Louisiana. Journal of Medical Virology. 1995;46:281–286. doi: 10.1002/jmv.1890460320. [DOI] [PubMed] [Google Scholar]
- 9.Levis S, Morzunov SP, Rowe JE, Enria D, Pini N, Calderon G, Sabattini M, St Jeor SC. Genetic diversity and epidemiology of hantaviruses in Argentina. Journal of Infectious Diseases. 1998;177:529–538. doi: 10.1086/514221. [DOI] [PubMed] [Google Scholar]
- 10.Lopez N, Padula P, Rossi C, Lazaro ME, Franze-Fernandez MT. Genetic identification of a new hantavirus causing severe pulmonary syndrome in Argentina. Virology. 1996;220:223–226. doi: 10.1006/viro.1996.0305. [DOI] [PubMed] [Google Scholar]
- 11.Lopez N, Padula P, Rossi C, Miguel S, Edelstein A, Ramirez E, Franze-Fernandez MT. Genetic characterization and phylogeny of Andes virus and variants from Argentina and Chile. Virus Research. 1997;50:77–84. doi: 10.1016/s0168-1702(97)00053-1. [DOI] [PubMed] [Google Scholar]
- 12.Morzunov SP, Feldmann H, Spiropoulou CF, Semenova VA, Rollin PE, Ksiazek TG, Peters CJ, Nichol ST. A newly recognized virus associated with a fatal case of hantavirus pulmonary syndrome in Louisiana. Journal of Virology. 1995;69:1980–1983. doi: 10.1128/jvi.69.3.1980-1983.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nichol ST, Spiropoulou CF, Morzunov S, Rollin PE, Ksiazek TG, Feldmann H, Sanchez A, Childs J, Zaki S, Peters CJ. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262:914–917. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- 14.Padula P, Della Valle MG, Alai MG, Cortada P, Villagra M, Gianella A. Andes virus and first case report of Bermejo virus causing fatal pulmonary syndrome. Emerging Infectious Diseases. 2002;8:437–439. doi: 10.3201/eid0804.010300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ravkov EV, Rollin PE, Ksiazek TG, Peters CJ, Nichol ST. Genetic and serologic analysis of Black Creek Canal virus and its association with human disease and Sigmodon hispidus infection. Virology. 1995;210:482–489. doi: 10.1006/viro.1995.1366. [DOI] [PubMed] [Google Scholar]
- 16.Rhodes LV, 3rd, Huang C, Sanchez AJ, Nichol ST, Zaki SR, Ksiazek TG, Humphreys JG, Freeman JJ, Knecht KR. Hantavirus pulmonary syndrome associated with Monongahela virus, Pennsylvania. Emerging Infectious Diseases. 2000;6:616–621. doi: 10.3201/eid0606.000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vincent MJ, Quiroz E, Gracia F, Sanchez AJ, Ksiazek TG, Kitsutani PT, Ruedas LA, Tinnin DS, Caceres L, Garcia A, Rollin PE, Mills JN, Peters CJ, Nichol ST. Hantavirus pulmonary syndrome in Panama: identification of novel hantaviruses and their likely reservoirs. Virology. 2000;277:14–19. doi: 10.1006/viro.2000.0563. [DOI] [PubMed] [Google Scholar]
- 18.Hooper JW, Larsen T, Custer DM, Schmaljohn CS. A lethal disease model for hantavirus pulmonary syndrome. Virology. 2001;289:6–14. doi: 10.1006/viro.2001.1133. [DOI] [PubMed] [Google Scholar]
- 19.Milazzo ML, Eyzaguirre EJ, Molina CP, Fulhorst CF. Maporal viral infection in the Syrian golden hamster: a model of hantavirus pulmonary syndrome. Journal of Infectious Diseases. 2002;186:1390–1395. doi: 10.1086/344735. [DOI] [PubMed] [Google Scholar]
- 20.Fulhorst CF, Cajimat MN, Utrera A, Milazzo ML, Duno GM. Maporal virus, a hantavirus associated with the fulvous pygmy rice rat (Oligoryzomys fulvescens) in western Venezuela. Virus Research. 2004;104:139–144. doi: 10.1016/j.virusres.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 21.Mackow ER, Gavrilovskaya IN. Cellular receptors and hantavirus pathogenesis. Current Topics in Microbiology and Immunology. 2001;256:91–115. doi: 10.1007/978-3-642-56753-7_6. [DOI] [PubMed] [Google Scholar]
