Abstract
Sentences and phrases that represent a certain meaning often exhibit patterns of variation where they differ from a basic structural form by one or two words. We present an algorithm that utilizes multiple sequence alignments (MSAs) to generate a representation of groups of phrases that possess the same semantic meaning but also share in common the same basic word sequence structure. The MSA enables the determination not only of the words that compose the basic word sequence, but also of the locations within the structure that exhibit variation. The algorithm can be utilized to generate patterns of text sequences that can be used as the basis for a pattern-based classifier, as a starting point to bootstrap the pattern building process for a regular expression-based classifiers, or serve to reveal the variation characteristics of sentences and phrases within a particular domain.
Introduction
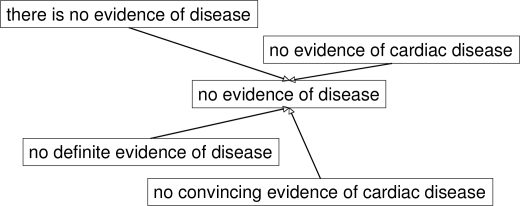
Sentences and phrases that represent a certain meaning often exhibit patterns of variation where they differ from a basic structural form by one or two words. As shown in Figure 1, the phrase, no evidence of cardiac disease, has many variations that basically mean that there was no cardiac disease found. It has also been noted that in order to express that a finding is negated, there are certain phrasal patterns, or signal phrases, usually found in the surrounding context of the finding that strongly indicate the presence of a negation 4, 16. We present an algorithm that utilizes multiple sequence alignments (MSAs) to generate a representation of groups of phrases that possess the same semantic meaning but also share in common the same basic word sequence structure. The MSA enables the determination not only of the words that compose the basic word sequence, but also of the locations within the structure that exhibit variation. The algorithm can be utilized to generate patterns of text sequences that can be used as the basis for a pattern-based classifier, as a starting point to bootstrap the pattern building process for regular expression-based classifiers, or serve to reveal the variation characteristics of sentences and phrases within a particular domain. In particular, it may be useful to know how a corpus of text varies with respect to the application at hand in order to understand the level of variation and the difficulty in modeling the text using approaches such as statistical classifiers. If the more commonly encountered examples could be easily grouped into clusters (e.g., the common 80%), then more effort could be concentrated on dealing with the numerous lesser-known and infrequent variations (e.g., the “long tail”). We hypothesize that for a given domain, there are a finite number of basic sequence structures that are used and that much of the variation will be based on these basic forms.
Figure 1.
Variations from the same basic phrase
Related Work
Multiple sequence alignments have been used throughout bioinformatics as a methodology for identifying commonalities and features of related genetic sequences. Methods for obtaining multiple sequence alignments include multidimensional dynamic programming, progressive methods such as Clustal W or Clustal X, 10 and hidden Markov models. Notredame 14 presents a survey paper describing details of existing multiple sequence alignment generation algorithms. Work has also been done to apply sequence alignment techniques to natural language processing problems. In Barzilay and Lee, 3 sequence alignment is used to determine sentence paraphrases in news articles and the work reported in Dolan et. al. 7 used edit distance with an added heuristic to determine sentence similarity for generating a large paraphrase corpus. Chiang and Yu 6 used sentence alignment to generate patterns that mine out gene product functions in biomedical literature. Meng et. al. 12 used approximate sequence alignment to determine similarity between sentences for generating clinical notes based on past clinical documents of the same patient. Whereas the work in Barzilay and Lee uses multiple sequence alignment to determine structural variations between several instances of entire sentences that have the same meaning, the goal of the methodology presented in this paper is to process large amounts of text data to find general variation patterns for phrases (or sub-sequences of sentences) of the same meaning.
There has been much work in fields of information extraction and information retrieval that tackle the issues pertaining to variations in natural language of text that represents the same meaning. Several information extraction approaches have developed algorithms to automatically generate extraction patterns from a corpus of text. For biomedical literature Nguyen et. al. 13 describe a system that utilizes extraction patterns to mine out relations between identified biological or genetic entities. Medical documents are retrieved using a phrase-based vector space model in Mao and Chu. 11 To avoid structural differences between sentences having the same semantic meaning, approaches have been developed to process sentences at the semantic level. 1 In medical informatics, the NegEx algorithm4 for identifying negated findings and the ConText algorithm5 for extracting contextual information are well-known examples of information extraction systems. Though the patterns generated by our algorithm can be utilized as extraction patterns, we intended the results to be used more generally, such as to characterize the variations inherent in the text of the data set. Since clinical language often exhibits particular patterns due to the use of templates or the constraints imposed by legal or insurance concerns, we feel multiple sequence alignments can better exploit these inherent regularities than general-purpose information extraction techniques. We also envision that multiple sequence alignments can be applied to other tasks such as enabling more efficient tagging of very large data sets. We describe possible applications of our approach in the discussion section at the end of the paper.
Handling negation within natural language text has been an active area of research, particularly within the medical informatics community. As previously mentioned, NegEx is a well-known regular expression-based system to identify negation in medical documents and has been proven to perform at a very high level. NegEx can be considered as a specialized information extraction system, where the negation phrases are extraction patterns similar to those described previously. Other approaches, such as that described in Yang and Lowe, 16 leverage the syntactic parse tree of a sentence, along with known phrases that signal negation, to automatically extract the finding being negated.
Methodology
We chose negation of findings in neuro-radiology documents as an example application of using multiple sequence alignments to determine phrase variation patterns because systems such as NegEx have already established a standard set of negation phrases, which can serve as a gold standard. A comparison of the results of our methodology with a set of negation phrases from NegEx is given in the results section. Our methodology proceeds in several steps: preprocessing documents by breaking them up into sentences and tokens, identifying the target phrases that are of interest, tagging those targets, generating context phrases that are the sequence of words surrounding the targets that affect the target’s meaning, generating multiple sequence alignments of the context phrases, and filtering the multiple sequence alignments to reduce redundancy and inaccuracies. Details are given for each step in this section.
Preprocessing Documents
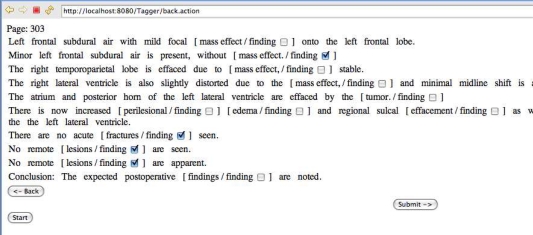
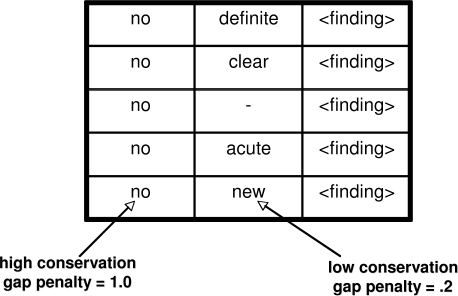
The data set was preprocessed in a standard way in order to facilitate storage and analysis. First, each document was decomposed into its constituent sentences using a simple sentence boundary detector. Next, each sentence was tokenized by using space characters as delimiters. Document meta-data, sentences, and tokens were all stored in a back-end relational database for easy access and fast retrieval. Because our chosen example application was detecting negated findings, all instances of phrases that represent findings were identified using a medical glossary developed at UCLA.15 We refer to phrases within a particular semantic class that is of interest to the application as targets. For instance, if the application required the identification of BMI measurements within a medical record, the targets would consist of all numerals present in those documents. We developed a term mapping algorithm that identified single or multi-word phrases within our sentences that were also present in the glossary. A simple precedence rule of allowing longer phrases to trump shorter phrases resolved most conflict issues. This functionality is similar to that provided by MetaMap for UMLS, 2 and we chose to use the UCLA glossary because we found it had very comprehensive coverage of terms for the domain covered by our corpus. We also developed a simple browser-based tagging tool that reads information from the relational database and allows a domain expert to tag targets (in this case, finding) using a predefined set of labels. As the example screenshot in Figure 2 shows, the user is able to indicate with a checkbox whether a given finding is negated or not.