- 22.Gavrilovskaya IN, Brown EJ, Ginsberg MH, Mackow ER. Cellular entry of hantaviruses which cause hemorrhagic fever with renal syndrome is mediated by beta3 integrins. Journal of Virology. 1999;73:3951–3959. doi: 10.1128/jvi.73.5.3951-3959.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gavrilovskaya IN, Shepley M, Shaw R, Ginsberg MH, Mackow ER. beta3 Integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:7074–7079. doi: 10.1073/pnas.95.12.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reynolds LE, Wyder L, Lively JC, Taverna D, Robinson SD, Huang X, Sheppard D, Hynes RO, Hodivala-Dilke KM. Enhanced pathological angiogenesis in mice lacking beta3 integrin or beta3 and beta5 integrins. Nature Medicine. 2002;8:27–34. doi: 10.1038/nm0102-27. [DOI] [PubMed] [Google Scholar]
- 25.Robinson SD, Reynolds LE, Wyder L, Hicklin DJ, Hodivala-Dilke KM. Beta3-integrin regulates vascular endothelial growth factor-A-dependent permeability. Arteriosclerosis, Thrombosis, and Vascular Biology. 2004;24:2108–2114. doi: 10.1161/01.ATV.0000143857.27408.de. [DOI] [PubMed] [Google Scholar]
- 26.Antic D, Kang CY, Spik K, Schmaljohn C, Vapalahti O, Vaheri A. Comparison of the deduced gene products of the L, M and S genome segments of hantaviruses. Virus Research. 1992;24:35–46. doi: 10.1016/0168-1702(92)90029-9. [DOI] [PubMed] [Google Scholar]
- 27.Sen N, Sen A, Mackow ER. Degrons at the C terminus of the pathogenic but not the nonpathogenic hantavirus G1 tail direct proteasomal degradation. Journal of Virology. 2007;81:4323–4330. doi: 10.1128/JVI.02279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geimonen E, LaMonica R, Springer K, Farooqui Y, Gavrilovskaya IN, Mackow ER. Hantavirus pulmonary syndrome-associated hantaviruses contain conserved and functional ITAM signaling elements. Journal of Virology. 2003;77:1638–1643. doi: 10.1128/JVI.77.2.1638-1643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geimonen E, Fernandez I, Gavrilovskaya IN, Mackow ER. Tyrosine residues direct the ubiquitination and degradation of the NY-1 hantavirus G1 cytoplasmic tail. Journal of Virology. 2003;77:10760–10868. doi: 10.1128/JVI.77.20.10760-10768.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alff PJ, Gavrilovskaya IN, Gorbunova E, Endriss K, Chong Y, Geimonen E, Sen N, Reich NC, Mackow ER. The pathogenic NY-1 hantavirus G1 cytoplasmic tail inhibits RIG-I- and TBK-1-directed interferon responses. Journal of Virology. 2006;80:9676–9686. doi: 10.1128/JVI.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leyssen P, De Clercq E, Neyts J. Molecular strategies to inhibit the replication of RNA viruses. Antiviral Research. 2008;78:9–25. doi: 10.1016/j.antiviral.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gowen BB, Wong MH, Jung KH, Sanders AB, Mendenhall M, Bailey KW, Furuta Y, Sidwell RW. In vitro and in vivo activities of T-705 against arenavirus and bunyavirus infections. Antimicrobial Agents and Chemotherapy. 2007;51:3168–3176. doi: 10.1128/AAC.00356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gowen BB, Wong MH, Jung KH, Smee DF, Morrey JD, Furuta Y. Efficacy of favipiravir (T-705) and T-1106 pyrazine derivatives in phlebovirus disease models. Antiviral Research. 2010;86:121–127. doi: 10.1016/j.antiviral.