Figure 2.
Browser-based Tagging Tool
Finding Context Phrases
Given a set of tagged sentences, the first step is to determine the relevant words surrounding each target that affect the meaning of that specific target. We call this set of words the context phrase of the target. For instance, the context phrase of the target finding1 in the sentence, There is no evidence of <finding1> but there is some history of <finding2>, could be considered as the phrase there is no evidence of <finding1>. The rest of the words in the sentence do not directly affect the meaning of finding1 in terms of whether it is negated or not. Put another way, the sequence of words preceding or following the context can vary and change yet the most likely interpretation is that finding1 is negated. In order to identify the context, it seems possible to utilize a syntactic parse tree’s groupings of words into phrasal structures. However, these groupings do not always enable a systematic methodology for identifying the words that are relevant to the meaning of the target. Our technique for identifying context phrases is based on the assumption that targets having the same meaning will tend to have surrounding words in common. If the common surrounding words can be extracted, sequences of these words can be considered as context phrases. We utilized approximate local sequence alignment techniques to determine the common words between sentences containing targets having the same meaning. Specifically, we used the Smith-Waterman approximate local alignment algorithm 8 which is frequently used within the bioinformatics community to compare and align genetic sequences. The algorithm determines similarity while allowing for variations between the sequences. It also finds the best aligning sub-sequences and does not require that all symbols in the original sequences need to participate in the final alignment.
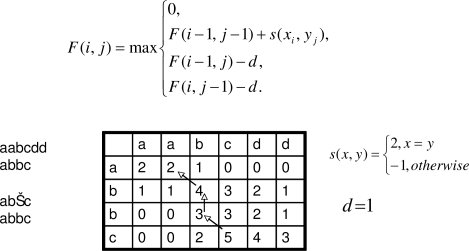
The following is a general overview of the Smith-Waterman algorithm. Figure 3 illustrates the main components and mechanisms of the algorithm: F is the score matrix, s(x,y) is the scoring function, and d is the gap penalty.
Figure 3.
Smith-Waterman Algorithm
Each cell in the matrix F corresponds with one token from each sequence being aligned. In the example shown in Figure 3, two sequence aabcdd and abbc are being locally aligned. The lower right hand cell of the matrix F corresponds to the last token of each sequence (i.e., d and c). The score contained in each cell indicates the best score for an alignment that includes its two corresponding tokens. Again in our example, the lower right hand cell indicates that a score of 3 is the best score for all local alignments that include the last token of each sequence. Links are associated with each cell to indicate which neighboring cell (either upper, left, or upper left) was used to derive that cell’s score. A diagonal link from the cell containing a score of 5 shows that the score for this cell was derived from the neighboring cell to its immediate upper left. To generate the final best local alignment, the cell containing the highest score within the entire matrix is first identified. The alignment is built in reverse by backtracking from the high score cell using the links and pre-pending each cell’s corresponding tokens to the alignment. In Figure 3, the high score cell is the one containing a score of 5. Thus, the alignment is generated by first adding the high score cell’s corresponding tokens (c and c). Following the cell’s link brings us to its adjacent upper left cell containing a score of 3. This cell’s corresponding tokens are pre-pended to the existing alignment. This process is repeated until a cell with the score of 0 is encountered. The final alignment is shown at the bottom left of Figure 3. A detailed and full explanation of the algorithm can be found in Durbin et. al.8
To focus each sentence on its particular target, a generic symbol that does not occur in normal English is used as a placeholder for the target (e.g., <finding>). In order to focus on one target at a time, a sentence containing multiple targets was duplicated once for each instance of a target. Thus, a sentence containing three targets will be duplicated three times, once for each target, where the target symbol is placed at the location of each of the three targets in turn. Further normalization of the sentences was accomplished by replacing phrases of certain semantic classes that were deemed to not influence the meaning of the target with generic symbols as well. As an example, we replaced anatomy phrases with the generic symbol <anat> and orientation phrases with <orientation>. We then aligned each normalized sentence against every other sentence using the Smith-Waterman algorithm tailored to always include the target in the resulting alignment by assigning a very high score to the target symbol within the scoring function. By limiting the number of gaps in the resulting alignments, we were able to control the level of variableness allowed between two sentences. An example of an alignment is shown in Figure 4. In this particular case, a gap (-) was added to allow for the best local alignment. The two phrases shaded in gray are taken as candidate context phrases extracted by our technique.