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furuta Y, Takahashi K, Fukuda Y, Kuno M, Kamiyama T, Kozaki K, Nomura N, Egawa H, Minami S, Watanabe Y, Narita H, Shiraki K. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrobial Agents and Chemotherapy. 2002;46:977–981. doi: 10.1128/AAC.46.4.977-981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sidwell RW, Barnard DL, Day CW, Smee DF, Bailey KW, Wong MH, Morrey JD, Furuta Y. Efficacy of Orally Administered T-705 on Lethal Avian Influenza A (H5N1) Virus Infections in Mice. Antimicrobial Agents and Chemotherapy. 2007;51:845–851. doi: 10.1128/AAC.01051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gowen BB, Smee DF, Wong MH, Hall JO, Jung KH, Bailey KW, Stevens JR, Furuta Y, Morrey JD. Treatment of late stage disease in a model of arenaviral hemorrhagic fever: T-705 efficacy and reduced toxicity suggests an alternative to ribavirin. PLoS ONE. 2008;3:e3725. doi: 10.1371/journal.pone.0003725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morrey JD, Taro B, Siddharthan V, Wang H, Smee DF, Christensen AJ, Furuta Y. Efficacy of orally administered T-705 pyrazine analog on lethal West Nile virus infection in rodents. Antiviral Research. 2008 doi: 10.1016/j.antiviral.2008.07.009. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Julander JG, Shafer K, Smee DF, Morrey JD, Furuta Y. Activity of T-705 in a hamster model of yellow fever virus infection in comparison with that of a chemically related compound, T-1106. Antimicrobial Agents and Chemotherapy. 2009;53:202–209. doi: 10.1128/AAC.01074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiso M, Takahashi K, Sakai-Tagawa Y, Shinya K, Sakabe S, Le QM, Ozawa M, Furuta Y, Kawaoka Y. T-705 (favipiravir) activity against lethal H5N1 influenza A viruses. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:882–887. doi: 10.1073/pnas.0909603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 41.Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. Evolving Genes and Proteins.
- 42.Sun Y, Chung DH, Chu YK, Jonsson CB, Parker WB. Activity of ribavirin against Hantaan virus correlates with production of ribavirin-5'-triphosphate, not with inhibition of IMP dehydrogenase. Antimicrobial Agents and Chemotherapy. 2007;51:84–88. doi: 10.1128/AAC.00790-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Severson WE, Schmaljohn CS, Javadian A, Jonsson CB. Ribavirin causes error catastrophe during Hantaan virus replication. Journal of Virology. 2003;77:481–488. doi: 10.1128/JVI.77.1.481-488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy ME, Kariwa H, Mizutani T, Yoshimatsu K, Arikawa J, Takashima I. In vitro antiviral activity of lactoferrin and ribavirin upon hantavirus. Archives of Virology. 2000;145:1571–1582. doi: 10.1007/s007050070077. [DOI] [PubMed] [Google Scholar]
- 45.Mogford JE, Davis GE, Platts SH, Meininger GA. Vascular smooth muscle alpha v beta 3 integrin mediates arteriolar vasodilation in response to RGD peptides. Circulation Research. 1996;79:821–826. doi: 10.1161/01.res.79.4.821. [DOI] [PubMed] [Google Scholar]
- 46.Platts SH, Mogford JE, Davis MJ, Meininger GA. Role of K+ channels in arteriolar vasodilation mediated by integrin interaction with RGD-containing peptide. American Journal of Physiology. 1998;275:H1449–1454. doi: 10.1152/ajpheart.1998.275.4.H1449. [DOI] [PubMed] [Google Scholar]