Figure 4.
Approximate Local Alignment Example
Generating Multiple Sequence Alignments of Context Phrases
In order to characterize the variability patterns of similar context phrases, we clustered phrases that differed slightly from each other into multiple sequence alignments. Aligning multiple phrases together into one multiple sequence alignment enables the identification of traits and characteristics that can be leveraged to determine whether these groups belong to the same family.
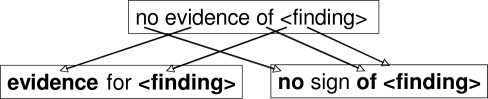
Each MSA generated was based on a particular sequence of noncontiguous words that all phrases in the MSA have in common. As an example, consider the context phrase no evidence of <finding> shown in Figure 5, where two possible noncontiguous word sequences generated from this phrase are shown below it. The sequence on the left consists of the word evidence, followed by an arbitrary word, followed by the target <finding> that is represented by the string evidence - <finding>. The gap symbol represents an arbitrary word.a Similarly, the MSA on the right is composed of all phrases having a sub-sequence consisting of the word no, followed by an arbitrary word, followed by the word of, followed by the target <finding>. To generate MSAs for the entire set of context phrases, each phrase takes a turn as the starting point on which the MSA will be based. The starting phrase is aligned against every other phrase, and those phrases that align with a low degree of variability are chosen to participate in the MSA. The entire MSA generation process is terminated when every phrase has been used as the starting point. An example MSA based on the word sequence no - <finding> is shown partially in Figure 6.
Figure 5.
Generating Multiple Sequence Alignments from Context Phrase
Figure 6.
Example Multiple Sequence Alignment
The Smith-Waterman algorithm was adapted to handle aligning a single sequence with an existing multiple sequence alignment, where a symbol from the single sequence is matched against a column of symbols in the MSA. The final score placed in the F matrix for a cell is the highest score between the single sequence’s symbol and any symbol within the MSA’s column. Once the F matrix is generated, the rest of the algorithm proceeds as usual. This procedure is shown in Figure 7, where the single sequence shown in the row dimension of the F matrix is being aligned against the MSA in the column dimension of the F matrix. MSA columns that contain multiple symbols are shown in a bracketed list and the best matching score value is shown in the F matrix for each cell. For instance, the second word of the single sequence clear exactly matches one of the words in the MSA’s second column, so the score in the corresponding cell represents an exact match.
Figure 7.
Aligning MSA with Single Sequence
Matching MSAs using Profiles
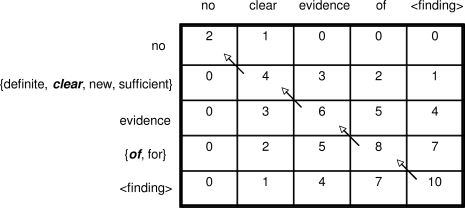
The main utility of a multiple sequence alignment is its ability to match against new sequences to determine whether the sequence exhibits the characteristics of the family represented by the MSA. In bioinformatics, a profile is used to summarize the more salient characteristics of the MSA, which can be represented using various techniques such as Hidden Markov Models or statistical classifiers.8 We used a simple profile that computed the frequency percentage of each token within a given column. To match a profile with a sequence, the match score for a token in the scoring function was derived from a token’s percentage within a column of the profile. Gap penalties were computed based on the overall conservation level of the column, where conservation was measured as the maximum percentage of any token in the column. A column with a high conservation level will be dominated by a few tokens whereas a column with low conservation will contain many low frequency words. Having gap penalties that are proportional to conservation levels reflects the notion that a column with a high level of variableness should not impose a high penalty if there are no exact matches. Figure 8 shows a simple MSA and highlights the different column conservation levels that can be reflected in the MSA’s profile.
Figure 8.
Column Conservation and Gap Penalty
Filtering MSAs
Because our MSA generation technique results in multiple MSAs that may be redundant, it was necessary to implement a filtering mechanism in order to eliminate unnecessary or inaccurate MSAs. We filtered MSAs in two steps: the first step removed MSAs that were inaccurate because they represented patterns of words which did not correspond to the specified meaning (e.g., negated findings) and the second step removed MSAs that were specific instances of more general MSAs. Because we generated the original set of MSAs using a tagged data set for negated findings, we used the tags a gold standard to compute the precision and recall of each MSA as if it were a regular expression classifier. Each MSA was matched against every finding target in the data set (regardless of whether it was tagged as negated or not) using its associated profile as described in the previous section. If the target and its surrounding context matched the profile with a very high similarity metric, it was marked as being negated according to this MSA. Only MSAs with very high precision (e.g., > 95%) were kept and all others were discarded. MSA overlap was determined by analyzing the target set that matched each MSA. If a MSA’s target set was a subset of another MSA’s set, the first MSA was eliminated because it is a specific instance of the second MSA.
Results
Our particular data set consisted of 549 neuro-radiology documents composed of 8,466 total sentences. In all, there were 14,317 references to findings, and after tagging for negated findings, we found there were 1,018 findings that were negated. It is these 1,018 finding instances that participated as targets in the MSA generation process. Overall, 600 MSAs representing variations on phrases that indicate negated findings were generated from our corpus after the first pass of the algorithm. This number was reduced to 104 after filtering was performed. Of the 1,018 negated targets, the 104 MSAs matched 805 of these, which is roughly 80% of the total negated finding targets. Thus, 203 negated finding cases were not matched, yielding a final total of 307 context phrases for negated findings in the data set. As a test for validity, the 104 MSAs were then used together as a classifier to identify negated findings in the data set and it was found to have a precision of 96% and a recall of 80%. Since this test was performed against the same data set from which the MSAs were generated, it was not meant to yield an absolute performance measure for the MSAs, but can be viewed as a way to determine the goodness of the MSAs. A cross validation or evaluation against a separate data set will be needed to determine proper performance metrics. These numbers and results are summarized in Table 1.
Table 1.
Summary of Results
| Number of documents | 548 |
| Number of sentences | 8,466 |
| References to findings | 14,317 |
| Negated findings | 1,018 |
| Original number of MSAs generated | 600 |
| Number of MSAs after filtering | 104 |
| Percentage of negated findings covered by MSAs | 80% |
| Precision of MSAs when identifying negated findings | 96% |
| Recall of MSAs when identifying negated findings | 80% |
Comparison with NegEx
The NegEx algorithm utilizes a very comprehensive set of phrases that indicate the negation of a finding. 4 In Table 2, we summarize how the MSAs generated by our algorithm compared with NegEx pre-negation phrases. As can be seen, most of NegEx’s phrases did not occur frequently enough in the data set for the algorithm to generate a corresponding MSA to represent the variations of that phrase. In the next phase of development, we plan to run the algorithm against a much larger data set in order to increase the probability of detecting more negation phrases.
Table 2.