- 47.Buranda T, Wu Y, Perez D, Jett SD, BonduHawkins V, Ye C, Edwards B, Hall P, Larson RS, Lopez GP, Sklar LA, Hjelle B. Recognition of decay accelerating factor and alpha(v)beta(3) by inactivated hantaviruses: Toward the development of high-throughput screening flow cytometry assays. Analytical Biochemistry. 2010;402:151–160. doi: 10.1016/j.ab.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krautkramer E, Zeier M. Hantavirus causing hemorrhagic fever with renal syndrome enters from the apical surface and requires decay-accelerating factor (DAF/CD55). Journal of Virology. 2008;82:4257–4264. doi: 10.1128/JVI.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chu JJ, Ng ML. Interaction of West Nile virus with alpha v beta 3 integrin mediates virus entry into cells. Journal of Biological Chemistry. 2004;279:54533–54541. doi: 10.1074/jbc.M410208200. [DOI] [PubMed] [Google Scholar]
- 50.Elliott RM. Emerging viruses: the Bunyaviridae. Molecular Medicine. 1997;3:572–577. [PMC free article] [PubMed] [Google Scholar]
- 51.Hjertqvist M, Klein SL, Ahlm C, Klingstrom J. Mortality rate patterns for hemorrhagic fever with renal syndrome caused by Puumala virus. Emerging Infectious Diseases. 2010;16:1584–1586. doi: 10.3201/eid1610.100242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monroe MC, Morzunov SP, Johnson AM, Bowen MD, Artsob H, Yates T, Peters CJ, Rollin PE, Ksiazek TG, Nichol ST. Genetic diversity and distribution of Peromyscus-borne hantaviruses in North America. Emerging Infectious Diseases. 1999;5:75–86. doi: 10.3201/eid0501.990109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plyusnin A, Morzunov SP. Virus evolution and genetic diversity of hantaviruses and their rodent hosts. Current Topics in Microbiology and Immunology. 2001;256:47–75. doi: 10.1007/978-3-642-56753-7_4. [DOI] [PubMed] [Google Scholar]
- 54.Furuta Y, Takahashi K, Kuno-Maekawa M, Sangawa H, Uehara S, Kozaki K, Nomura N, Egawa H, Shiraki K. Mechanism of action of T-705 against influenza virus. Antimicrobial Agents and Chemotherapy. 2005;49:981–986. doi: 10.1128/AAC.49.3.981-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mendenhall M, Russell A, Juelich T, Messina EL, Smee DF, Freiberg AN, Holbrook MR, Furuta Y, de la Torre JC, Nunberg JH, Gowen BB. T-705 (Favipiravir) Inhibition of Arenavirus Replication in Cell Culture. Antimicrobial Agents and Chemotherapy. 2010 doi: 10.1128/AAC.01219-10. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim GR, McKee KT., Jr Pathogenesis of Hantaan virus infection in suckling mice: clinical, virologic, and serologic observations. American Journal of Tropical Medicine and Hygiene. 1985;34:388–395. doi: 10.4269/ajtmh.1985.34.388. [DOI] [PubMed] [Google Scholar]
- 57.McKee KT, Jr., Kim GR, Green DE, Peters CJ. Hantaan virus infection in suckling mice: virologic and pathologic correlates. Journal of Medical Virology. 1985;17:107–117. doi: 10.1002/jmv.1890170203. [DOI] [PubMed] [Google Scholar]
- 58.Engelthaler DM, Mosley DG, Cheek JE, Levy CE, Komatsu KK, Ettestad P, Davis T, Tanda DT, Miller L, Frampton JW, Porter R, Bryan RT. Climatic and environmental patterns associated with hantavirus pulmonary syndrome, Four Corners region, United States. Emerging Infectious Diseases. 1999;5:87–94. doi: 10.3201/eid0501.990110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wenzel RP. A new hantavirus infection in North America. New England Journal of Medicine. 1994;330:1004–1005. doi: 10.1056/NEJM199404073301410. [DOI] [PubMed] [Google Scholar]