Comparison with NegEx Pre-Negation Phrases
| NegEx Pre-Negation Phrases | Results |
|---|---|
| no, no abnormal, no evidence, no new evidence, no other evidence, no mammographic evidence of, no new, no radiographic evidence of, no significant, no suspicious, without | Covered by MSAs |
| absence of, cannot, cannot see, checked for, declined, declines, denied, denies, denying, evaluate for, fails to reveal, free of, negative for, never developed, never had, no cause of, no complaints of, no evidence to suggest, no findings of, no findings to indicate, no sign(s) of, no suggestion of, not, not appreciate, not associated with, not complain of, not demonstrate, not exhibit, not feel, not had, not have, not know of, not known to have, not reveal, not see, not to be, resolved, with no, without any evidence of, without evidence of, without indication of, without sign of, (all phrases containing “rules” or “ruled”) | Too few or no occurrences of phrase in data set |
| not appear, rather than, unremarkable | Sufficient number of occurrences of phrase in data set, but not covered by MSAs |
Error Analysis
As reported in Table 1, the filtered set of MSAs performed at a very high level of precision and an acceptable level of recall. The few false positives were mostly due to the fact that the phrase structure of the non-negation was very similar to that of a real negation and the negation was actually associated with a property of the finding (e.g., no increase of <finding>). False negatives were more abundant and these occurred mainly because the negation phrase did not occur frequently enough in the data set and there were not enough training examples to build a multiple sequence alignment. False negatives also occurred when the negation phrase did not match an MSA with a sufficiently high score. These results are summarized in Table 3, which also shows some examples of false positives and false negatives instances from the data set.
Table 3.
Error Analysis
| Error Type | Reason | Example(s) |
|---|---|---|
| False Positive | Phrase structure very similar to negation | No appreciable enlargement of these <finding> is noted. No convincing increase of <finding> is seen. |
| False Negative | Phrase occurred too infrequently | Absence of <finding>. <finding> is resolved. |
| False Negative | Phrase did not match any MSA profile with sufficiently high score | no radiographic evidence suggestive of <finding> |
Example MSAs
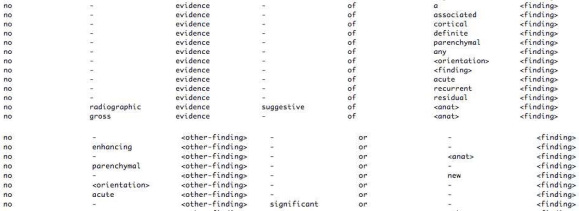
Figure 9 shows excerpts of two example MSAs generated for negated findings from our data set representing word sequences no – evidence – of – <finding> and no – <other finding> – or – <finding> respectively.
Figure 9.
Example MSAs
Discussion
Though this methodology seems promising for automatically determining variation patterns in phrases having representing negated findings, it still needs to be determined whether it will work well for other applications. Negated findings have the advantage that there exist specific signal phrases and that there are not too many basic ways of expressing negation in English. However, consider generating MSAs for positive findings where there are no signal phrases. Patterns such as there is - <finding> do not represent a positive finding with high accuracy because the gap symbol could represent negation words such as no or not. Thus, there may be the need to introduce weights to words and phrases depending on whether they help or hinder the MSA pattern. Another issue is the ability to increase the recall level, where the higher the recall level, the more complexity has been reduced in analyzing the data set. Pushing the recall level higher than 80% will be a focus of our research efforts in the future.
We also plan to apply MSAs to the problem of tagging very large data sets. Halevy et. al 9 have shown that a large increase in the amount of tagged data will improve the performance of standard machine learning algorithms. However, the results reported depended mostly on gathering large amounts of tagged data in the wild (e.g., parallel corpora from multi-lingual government documents). The amount of electronic medical records will only increase in the near future, and institutions such as the VA already have very large amounts of patient data in electronic form. Having large data sets enables the potential of high performance machine learning systems for medical applications. However, tagging such large data sets will become more laborious if efforts are not focused on improving the efficiency of the tagging process. We believe clustering similar phrases using techniques such as MSAs can help to boost the efficiency of the tagging process by grouping similar phrases together and indirectly tagging large groups of examples through inference techniques.
It has been noted that neuro-radiology documents may exhibit a high level of language and grammatical regularity, thus making the problem easier as opposed to using other types of medical documents that are less structured. We believe our technique will be able to handle documents with greater language variation due to the fact that the MSAs focus on the context phrases for the targets not the entire sentences themselves. Higher language variability will generate more MSAs and may result in more false negatives because there may be too few examples to generate an MSA for every context phrase family. Generating regular expression patterns by hand for systems like NegEx will also encounter the same problems and the goal of this work as stated previously is to formulate a technique to help bootstrap those types of systems and to provide a tool for analyzing the variation of the data set.
Conclusion
We presented a methodology for determining phrase variation patterns from a tagged data set using multiple sequence alignments. Given a tagged data set, the methodology first determined context phrases, the sequence of words surrounding the target phrase that have an affect on the target’s meaning. In our example application, the targets were phrases representing findings and they were tagged according to whether they were negated or not. These context phrases were then aligned using a standard approximate local alignment into MSAs. The resulting MSAs were then filtered resulting in a set of MSAs that best represent the variation patterns of commonly occurring phrases expressing negated findings. We believe this technique has many useful applications such as for analyzing the variation patterns and complexity of a given data set, or for seeding pattern-based classifier and extraction systems.
Footnotes
The * symbol could have been used here to follow regular expression conventions, but a gap better corresponds with bioinformatics notation
References
- 1.Achananuparp P, Hu X, Yang CC. Addressing the variability of natural language expression in sentence similarity with semantic structure of the sentences. Proceedings of the 13th Pacific-Asia Conference on Advances in Knowledge Discovery and Data Mining (PAKDD ’09); 2009. [Google Scholar]
- 2.Aronson AR. Effective mapping of biomedical text to the umls metathesaurus: the metamap program. 2001. [PMC free article] [PubMed]
- 3.Barzilay R, Lee L. Learning to paraphrase: an unsupervised approach using multiple-sequence alignment. Proceedings of NAACL-HLT; 2003. [Google Scholar]
- 4.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001 Oct;34(5):301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 5.Chapman WW, Chu D, Dowling JN. ConText: An algorithm for identifying contextual features from clinical text. Proceedings of the ACL Workshop on BioNLP 2007; 2007. pp. 81–88. [Google Scholar]
- 6.Chiang JH, Yu HC. MeKE: discovering the functions of gene products from biomedical literature via sentence alignment. Bioinformatics. 2003 Jul 22;19(11):1417–22. doi: 10.1093/bioinformatics/btg160. [DOI] [PubMed] [Google Scholar]
- 7.Dolan W, Quirk C, Brockett C. Unsupervised construction of large paraphrase corpora: Exploiting massively parallel new sources. Proceedings of the 20th International Conference on Computational Linguistics; 2004. [Google Scholar]
- 8.Durbin R, Eddy S, Krogh A, Mitchison G. Biological sequence analysis: probabilistic models of proteins and nucleic acids. Cambridge University; 1998. [Google Scholar]
- 9.Halevy A, Norvig P, Pereira F. The unreasonable effectiveness of data. IEEE Intelligent Systems. 2009;24(2):8–12. [Google Scholar]
- 10.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal w and clustal x version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 11.Mao W, Chu WW. Free-text medical document retrieval via phrase-based vector space model. Proc AMIA Symp. 2002:489–93. [PMC free article] [PubMed] [Google Scholar]
- 12.Meng F, Taira RK, Bui AA, Kangarloo H, Churchill BM. Automatic generation of repeated patient information for tailoring clinical notes. Int J Med Inform. 2005;74(7–8):663–673. doi: 10.1016/j.ijmedinf.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen QL, Tikk D, Leser U. Simple tricks for improving pattern-based information extraction from the biomedical literature. J Biomed Semantics. 2010 Sep 24;1(1):9. doi: 10.1186/2041-1480-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Notredame C. Recent evolutions of multiple sequence alignment algorithms. PLoS Comput Biol. 2007;3(8):e123. doi: 10.1371/journal.pcbi.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taira RK, Soderland SG, Jakobovits RM. Automatic structuring of radiology free text reports. Radiographics. 2001;21:237–245. doi: 10.1148/radiographics.21.1.g01ja18237. [DOI] [PubMed] [Google Scholar]
- 16.Yang H, Lowe HJ. A novel hybrid approach to automated negation detection in clinical radiology reports. JAMIA. 2007;14:304–311. doi: 10.1197/jamia.M2284. [DOI] [PMC free article] [PubMed] [Google Scholar]